Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment
Abstract
1. Introduction
2. The Potential of Metabolomics in Multiple Sclerosis (MS)
3. Evidence from the Literature
3.1. The Potential of Metabolomics as a Diagnostic Tool
3.2. Biochemical Differences According to Disease Stage
3.3. Insights into MS Molecular Mechanisms
3.4. Potential Biomarkers for MS and Patients’ Symptoms: Is There an Association?
3.5. Integration of Metabolomics and Neuroimaging Data
3.6. Integration of Metabolomics and Gene Expression Data
3.7. Evidence from the Use of Multivariate Approaches
3.8. Differences between Multiple Sclerosis and Other Demyelinating Disorders
3.9. The Potential of Metabolomics in Monitoring Treatment Effects
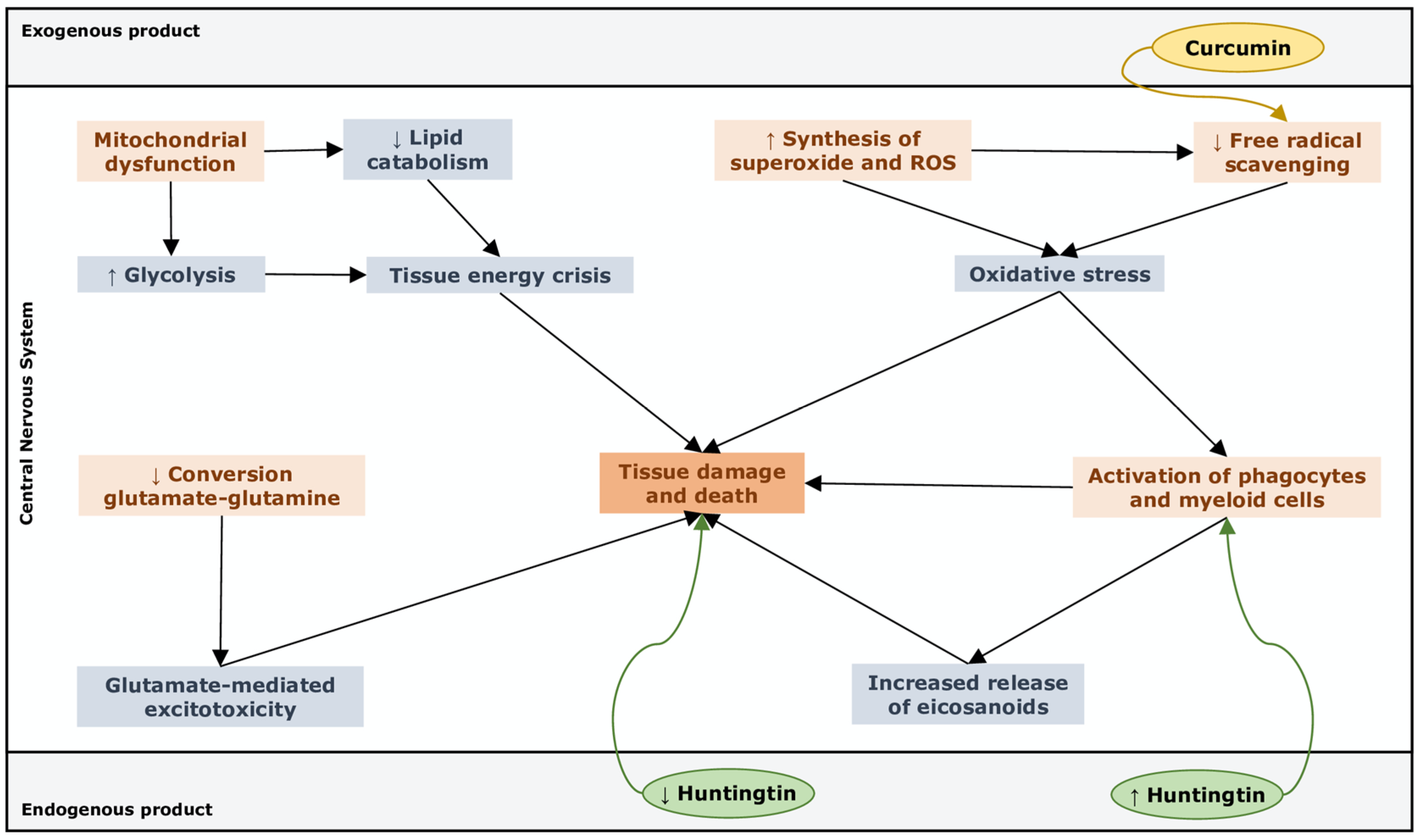
4. Pathway Analysis
- ▪
- “Free radical scavenging”, highly activated through upregulation of superoxide and ROS production, which suggests that cells undergo oxidative stress that feeds demyelination and neuronal damage [97].
- ▪
- “Small molecule biochemistry”, upregulated by an increased release of hormones, eicosanoids and neurotransmitters, a higher synthesis of ROS and NO, an increased concentration of glutamate and a metabolic switch toward glycolysis with reduced lipid catabolism and weak GSH synthesis.
- ▪
- “Cell death and survival”, which refers to apoptosis of astrocytes and other unspecified neuronal cells in the CNS.
- ▪
- “Cell-to-cell signalling”, remarked with the activation of macrophages, phagocytes and myeloid cells and concomitant reduced capacity of providing powerful responses to stress, thus supporting the inner link with the immune system in the pathogenesis of MS.
- ▪
- “Metabolic disease”, which comes from metabolic features that concur to create a pathogenic phenotype similar to that of hepatic steatosis.
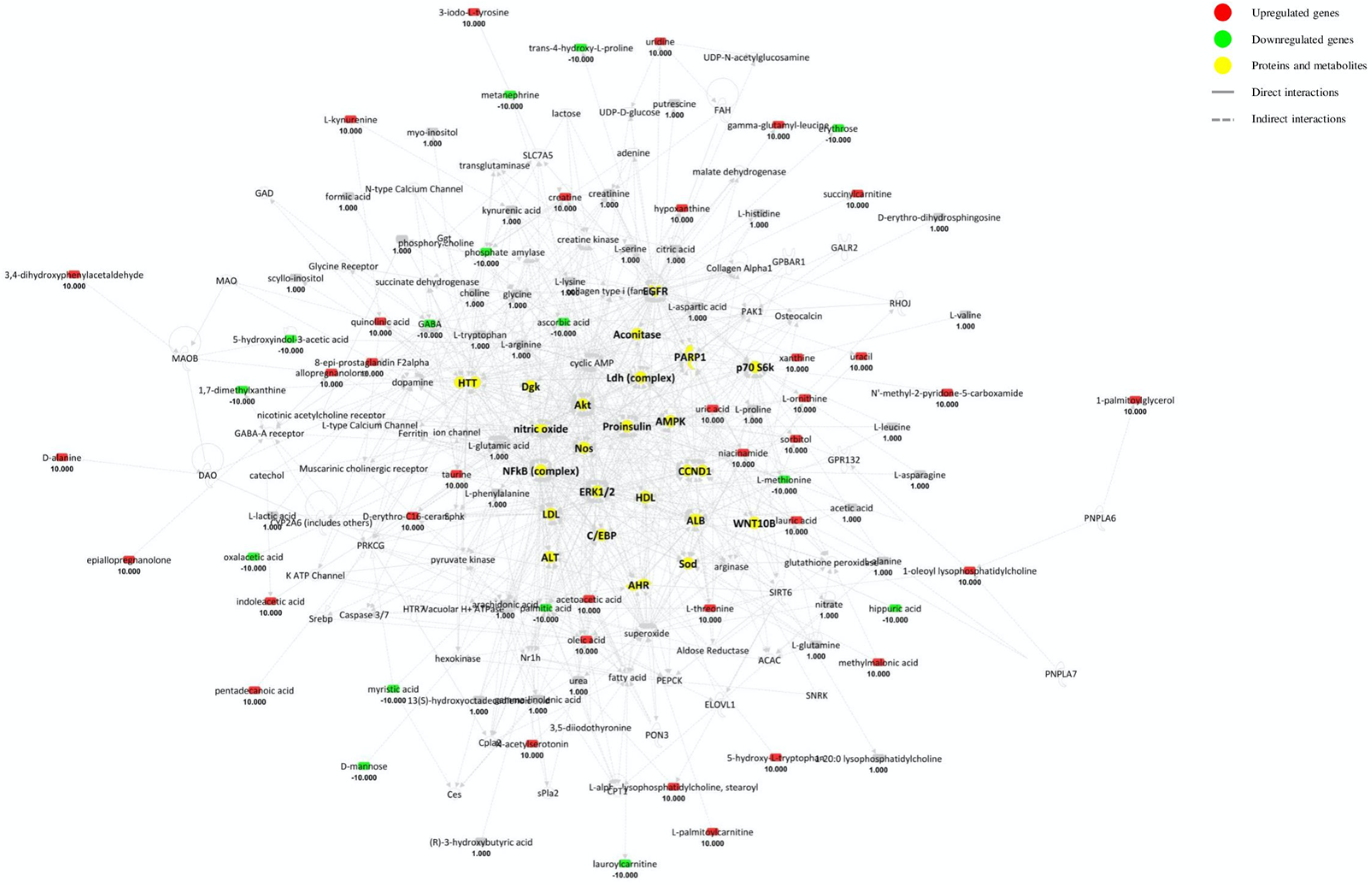
4.1. Cerebrospinal Fluid (CSF) Pathway Analysis
- ▪
- “Small molecule biochemistry” is activated through the upregulation of glutamine and other amino acid uptake, the increased concentration of neurotoxic glutamate and triacylglycerols, the release of eicosanoids and ROS, and the concomitant inhibition of nucleotides synthesis, neurotransmitter release and peroxidation of lipids.
