Translational Applications of Linear and Circular Long Noncoding RNAs in Endometriosis
Abstract
1. Introduction
2. Biogenesis, Structure, and Function of Linear and Circular lncRNAs
2.1. Biogenesis, Structure, and Function of Linear lncRNAs
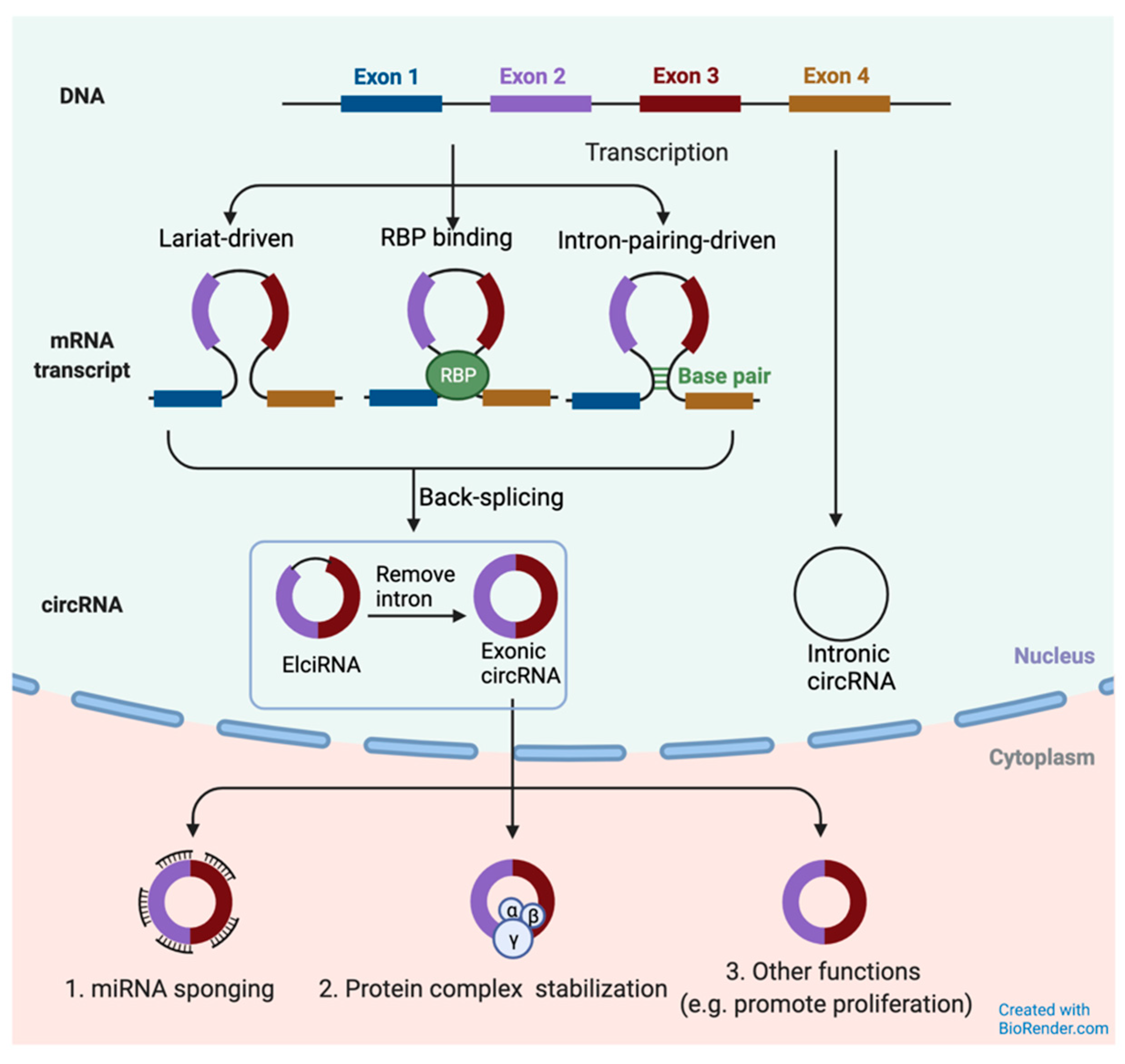
2.2. CircRNA Biogenesis and Structure
3. Approaches for Discovering lncRNAs
3.1. ncRNAs Databases
3.2. High Throughput Identification
3.3. Experimental Validation
4. The Importance of Noninvasive Biomarkers in Endometriosis
5. Therapeutic Opportunities for lncRNAs
6. Challenges to Clinical Application and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Bulun, S.E. Endometriosis. N. Engl. J. Med. 2009, 360, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Giudice, L.C.; Kao, L.C. Endometriosis. Lancet 2004, 364, 1789–1799. [Google Scholar] [CrossRef]
- Holoch, K.J.; Lessey, B.A. Endometriosis and infertility. Clin. Obs. Gynecol. 2010, 53, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.L.; Khachikyan, I.; Stratton, P. Invasive and noninvasive methods for the diagnosis of endometriosis. Clin. Obs. Gynecol. 2010, 53, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.S.; Kotlyar, A.M.; Flores, V.A. Endometriosis is a chronic systemic disease: Clinical challenges and novel innovations. Lancet 2021, 397, 839–852. [Google Scholar] [CrossRef]
- Falcone, T.; Flyckt, R. Clinical Management of Endometriosis. Obs. Gynecol. 2018, 131, 557–571. [Google Scholar] [CrossRef]
- Somigliana, E.; Vercellini, P.; Vigano, P.; Benaglia, L.; Crosignani, P.G.; Fedele, L. Non-invasive diagnosis of endometriosis: The goal or own goal? Hum. Reprod. 2010, 25, 1863–1868. [Google Scholar] [CrossRef]
- Practice bulletin, no. 114: Management of endometriosis. Obs. Gynecol. 2010, 116, 223–236. [Google Scholar]
- Arruda, M.S.; Petta, C.A.; Abrao, M.S.; Benetti-Pinto, C.L. Time elapsed from onset of symptoms to diagnosis of endometriosis in a cohort study of Brazilian women. Hum. Reprod. 2003, 18, 756–759. [Google Scholar] [CrossRef]
- Brown, J.; Crawford, T.J.; Allen, C.; Hopewell, S.; Prentice, A. Nonsteroidal anti-inflammatory drugs for pain in women with endometriosis. Cochrane Database Syst Rev. 2017, 1, CD004753. [Google Scholar] [CrossRef]
- Taylor, H.S.; Giudice, L.C.; Lessey, B.A.; Abrao, M.S.; Kotarski, J.; Archer, D.F.; Diamond, M.P.; Surrey, E.; Johnson, N.P.; Watts, N.B.; et al. Treatment of Endometriosis-Associated Pain with Elagolix, an Oral GnRH Antagonist. N. Engl. J. Med. 2017, 377, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, C. Endometriosis. BMJ 2007, 334, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Yang, H.; Du, E.X.; Kelley, C.; Winkel, C. The direct and indirect costs associated with endometriosis: A systematic literature review. Hum. Reprod. 2016, 31, 712–722. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.R.H.; Wang, X.; Hawkins, S.M. The Endometriotic Tumor Microenvironment in Ovarian Cancer. Cancers 2018, 10, 261. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Ghaffari, S.H. Why have microRNA biomarkers not been translated from bench to clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, S.; Taylor, H.S. MicroRNAs in endometriosis: Biological function and emerging biomarker candidatesdagger. Biol. Reprod. 2019, 100, 1135–1146. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Charles Richard, J.L.; Eichhorn, P.J.A. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018, 23, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Liu, X.; Wei, B.; Zhan, L. Long noncoding RNAs in endometriosis: Biological functions, expressions, and mechanisms. J. Cell Physiol. 2021, 236, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e3. [Google Scholar] [CrossRef]
- Pertea, M. The human transcriptome: An unfinished story. Genes 2012, 3, 344–360. [Google Scholar] [CrossRef]
- Hube, F.; Francastel, C. Coding and Non-coding RNAs, the Frontier Has Never Been So Blurred. Front. Genet. 2018, 9, 140. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. Gencode 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- van Rossum, D.; Verheijen, B.M.; Pasterkamp, R.J. Circular RNAs: Novel Regulators of Neuronal Development. Front. Mol. Neurosci 2016, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef]
- Sun, M.; Kraus, W.L. From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Pan, X.; Liu, M.; Wang, S.; Huang, T.; Cai, Y.D. Tissue Expression Difference between mRNAs and lncRNAs. Int. J. Mol. Sci. 2018, 19, 3416. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Yang, Z.; Jia, R.; Ge, S. The novel roles of circRNAs in human cancer. Mol. Cancer 2019, 18, 6. [Google Scholar] [CrossRef]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Chen, Y.Q. Principles and innovative technologies for decrypting noncoding RNAs: From discovery and functional prediction to clinical application. J. Hematol. Oncol. 2020, 13, 109. [Google Scholar] [CrossRef]
- Shoemaker, D.D.; Schadt, E.E.; Armour, C.D.; He, Y.D.; Garrett-Engele, P.; McDonagh, P.D.; Loerch, P.M.; Leonardson, A.; Lum, P.Y.; Cavet, G.; et al. Experimental annotation of the human genome using microarray technology. Nature 2001, 409, 922–927. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ilott, N.E.; Ponting, C.P. Predicting long non-coding RNAs using RNA sequencing. Methods 2013, 63, 50–59. [Google Scholar] [CrossRef]
- Bertone, P.; Stolc, V.; Royce, T.E.; Rozowsky, J.S.; Urban, A.E.; Zhu, X.; Rinn, J.L.; Tongprasit, W.; Samanta, M.; Weissman, S.; et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, 2242–2246. [Google Scholar] [CrossRef]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T.; et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef]