- ▪
- “Molecular transport”, “Lipid metabolism”, “Amino acid metabolism” appear activated as well.
- ▪
- “Organismal injury and abnormalities” is upregulated by the overt neuroinflammation.
- ▪
- “Cell death and survival” and “cell-to-cell signalling” are matched with neuronal death and inhibition of orexin neurons and other cells.
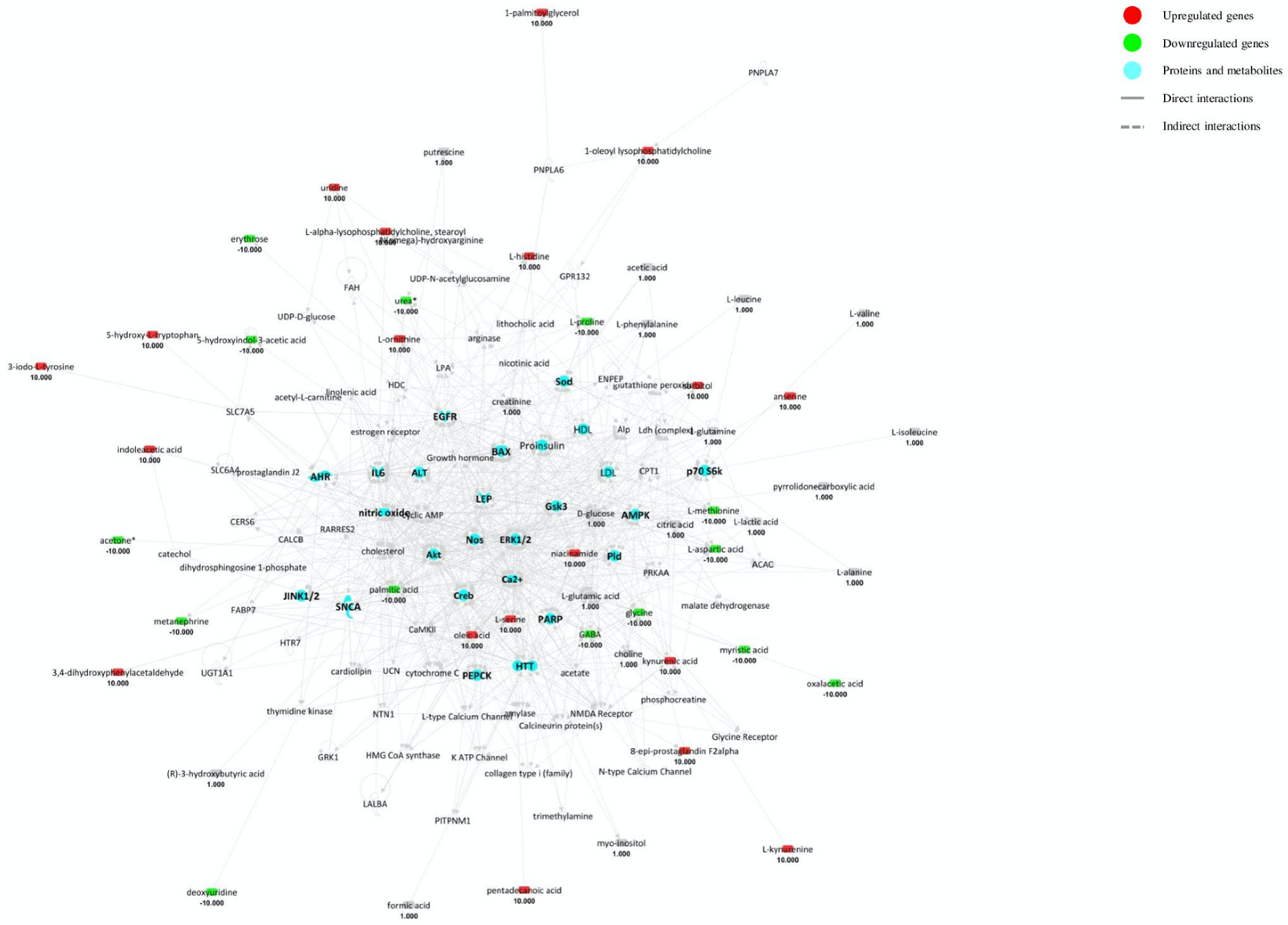
4.2. Blood Pathway Analysis
- ▪
- “Small molecule biochemistry” is activated by the upregulation of steroid/amino acid metabolism, lipid release and NO synthesis and by the downregulation of lipid/hormone binding, oxidation of lipids and amino acid uptake.
- ▪
- “Molecular transport” is highlighted by the upregulation of quantity of calcium and other metal ions.
- ▪
- “Lipid metabolism”, “Amino acid metabolism”, “Cell signalling” and “Free radical scavenging” are activated as well.
- ▪
- “Cell death and survival” is activated with the upregulation of cell viability and neuronal cell function and the downregulation of necrosis and cell death of CNS cells.
- ▪
- “Haematological system” is activated through the upregulation of myeloid cells, phagocytes, leucocytes and blood cell activation.
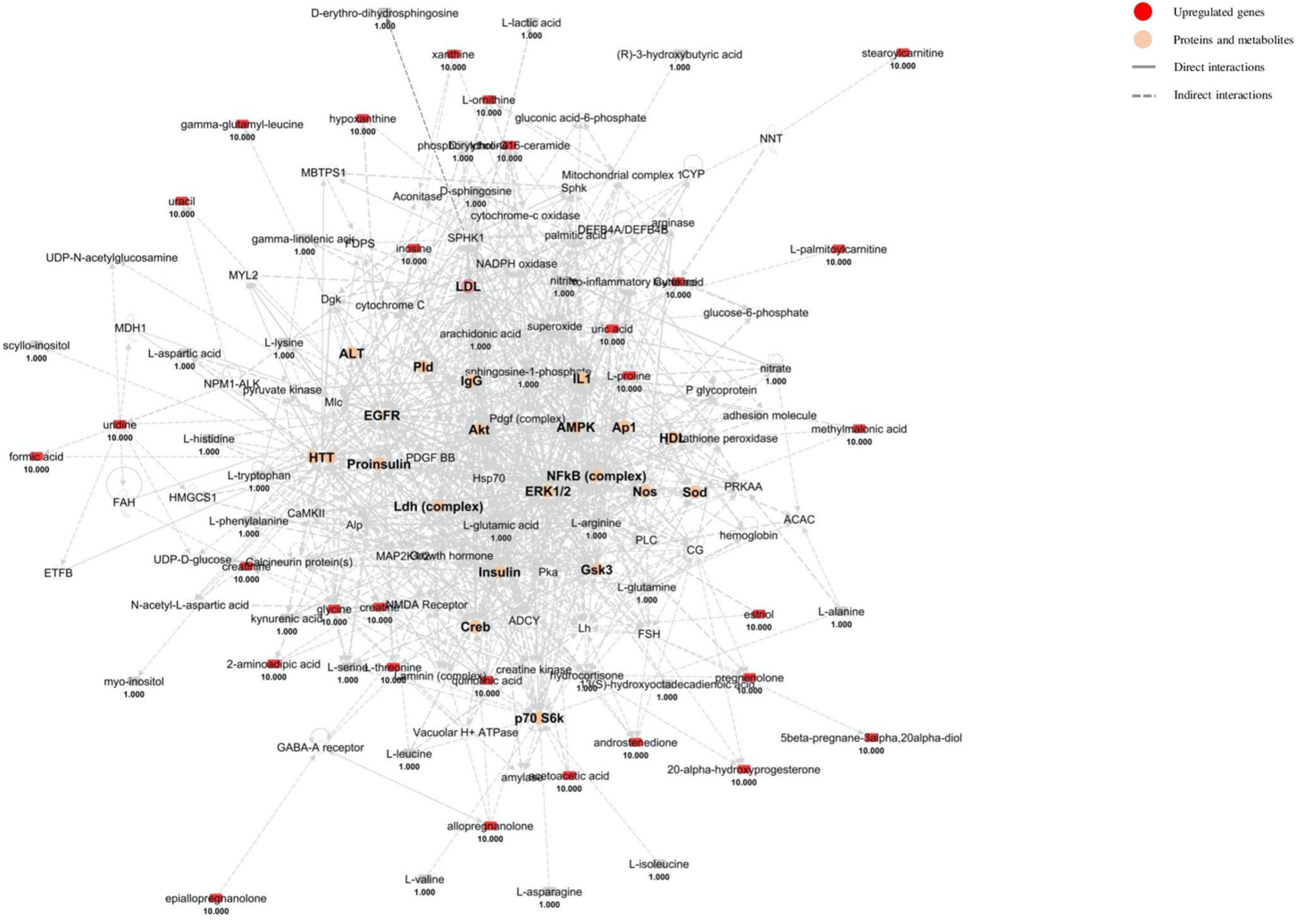
4.3. Urine Pathway Analysis
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boersma, M.G.; Solyanikova, I.P.; Van Berkel, W.J.; Vervoort, J.; Golovleva, L.A.; Rietjens, I.M. 19F NMR metabolomics for the elucidation of microbial degradation pathways of fluorophenols. J. Ind. Microbiol. Biotechnol. 2001, 26, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Smolinska, A.; Blanchet, L.; Buydens, L.M.; Wijmenga, S.S. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal. Chim. Acta 2012, 750, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Botas, A.; Campbell, H.M.; Han, X.; Maletic-Savatic, M. Metabolomics of neurodegenerative diseases. Int. Rev. Neurobiol. 2015, 122, 53–80. [Google Scholar] [CrossRef]
- Wishart, D.S. Current progress in computational metabolomics. Brief. Bioinform. 2007, 8, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Zhu, L.; Lu, J.; Zhang, X. Application of metabolomics in autoimmune diseases: Insight into biomarkers and pathology. J. Neuroimmunol. 2015, 279, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Dumas, M.E.; Maibaum, E.C.; Teague, C.; Ueshima, H.; Zhou, B.; Lindon, J.C.; Nicholson, J.K.; Stamler, J.; Elliott, P.; Chan, Q.; et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: The INTERMAP Study. Anal. Chem. 2006, 78, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Weljie, A.M. NMR Spectroscopy-Based Metabolic Profiling of Biospecimens. Curr. Protoc. Protein Sci. 2019, 98, e98. [Google Scholar] [CrossRef] [PubMed]
- Hassan-Smith, G.; Wallace, G.R.; Douglas, M.R.; Sinclair, A.J. The role of metabolomics in neurological disease. J. Neuroimmunol. 2012, 248, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Xu, G. Current state-of-the-art of nontargeted metabolomics based on liquid chromatography-mass spectrometry with special emphasis in clinical applications. J. Chromatogr. 2014, 1374, 1–13. [Google Scholar] [CrossRef]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Fitzgerald, K.C.; Calabresi, P.A.; Mowry, E.M. Metabolic alterations in multiple sclerosis and the impact of vitamin D supplementation. JCI Insight 2017, 2, e95302. [Google Scholar] [CrossRef]
- Alonso, A.; Marsal, S.; Julià, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef]
- Yoon, H.R. Screening newborns for metabolic disorders based on targeted metabolomics using tandem mass spectrometry. Ann. Pediatr. Endocrinol. Metab. 2015, 20, 119–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aboud, O.A.; Weiss, R.H. New opportunities from the cancer metabolome. Clin. Chem. 2013, 59, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Claudino, W.M.; Quattrone, A.; Biganzoli, L.; Pestrin, M.; Bertini, I.; Di Leo, A. Metabolomics: Available results, current research projects in breast cancer, and future applications. J. Clin. Oncol. 2007, 25, 2840–2846. [Google Scholar] [CrossRef] [PubMed]
- Klupczyńska, A.; Dereziński, P.; Kokot, Z.J. Metabolomics in medical sciences—Trends, challenges and perspectives. Acta Pol. Pharm. 2015, 72, 629–641. [Google Scholar] [PubMed]
- Cilingir, V.; Batur, M. Axonal Degeneration Independent of Inflammatory Activity: Is It More Intense in the Early Stages of Relapsing-Remitting Multiple Sclerosis Disease? Eur. Neurol. 2020, 83, 508–516. [Google Scholar] [CrossRef]
- Tremlett, H.; Zhao, Y.; Rieckmann, P.; Hutchinson, M. New perspectives in the natural history of multiple sclerosis. Neurology 2010, 74, 2004–2015. [Google Scholar] [CrossRef] [PubMed]
- Aasly, J.; Gårseth, M.; Sonnewald, U.; Zwart, J.A.; White, L.R.; Unsgård, G. Cerebrospinal fluid lactate and glutamine are reduced in multiple sclerosis. Acta Neurol. Scand. 1997, 95, 9–12. [Google Scholar] [CrossRef]
- Regenold, W.T.; Phatak, P.; Makley, M.J.; Stone, R.D.; Kling, M.A. Cerebrospinal fluid evidence of increased extra-mitochondrial glucose metabolism implicates mitochondrial dysfunction in multiple sclerosis disease progression. J. Neurol. Sci. 2008, 275, 106–112. [Google Scholar] [CrossRef]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.J.; Guillemin, G.J. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef] [PubMed]
- Mehrpour, M.; Kyani, A.; Tafazzoli, M.; Fathi, F.; Joghataie, M.T. A metabonomics investigation of multiple sclerosis by nuclear magnetic resonance. Magn. Reson. Chem. 2013, 51, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, V.K.; Jin, W.; Mochizuki, A.; Chitou, B.; Goodenowe, D.B. Metabolic dysfunctions in multiple sclerosis: Implications as to causation, early detection, and treatment, a case control study. BMC Neurol. 2015, 15, 154. [Google Scholar] [CrossRef] [PubMed]
- Herman, S.; Khoonsari, P.E.; Tolf, A.; Steinmetz, J.; Zetterberg, H.; Åkerfeldt, T.; Jakobsson, P.J.; Larsson, A.; Spjuth, O.; Burman, J.; et al. Integration of magnetic resonance imaging and protein and metabolite CSF measurements to enable early diagnosis of secondary progressive multiple sclerosis. Theranostics 2018, 8, 4477–4490. [Google Scholar] [CrossRef]
- Dickens, A.M.; Larkin, J.R.; Griffin, J.L.; Cavey, A.; Matthews, L.; Turner, M.R.; Wilcock, G.K.; Davis, B.G.; Claridge, T.D.; Palace, J.; et al. A type 2 biomarker separates relapsing-remitting from secondary progressive multiple sclerosis. Neurology 2014, 83, 1492–1499. [Google Scholar] [CrossRef]
- Smolinska, A.; Blanchet, L.; Coulier, L.; Ampt, K.A.; Luider, T.; Hintzen, R.Q.; Wijmenga, S.S.; Buydens, L.M. Interpretation and visualization of non-linear data fusion in kernel space: Study on metabolomic characterization of progression of multiple sclerosis. PLoS ONE 2012, 7, e38163. [Google Scholar] [CrossRef]
- Stoessel, D.; Stellmann, J.P.; Willing, A.; Behrens, B.; Rosenkranz, S.C.; Hodecker, S.C.; Stürner, K.H.; Reinhardt, S.; Fleischer, S.; Deuschle, C.; et al. Metabolomic Profiles for Primary Progressive Multiple Sclerosis Stratification and Disease Course Monitoring. Front. Hum. Neurosci. 2018, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Lorefice, L.; Murgia, F.; Fenu, G.; Frau, J.; Coghe, G.; Murru, M.R.; Tranquilli, S.; Visconti, A.; Marrosu, M.G.; Atzori, L.; et al. Assessing the Metabolomic Profile of Multiple Sclerosis Patients Treated with Interferon Beta 1a by. Neurotherapeutics 2019, 16, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Signoriello, E.; Iardino, P.; Casertano, S.; De Lucia, D.; Pucciarelli, A.; Puoti, G.; Chiosi, E.; Lus, G. 12-months prospective Pentraxin-3 and metabolomic evaluation in multiple sclerosis patients treated with glatiramer acetate. J. Neuroimmunol. 2020, 348, 577385. [Google Scholar] [CrossRef] [PubMed]
- Lutz, N.W.; Viola, A.; Malikova, I.; Confort-Gouny, S.; Audoin, B.; Ranjeva, J.P.; Pelletier, J.; Cozzone, P.J. Inflammatory multiple-sclerosis plaques generate characteristic metabolic profiles in cerebrospinal fluid. PLoS ONE 2007, 2, e595. [Google Scholar] [CrossRef] [PubMed]
- Del Boccio, P.; Rossi, C.; di Ioia, M.; Cicalini, I.; Sacchetta, P.; Pieragostino, D. Integration of metabolomics and proteomics in multiple sclerosis: From biomarkers discovery to personalized medicine. Proteom. Clin. Appl. 2016, 10, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Cree, B.A.; Magnusson, B.; Rouyrre, N.; Fox, R.J.; Giovannoni, G.; Vermersch, P.; Bar-Or, A.; Gold, R.; Piani Meier, D.; Karlsson, G.; et al. Siponimod: Disentangling disability and relapses in secondary progressive multiple sclerosis. Mult. Scler. 2020, 1352458520971819. [Google Scholar] [CrossRef]
- Freedman, M.S.; Brod, S.; Singer, B.A.; Cohen, B.A.; Hayward, B.; Dangond, F.; Coyle, P.K. Clinical and MRI efficacy of sc IFN β-1a tiw in patients with relapsing MS appearing to transition to secondary progressive MS: Post hoc analyses of PRISMS and SPECTRIMS. J. Neurol. 2020, 267, 64–75. [Google Scholar] [CrossRef]
- Miller, D.H.; Leary, S.M. Primary-progressive multiple sclerosis. Lancet Neurol. 2007, 6, 903–912. [Google Scholar] [CrossRef]
- Montalban, X.; Hauser, S.L.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Comi, G.; de Seze, J.; Giovannoni, G.; Hartung, H.P.; Hemmer, B.; et al. Ocrelizumab versus Placebo in Primary Progressive Multiple Sclerosis. N. Engl. J. Med. 2017, 376, 209–220. [Google Scholar] [CrossRef]
- Lynch, J.; Peeling, J.; Auty, A.; Sutherland, G.R. Nuclear magnetic resonance study of cerebrospinal fluid from patients with multiple sclerosis. Can. J. Neurol. Sci. 1993, 20, 194–198. [Google Scholar]
- Koschorek, F.; Offermann, W.; Stelten, J.; Braunsdorf, W.E.; Steller, U.; Gremmel, H.; Leibfritz, D. High-resolution 1H NMR spectroscopy of cerebrospinal fluid in spinal diseases. Neurosurg. Rev. 1993, 16, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, H.; Brieva, L.; Tatzber, F.; Jové, M.; Cacabelos, D.; Cassanyé, A.; Lanau-Angulo, L.; Boada, J.; Serrano, J.C.; González, C.; et al. Lipidome analysis in multiple sclerosis reveals protein lipoxidative damage as a potential pathogenic mechanism. J. Neurochem. 2012, 123, 622–634. [Google Scholar] [CrossRef]
- A’t Hart, B.; Vogels, J.T.; Spijksma, G.; Brok, H.P.; Polman, C.; van der Greef, J. 1H-NMR spectroscopy combined with pattern recognition analysis reveals characteristic chemical patterns in urines of MS patients and non-human primates with MS-like disease. J. Neurol. Sci. 2003, 212, 21–30. [Google Scholar] [CrossRef]
- Gebregiworgis, T.; Nielsen, H.H.; Massilamany, C.; Gangaplara, A.; Reddy, J.; Illes, Z.; Powers, R. A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. J. Proteome Res. 2016, 15, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Cicalini, I.; Rossi, C.; Pieragostino, D.; Agnifili, L.; Mastropasqua, L.; di Ioia, M.; De Luca, G.; Onofrj, M.; Federici, L.; Del Boccio, P. Integrated Lipidomics and Metabolomics Analysis of Tears in Multiple Sclerosis: An Insight into Diagnostic Potential of Lacrimal Fluid. Int. J. Mol. Sci. 2019, 20, 1265. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Calabresi, P.A. Metabolomics in multiple sclerosis. Mult. Scler. 2016, 22, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, G.A.; Baig, M.S. Quantitation of free amino acids in biological samples by high-performance liquid chromatography. Application of the method in evaluating amino acid levels in cerebrospinal fluid and plasma of patients with multiple sclerosis. J. Chromatogr. 1988, 459, 237–244. [Google Scholar] [CrossRef]
- Wood, J.H. Neurochemical analysis of cerebrospinal fluid. Neurology 1980, 30, 645–651. [Google Scholar] [CrossRef]
- Reinke, S.N.; Broadhurst, D.L.; Sykes, B.D.; Baker, G.B.; Catz, I.; Warren, K.G.; Power, C. Metabolomic profiling in multiple sclerosis: Insights into biomarkers and pathogenesis. Mult. Scler. 2014, 20, 1396–1400. [Google Scholar] [CrossRef]
- Cocco, E.; Murgia, F.; Lorefice, L.; Barberini, L.; Poddighe, S.; Frau, J.; Fenu, G.; Coghe, G.; Murru, M.R.; Murru, R.; et al. (1)H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e185. [Google Scholar] [CrossRef] [PubMed]
- Kasakin, M.F.; Rogachev, A.D.; Predtechenskaya, E.V.; Zaigraev, V.J.; Koval, V.V.; Pokrovsky, A.G. Targeted metabolomics approach for identification of relapsing-remitting multiple sclerosis markers and evaluation of diagnostic models. MedChemComm 2019, 10, 1803–1809. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, E.M.L.; Montani, D.A.; Oliveira-Silva, D.; Rodrigues-Oliveira, A.F.; Matas, S.L.A.; Fernandes, G.B.P.; Silva, I.D.C.G.; Lo Turco, E.G. Multiple sclerosis has a distinct lipid signature in plasma and cerebrospinal fluid. Arq. Neuropsiquiatr. 2019, 77, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Poddighe, S.; Murgia, F.; Lorefice, L.; Liggi, S.; Cocco, E.; Marrosu, M.G.; Atzori, L. Metabolomic analysis identifies altered metabolic pathways in Multiple Sclerosis. Int. J. Biochem. Cell Biol. 2017, 93, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Tsoukalas, D.; Fragoulakis, V.; Sarandi, E.; Docea, A.O.; Papakonstaninou, E.; Tsilimidos, G.; Anamaterou, C.; Fragkiadaki, P.; Aschner, M.; Tsatsakis, A.; et al. Targeted Metabolomic Analysis of Serum Fatty Acids for the Prediction of Autoimmune Diseases. Front. Mol. Biosci. 2019, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.J.; Viant, M.R.; Ball, A.K.; Burdon, M.A.; Walker, E.A.; Stewart, P.M.; Rauz, S.; Young, S.P. NMR-based metabolomic analysis of cerebrospinal fluid and serum in neurological diseases—A diagnostic tool? NMR Biomed. 2010, 23, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Lazzarino, G.; Amorini, A.M.; Petzold, A.; Gasperini, C.; Ruggieri, S.; Quartuccio, M.E.; Di Stasio, E.; Tavazzi, B. Serum Compounds of Energy Metabolism Impairment Are Related to Disability, Disease Course and Neuroimaging in Multiple Sclerosis. Mol. Neurobiol. 2017, 54, 7520–7533. [Google Scholar] [CrossRef] [PubMed]
- Nicoli, F.; Vion-Dury, J.; Confort-Gouny, S.; Maillet, S.; Gastaut, J.L.; Cozzone, P.J. Cerebrospinal fluid metabolic profiles in multiple sclerosis and degenerative dementias obtained by high resolution proton magnetic resonance spectroscopy. C. R. Acad. Sci. III 1996, 319, 623–631. [Google Scholar] [PubMed]
- Norenberg, M.D.; Martinez-Hernandez, A. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res. 1979, 161, 303–310. [Google Scholar] [CrossRef]
- Tarp, U. Selenium and the selenium-dependent glutathione peroxidase in rheumatoid arthritis. Dan Med. Bull. 1994, 41, 264–274. [Google Scholar]
- De Toledo, J.H.D.S.; Fraga-Silva, T.F.C.; Borim, P.A.; de Oliveira, L.R.C.; Oliveira, E.D.S.; Périco, L.L.; Hiruma-Lima, C.A.; de Souza, A.A.L.; de Oliveira, C.A.F.; Padilha, P.M.; et al. Organic Selenium Reaches the Central Nervous System and Downmodulates Local Inflammation: A Complementary Therapy for Multiple Sclerosis? Front. Immunol. 2020, 11, 571844. [Google Scholar] [CrossRef] [PubMed]
- Simone, I.L.; Federico, F.; Trojano, M.; Tortorella, C.; Liguori, M.; Giannini, P.; Picciola, E.; Natile, G.; Livrea, P. High resolution proton MR spectroscopy of cerebrospinal fluid in MS patients. Comparison with biochemical changes in demyelinating plaques. J. Neurol. Sci. 1996, 144, 182–190. [Google Scholar] [CrossRef]
- Tavazzi, B.; Batocchi, A.P.; Amorini, A.M.; Nociti, V.; D’Urso, S.; Longo, S.; Gullotta, S.; Picardi, M.; Lazzarino, G. Serum metabolic profile in multiple sclerosis patients. Mult. Scler. Int. 2011, 2011, 167156. [Google Scholar] [CrossRef] [PubMed]
- Podlecka-Piętowska, A.; Kacka, A.; Zakrzewska-Pniewska, B.; Nojszewska, M.; Zieminska, E.; Chalimoniuk, M.; Toczylowska, B. Altered Cerebrospinal Fluid Concentrations of Hydrophobic and Hydrophilic Compounds in Early Stages of Multiple Sclerosis-Metabolic Profile Analyses. J. Mol. Neurosci. 2019, 69, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.A.; Bassilian, S.; Lim, S.; Paul Lee, W.N. Coordination of peroxisomal beta-oxidation and fatty acid elongation in HepG2 cells. J. Biol. Chem. 2004, 279, 41302–41309. [Google Scholar] [CrossRef] [PubMed]
- Murgia, F.; Lorefice, L.; Poddighe, S.; Fenu, G.; Secci, M.A.; Marrosu, M.G.; Cocco, E.; Atzori, L. Multi-Platform Characterization of Cerebrospinal Fluid and Serum Metabolome of Patients Affected by Relapsing-Remitting and Primary Progressive Multiple Sclerosis. J. Clin. Med. 2020, 9, 863. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.S.; Hemmer, B.; Cepok, S. The role of antibodies in multiple sclerosis. Biochim. Biophys. Acta 2011, 1812, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Kallaur, A.P.; Reiche, E.M.V.; Oliveira, S.R.; Simão, A.N.C.; Pereira, W.L.C.J.; Alfieri, D.F.; Flauzino, T.; Proença, C.M.; Lozovoy, M.A.B.; Kaimen-Maciel, D.R.; et al. Genetic, Immune-Inflammatory, and Oxidative Stress Biomarkers as Predictors for Disability and Disease Progression in Multiple Sclerosis. Mol. Neurobiol. 2017, 54, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Kerr, S.J.; Smythe, G.A.; Smith, D.G.; Kapoor, V.; Armati, P.J.; Croitoru, J.; Brew, B.J. Kynurenine pathway metabolism in human astrocytes: A paradox for neuronal protection. J. Neurochem. 2001, 78, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J. Quinolinic acid: Neurotoxicity. FEBS J. 2012, 279, 1355. [Google Scholar] [CrossRef]
- Herman, S.; Åkerfeldt, T.; Spjuth, O.; Burman, J.; Kultima, K. Biochemical Differences in Cerebrospinal Fluid between Secondary Progressive and Relapsing—Remitting Multiple Sclerosis. Cells 2019, 8, 84. [Google Scholar] [CrossRef]
- Hartai, Z.; Klivenyi, P.; Janaky, T.; Penke, B.; Dux, L.; Vecsei, L. Kynurenine metabolism in multiple sclerosis. Acta Neurol. Scand. 2005, 112, 93–96. [Google Scholar] [CrossRef]
- Del Boccio, P.; Pieragostino, D.; Di Ioia, M.; Petrucci, F.; Lugaresi, A.; De Luca, G.; Gambi, D.; Onofrj, M.; Di Ilio, C.; Sacchetta, P.; et al. Lipidomic investigations for the characterization of circulating serum lipids in multiple sclerosis. J. Proteom. 2011, 74, 2826–2836. [Google Scholar] [CrossRef]
- Castro, K.; Ntranos, A.; Amatruda, M.; Petracca, M.; Kosa, P.; Chen, E.Y.; Morstein, J.; Trauner, D.; Watson, C.T.; Kiebish, M.A.; et al. Body Mass Index in Multiple Sclerosis modulates ceramide-induced DNA methylation and disease course. EBioMedicine 2019, 43, 392–410. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.; Bandaru, V.V.; Calabresi, P.A.; Nath, A.; Haughey, N.J. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain 2008, 131, 3092–3102. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Rossi, C.; Zucchelli, M.; Urbani, A.; Di Ilio, C.; Lugaresi, A.; Sacchetta, P.; Del Boccio, P. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol. Biosyst. 2015, 11, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; Cicalini, I.; Lanuti, P.; Ercolino, E.; di Ioia, M.; Zucchelli, M.; Zappacosta, R.; Miscia, S.; Marchisio, M.; Sacchetta, P.; et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of Multiple Sclerosis patients. Sci. Rep. 2018, 8, 3071. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, D.A.; Slupsky, C.M.; Aviv, R.I.; Swardfager, W.; Taha, A.Y. Untargeted metabolomic analysis of plasma from relapsing-remitting multiple sclerosis patients reveals changes in metabolites associated with structural changes in brain. Brain Res. 2020, 1732, 146589. [Google Scholar] [CrossRef]
- Liao, R.J.; Jiang, L.; Wang, R.R.; Zhao, H.W.; Chen, Y.; Li, Y.; Wang, L.; Jie, L.Y.; Zhou, Y.D.; Zhang, X.N.; et al. Histidine provides long-term neuroprotection after cerebral ischemia through promoting astrocyte migration. Sci. Rep. 2015, 5, 15356. [Google Scholar] [CrossRef] [PubMed]
- Politi, V.; Lavaggi, M.V.; Di Stazio, G.; Margonelli, A. Indole-3-pyruvic acid as a direct precursor of kynurenic acid. Adv. Exp. Med. Biol. 1991, 294, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.L.; Briggs, F.B.S.; Winnike, J.H.; Natanzon, Y.; Maichle, S.; Knagge, K.J.; Newby, L.K.; Gregory, S.G. Metabolome-based signature of disease pathology in MS. Mult. Scler. Relat. Disord. 2019, 31, 12–21. [Google Scholar] [CrossRef]
- Friese, M.A.; Schattling, B.; Fugger, L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 2014, 10, 225–238. [Google Scholar] [CrossRef]
- Kanceva, R.; Stárka, L.; Kancheva, L.; Hill, M.; Veliková, M.; Havrdová, E. Increased serum levels of C21 steroids in female patients with multiple sclerosis. Physiol. Res. 2015, 64, S247–S254. [Google Scholar] [CrossRef] [PubMed]
- Melcangi, R.C.; Giatti, S.; Calabrese, D.; Pesaresi, M.; Cermenati, G.; Mitro, N.; Viviani, B.; Garcia-Segura, L.M.; Caruso, D. Levels and actions of progesterone and its metabolites in the nervous system during physiological and pathological conditions. Prog. Neurobiol. 2014, 113, 56–69. [Google Scholar] [CrossRef]
- Rossi, C.; Cicalini, I.; Zucchelli, M.; di Ioia, M.; Onofrj, M.; Federici, L.; Del Boccio, P.; Pieragostino, D. Metabolomic Signature in Sera of Multiple Sclerosis Patients during Pregnancy. Int. J. Mol. Sci. 2018, 19, 3589. [Google Scholar] [CrossRef]
- Yeo, T.; Sealey, M.; Zhou, Y.; Saldana, L.; Loveless, S.; Claridge, T.D.W.; Robertson, N.; DeLuca, G.; Palace, J.; Anthony, D.C.; et al. A blood-based metabolomics test to distinguish relapsing-remitting and secondary progressive multiple sclerosis: Addressing practical considerations for clinical application. Sci. Rep. 2020, 10, 12381. [Google Scholar] [CrossRef] [PubMed]
- Villoslada, P.; Alonso, C.; Agirrezabal, I.; Kotelnikova, E.; Zubizarreta, I.; Pulido-Valdeolivas, I.; Saiz, A.; Comabella, M.; Montalban, X.; Villar, L.; et al. Metabolomic signatures associated with disease severity in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e321. [Google Scholar] [CrossRef] [PubMed]
- Moussallieh, F.M.; Elbayed, K.; Chanson, J.B.; Rudolf, G.; Piotto, M.; De Seze, J.; Namer, I.J. Serum analysis by 1H nuclear magnetic resonance spectroscopy: A new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult. Scler. 2014, 20, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jeong, I.H.; Kong, B.S.; Lee, J.E.; Kim, K.H.; Lee, D.Y.; Kim, H.J. Disease Type- and Status-Specific Alteration of CSF Metabolome Coordinated with Clinical Parameters in Inflammatory Demyelinating Diseases of CNS. PLoS ONE 2016, 11, e0166277. [Google Scholar] [CrossRef] [PubMed]
- Jurynczyk, M.; Probert, F.; Yeo, T.; Tackley, G.; Claridge, T.D.W.; Cavey, A.; Woodhall, M.R.; Arora, S.; Winkler, T.; Schiffer, E.; et al. Metabolomics reveals distinct, antibody-independent, molecular signatures of MS, AQP4-antibody and MOG-antibody disease. Acta Neuropathol. Commun. 2017, 5, 95. [Google Scholar] [CrossRef]
- Kim, H.H.; Jeong, I.H.; Hyun, J.S.; Kong, B.S.; Kim, H.J.; Park, S.J. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS ONE 2017, 12, e0181758. [Google Scholar] [CrossRef] [PubMed]
- Navab, M.; Anantharamaiah, G.M.; Fogelman, A.M. The role of high-density lipoprotein in inflammation. Trends Cardiovasc. Med. 2005, 15, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Waddington, K.E.; Papadaki, A.; Coelewij, L.; Adriani, M.; Nytrova, P.; Kubala Havrdova, E.; Fogdell-Hahn, A.; Farrell, R.; Dönnes, P.; Pineda-Torra, I.; et al. Using Serum Metabolomics to Predict Development of Anti-drug Antibodies in Multiple Sclerosis Patients Treated with IFNβ. Front. Immunol. 2020, 11, 1527. [Google Scholar] [CrossRef] [PubMed]
- Sedic, M.; Gethings, L.A.; Vissers, J.P.; Shockcor, J.P.; McDonald, S.; Vasieva, O.; Lemac, M.; Langridge, J.I.; Batinić, D.; Pavelić, S.K. Label-free mass spectrometric profiling of urinary proteins and metabolites from paediatric idiopathic nephrotic syndrome. Biochem. Biophys. Res. Commun. 2014, 452, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Wang, K.; Zhang, S.; Yuan, J.; Liao, X.; Wu, C.; Zou, Y.; Ha, Y.; Shen, Z.; Guo, J.; et al. Ingenuity pathway analysis of differentially expressed genes involved in signaling pathways and molecular networks in RhoE gene-edited cardiomyocytes. Int. J. Mol. Med. 2020, 46, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Ohl, K.; Tenbrock, K.; Kipp, M. Oxidative stress in multiple sclerosis: Central and peripheral mode of action. Exp. Neurol. 2016, 277, 58–67. [Google Scholar] [CrossRef]
- Bak, L.K.; Schousboe, A.; Waagepetersen, H.S. The glutamate/GABA-glutamine cycle: Aspects of transport, neurotransmitter homeostasis and ammonia transfer. J. Neurochem. 2006, 98, 641–653. [Google Scholar] [CrossRef]
- Jia, M.; Njapo, S.A.; Rastogi, V.; Hedna, V.S. Taming glutamate excitotoxicity: Strategic pathway modulation for neuroprotection. CNS Drugs 2015, 29, 153–162. [Google Scholar] [CrossRef]
- Malekzadeh Shafaroudi, M.; Zarei, H.; Malekzadeh Shafaroudi, A.; Karimi, N.; Abedini, M. The Relationship between Glutamate and Multiple Sclerosis. Int. Biol. Biomed. J. 2018, 4, 1–13. [Google Scholar]
- Cawley, N.; Solanky, B.S.; Muhlert, N.; Tur, C.; Edden, R.A.; Wheeler-Kingshott, C.A.; Miller, D.H.; Thompson, A.J.; Ciccarelli, O. Reduced gamma-aminobutyric acid concentration is associated with physical disability in progressive multiple sclerosis. Brain 2015, 138, 2584–2595. [Google Scholar] [CrossRef] [PubMed]
- Algahtani, H.; Alameer, S.; Marzouk, Y.; Shirah, B. Urea cycle disorder misdiagnosed as multiple sclerosis: A case report and review of the literature. Neuroradiol. J. 2018, 31, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Kotha, R.R.; Luthria, D.L. Curcumin: Biological, Pharmaceutical, Nutraceutical, and Analytical Aspects. Molecules 2019, 24, 2930. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, W.; Zhu, H.; Zhang, X.; Feng, Y.; Chen, Y.; Feng, H.; Lin, J. Curcumin attenuates blood-brain barrier disruption after subarachnoid hemorrhage in mice. J. Surg. Res. 2017, 207, 85–91. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, G.C.; Farag, S.H.; Ostroff, G.R.; Tabrizi, S.J.; Andre, R. Wild-type huntingtin regulates human macrophage function. Sci. Rep. 2020, 10, 17269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Drozda, M.; Chen, M.; Ren, S.; Mejia Sanchez, R.O.; Leavitt, B.R.; Cattaneo, E.; Ferrante, R.J.; Hayden, M.R.; et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J. Neurochem. 2003, 87, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Black, J.O. Xeroderma Pigmentosum. Head Neck Pathol. 2016, 10, 139–144. [Google Scholar] [CrossRef]
- Meira, M.; Sievers, C.; Hoffmann, F.; Bodmer, H.; Derfuss, T.; Kuhle, J.; Haghikia, A.; Kappos, L.; Lindberg, R.L. PARP-1 deregulation in multiple sclerosis. Mult. Scler. J. Exp. Transl. Clin. 2019, 5, 2055217319894604. [Google Scholar] [CrossRef]
- Qin, L.Y.; Zhao, L.G.; Chen, X.; Li, P.; Yang, Z.; Mo, W.N. The CCND1 G870A gene polymorphism and brain tumor risk: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 3607–3612. [Google Scholar] [CrossRef]
- Džoljić, E.; Grbatinić, I.; Kostić, V. Why is nitric oxide important for our brain? Funct. Neurol. 2015, 30, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Chan, J.Y. Brain stem NOS and ROS in neural mechanisms of hypertension. Antioxid. Redox Signal. 2014, 20, 146–163. [Google Scholar] [CrossRef] [PubMed]
- Kempuraj, D.; Thangavel, R.; Natteru, P.A.; Selvakumar, G.P.; Saeed, D.; Zahoor, H.; Zaheer, S.; Iyer, S.S.; Zaheer, A. Neuroinflammation Induces Neurodegeneration. J. Neurol. Neurosurg. Spine 2016, 1, 1003. [Google Scholar] [PubMed]
- Peixoto, C.A.; Oliveira, W.H.; Araújo, S.M.D.R.; Nunes, A.K.S. AMPK activation: Role in the signaling pathways of neuroinflammation and neurodegeneration. Exp. Neurol. 2017, 298, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Das, B.C. Involvement of AMP-activated protein kinase in neuroinflammation and neurodegeneration in the adult and developing brain. Int. J. Dev. Neurosci. 2019, 77, 48–59. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, L.; Hu, Z. Cerebral insulin, insulin signaling pathway, and brain angiogenesis. Neurol. Sci. 2016, 37, 9–16. [Google Scholar] [CrossRef]
- Mammana, S.; Bramanti, P.; Mazzon, E.; Cavalli, E.; Basile, M.S.; Fagone, P.; Petralia, M.C.; McCubrey, J.A.; Nicoletti, F.; Mangano, K. Preclinical evaluation of the PI3K/Akt/mTOR pathway in animal models of multiple sclerosis. Oncotarget 2018, 9, 8263–8277. [Google Scholar] [CrossRef]
- LeVine, S.M. Albumin and multiple sclerosis. BMC Neurol. 2016, 16, 47. [Google Scholar] [CrossRef]
- Jorissen, W.; Wouters, E.; Bogie, J.F.; Vanmierlo, T.; Noben, J.P.; Sviridov, D.; Hellings, N.; Somers, V.; Valcke, R.; Vanwijmeersch, B.; et al. Relapsing-remitting multiple sclerosis patients display an altered lipoprotein profile with dysfunctional HDL. Sci. Rep. 2017, 7, 43410. [Google Scholar] [CrossRef] [PubMed]
- Birkner, K.; Wasser, B.; Loos, J.; Plotnikov, A.; Seger, R.; Zipp, F.; Witsch, E.; Bittner, S. The Role of ERK Signaling in Experimental Autoimmune Encephalomyelitis. Int. J. Mol. Sci. 2017, 18, 1990. [Google Scholar] [CrossRef] [PubMed]
- Zarobkiewicz, M.K.; Kowalska, W.; Slawinski, M.; Rolinski, J.; Bojarska-Junak, A. The role of interleukin 22 in multiple sclerosis and its association with c-Maf and AHR. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech. Repub. 2019, 163, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T. Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol. Res. 2017, 39, 73–82. [Google Scholar] [CrossRef]
- Vallée, A.; Vallée, J.N.; Guillevin, R.; Lecarpentier, Y. Interactions between the Canonical WNT/Beta-Catenin Pathway and PPAR Gamma on Neuroinflammation, Demyelination, and Remyelination in Multiple Sclerosis. Cell Mol. Neurobiol. 2018, 38, 783–795. [Google Scholar] [CrossRef]
- Nakano, T. Roles of lipid-modulating enzymes diacylglycerol kinase and cyclooxygenase under pathophysiological conditions. Anat. Sci. Int. 2015, 90, 22–32. [Google Scholar] [CrossRef]
- Iñarrea, P.; Alarcia, R.; Alava, M.A.; Capablo, J.L.; Casanova, A.; Iñiguez, C.; Iturralde, M.; Larrodé, P.; Martín, J.; Mostacero, E.; et al. Mitochondrial complex enzyme activities and cytochrome C expression changes in multiple sclerosis. Mol. Neurobiol. 2014, 49, 1–9. [Google Scholar] [CrossRef]
- Beinert, H.; Kennedy, M.C. Aconitase, a two-faced protein: Enzyme and iron regulatory factor. FASEB J. 1993, 7, 1442–1449. [Google Scholar] [CrossRef] [PubMed]
- Ropele, S.; Enzinger, C.; Fazekas, F. Iron Mapping in Multiple Sclerosis. Neuroimag. Clin. N. Am. 2017, 27, 335–342. [Google Scholar] [CrossRef]
- Elkamhawy, A.; Hassan, A.H.E.; Paik, S.; Sup Lee, Y.; Lee, H.H.; Shin, J.S.; Lee, K.T.; Roh, E.J. EGFR inhibitors from cancer to inflammation: Discovery of 4-fluoro-N-(4-(3-(trifluoromethyl)phenoxy)pyrimidin-5-yl)benzamide as a novel anti-inflammatory EGFR inhibitor. Bioorg. Chem. 2019, 86, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Berven, L.A.; Willard, F.S.; Crouch, M.F. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell Res. 2004, 296, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.F.; Huang, X.H.; Shen, A.Z.; Li, J.; Sun, Y.H. Association between circulating leptin levels and multiple sclerosis: A systematic review and meta-analysis. Postgrad. Med. J. 2018, 94, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, F.; Wang, Z.J. CaMKIIα Mediates the Effect of IL-17 To Promote Ongoing Spontaneous and Evoked Pain in Multiple Sclerosis. J. Neurosci. 2018, 38, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Kroll, J.R.; Saras, A.; Tanouye, M.A. Drosophila sodium channel mutations: Contributions to seizure-susceptibility. Exp. Neurol. 2015, 274, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, U.K.; Kini, S.G.; Garg, V.; Agrawal, S.; Tomar, P.K.; Pathak, P.; Chaudhary, A.; Gupta, P.; Malik, A. JNK pathway signaling: A novel and smarter therapeutic targets for various biological diseases. Future Med. Chem. 2015, 7, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.I.; Wang, Y.; Wakelam, M.J.O.; Bankaitis, V.A. Mammalian phospholipase D: Function, and therapeutics. Prog. Lipid Res. 2020, 78, 101018. [Google Scholar] [CrossRef] [PubMed]
- Kular, L.; Needhamsen, M.; Adzemovic, M.Z.; Kramarova, T.; Gomez-Cabrero, D.; Ewing, E.; Piket, E.; Tegnér, J.; Beck, S.; Piehl, F.; et al. Neuronal methylome reveals CREB-associated neuro-axonal impairment in multiple sclerosis. Clin. Epigenet. 2019, 11, 86. [Google Scholar] [CrossRef]
- Keating, G.M. Afatinib: A Review in Advanced Non-Small Cell Lung Cancer. Target. Oncol. 2016, 11, 825–835. [Google Scholar] [CrossRef]
- Bagherpour, B.; Salehi, M.; Jafari, R.; Bagheri, A.; Kiani-Esfahani, A.; Edalati, M.; Kardi, M.T.; Shaygannejad, V. Promising effect of rapamycin on multiple sclerosis. Mult. Scler. Relat. Disord. 2018, 26, 40–45. [Google Scholar] [CrossRef]
- Trabjerg, M.S.; Mørkholt, A.S.; Lichota, J.; Oklinski, M.K.E.; Andersen, D.C.; Jønsson, K.; Mørk, K.; Skjønnemand, M.N.; Kroese, L.J.; Pritchard, C.E.J.; et al. Dysregulation of metabolic pathways by carnitine palmitoyl-transferase 1 plays a key role in central nervous system disorders: Experimental evidence based on animal models. Sci. Rep. 2020, 10, 15583. [Google Scholar] [CrossRef]
- Evangelopoulos, M.E.; Koutsis, G.; Markianos, M. Serum leptin levels in treatment-naive patients with clinically isolated syndrome or relapsing-remitting multiple sclerosis. Autoimmune Dis. 2014, 2014, 486282. [Google Scholar] [CrossRef]
- Renoux, F.; Stellato, M.; Haftmann, C.; Vogetseder, A.; Huang, R.; Subramaniam, A.; Becker, M.O.; Blyszczuk, P.; Becher, B.; Distler, J.H.W.; et al. The AP1 Transcription Factor Fosl2 Promotes Systemic Autoimmunity and Inflammation by Repressing Treg Development. Cell Rep. 2020, 31, 107826. [Google Scholar] [CrossRef]
- Mendiola, A.S.; Cardona, A.E. The IL-1β phenomena in neuroinflammatory diseases. J. Neural Transm. 2018, 125, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Graner, M.; Kennedy, P.G.E.; Liu, Y. The Role of Antibodies in the Pathogenesis of Multiple Sclerosis. Front. Neurol. 2020, 11, 533388. [Google Scholar] [CrossRef] [PubMed]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Barreiro, L.; Eixarch, H.; Montalban, X.; Espejo, C. Combined therapies to treat complex diseases: The role of the gut microbiota in multiple sclerosis. Autoimmun. Rev. 2018, 17, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ma, D.; Li, L.; Zhang, L. Progress in the Application of Drugs for the Treatment of Multiple Sclerosis. Front. Pharmacol. 2021, 12, 724718. [Google Scholar] [CrossRef]
- Chitnis, T.; Prat, A. A roadmap to precision medicine for multiple sclerosis. Mult. Scler. 2020, 26, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Bose, G.; Freedman, M.S. Precision medicine in the multiple sclerosis clinic: Selecting the right patient for the right treatment. Mult. Scler. 2020, 26, 540–547. [Google Scholar] [CrossRef]
- Barkhof, F. The clinico-radiological paradox in multiple sclerosis revisited. Curr. Opin. Neurol. 2002, 15, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Habek, M.; Borovecki, F.; Brinar, V.V. Genomics in multiple sclerosis. Clin. Neurol. Neurosurg. 2010, 112, 621–624. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kulakova, O.G.; Kabilov, M.R.; Danilova, L.V.; Popova, E.V.; Baturina, O.A.; Tsareva, E.Y.; Baulina, N.M.; Kiselev, I.S.; Boyko, A.N.; Favorov, A.V.; et al. Whole-Genome DNA Methylation Analysis of Peripheral Blood Mononuclear Cells in Multiple Sclerosis Patients with Different Disease Courses. Acta Nat. 2016, 8, 103–110. [Google Scholar] [CrossRef]
- Srinivasan, S.; Di Dario, M.; Russo, A.; Menon, R.; Brini, E.; Romeo, M.; Sangalli, F.; Costa, G.D.; Rodegher, M.; Radaelli, M.; et al. Dysregulation of MS risk genes and pathways at distinct stages of disease. Neurol. Neuroimmunol. Neuroinflamm. 2017, 4, e337. [Google Scholar] [CrossRef] [PubMed]
- Probert, F.; Yeo, T.; Zhou, Y.; Sealey, M.; Arora, S.; Palace, J.; Claridge, T.D.W.; Hillenbrand, R.; Oechtering, J.; Leppert, D.; et al. Integrative biochemical, proteomics and metabolomics cerebrospinal fluid biomarkers predict clinical conversion to multiple sclerosis. Brain Commun. 2021, 3, fcab084. [Google Scholar] [CrossRef] [PubMed]
- Tintore, M.; Rovira, À.; Río, J.; Otero-Romero, S.; Arrambide, G.; Tur, C.; Comabella, M.; Nos, C.; Arévalo, M.J.; Negrotto, L.; et al. Defining high, medium and low impact prognostic factors for developing multiple sclerosis. Brain 2015, 138, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Roxburgh, R.H.; Seaman, S.R.; Masterman, T.; Hensiek, A.E.; Sawcer, S.J.; Vukusic, S.; Achiti, I.; Confavreux, C.; Coustans, M.; le Page, E.; et al. Multiple Sclerosis Severity Score: Using disability and disease duration to rate disease severity. Neurology 2005, 64, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rispoli, M.G.; Valentinuzzi, S.; De Luca, G.; Del Boccio, P.; Federici, L.; Di Ioia, M.; Digiovanni, A.; Grasso, E.A.; Pozzilli, V.; Villani, A.; et al. Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment. Int. J. Mol. Sci. 2021, 22, 11112. https://doi.org/10.3390/ijms222011112
Rispoli MG, Valentinuzzi S, De Luca G, Del Boccio P, Federici L, Di Ioia M, Digiovanni A, Grasso EA, Pozzilli V, Villani A, et al. Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment. International Journal of Molecular Sciences. 2021; 22(20):11112. https://doi.org/10.3390/ijms222011112
Chicago/Turabian StyleRispoli, Marianna Gabriella, Silvia Valentinuzzi, Giovanna De Luca, Piero Del Boccio, Luca Federici, Maria Di Ioia, Anna Digiovanni, Eleonora Agata Grasso, Valeria Pozzilli, Alessandro Villani, and et al. 2021. "Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment" International Journal of Molecular Sciences 22, no. 20: 11112. https://doi.org/10.3390/ijms222011112
APA StyleRispoli, M. G., Valentinuzzi, S., De Luca, G., Del Boccio, P., Federici, L., Di Ioia, M., Digiovanni, A., Grasso, E. A., Pozzilli, V., Villani, A., Chiarelli, A. M., Onofrj, M., Wise, R. G., Pieragostino, D., & Tomassini, V. (2021). Contribution of Metabolomics to Multiple Sclerosis Diagnosis, Prognosis and Treatment. International Journal of Molecular Sciences, 22(20), 11112. https://doi.org/10.3390/ijms222011112