- Mumbach, M.R.; Granja, J.M.; Flynn, R.A.; Roake, C.M.; Satpathy, A.T.; Rubin, A.J.; Qi, Y.; Jiang, Z.; Shams, S.; Louie, B.H.; et al. HiChIRP reveals RNA-associated chromosome conformation. Nat. Methods 2019, 16, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D.; Wang, C.I.; Kharchenko, P.V.; West, J.A.; Chapman, B.A.; Alekseyenko, A.A.; Borowsky, M.L.; Kuroda, M.I.; Kingston, R.E. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20497–20502. [Google Scholar] [CrossRef] [PubMed]
- Stadhouders, R.; Kolovos, P.; Brouwer, R.; Zuin, J.; van den Heuvel, A.; Kockx, C.; Palstra, R.J.; Wendt, K.S.; Grosveld, F.; van Ijcken, W.; et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protoc. 2013, 8, 509–524. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Ule, J.; Hwang, H.W.; Darnell, R.B. The Future of Cross-Linking and Immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol. 2018, 10, a032243. [Google Scholar] [CrossRef] [PubMed]
- Friedersdorf, M.B.; Keene, J.D. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014, 15, R2. [Google Scholar] [CrossRef]
- McHugh, C.A.; Russell, P.; Guttman, M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014, 15, 203. [Google Scholar] [CrossRef]
- Rio, D.C. 5′-end labeling of RNA with [gamma-32P]ATP and T4 polynucleotide kinase. Cold Spring Harb. Protoc. 2014, 2014, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.V.; Chekanova, J.A. An Overview of Methodologies in Studying lncRNAs in the High-Throughput Era: When Acronyms ATTACK! Methods Mol. Biol. 2019, 1933, 1–30. [Google Scholar]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Marran, K.; Valentine, A.; Hannon, G.J. RNAi in cultured mammalian cells using synthetic siRNAs. Cold Spring Harb. Protoc. 2012, 2012, 957–961. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Birmingham, A.; Wiemann, S.; Beijersbergen, R.L.; Hornung, V.; Smith, A. Advances in CRISPR-Cas9 genome engineering: Lessons learned from RNA interference. Nucleic Acids Res. 2015, 43, 3407–3419. [Google Scholar] [CrossRef]
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Considerations when investigating lncRNA function in vivo. Elife 2014, 3, e03058. [Google Scholar] [CrossRef]
- Aparicio-Prat, E.; Arnan, C.; Sala, I.; Bosch, N.; Guigo, R.; Johnson, R. DECKO: Single-oligo, dual-CRISPR deletion of genomic elements including long non-coding RNAs. BMC Genom. 2015, 16, 846. [Google Scholar] [CrossRef]
- Goyal, A.; Myacheva, K.; Gross, M.; Klingenberg, M.; Duran Arque, B.; Diederichs, S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017, 45, e12. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Maracaja-Coutinho, V.; Paschoal, A.R.; Caris-Maldonado, J.C.; Borges, P.V.; Ferreira, A.J.; Durham, A.M. Noncoding RNAs Databases: Current Status and Trends. Methods Mol. Biol. 2019, 1912, 251–285. [Google Scholar] [PubMed]
- Ma, H.; Hao, Y.; Dong, X.; Gong, Q.; Chen, J.; Zhang, J.; Tian, W. Molecular mechanisms and function prediction of long noncoding RNA. ScientificWorldJournal 2012, 2012, 541786. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S. High-Throughput Methods to Detect Long Non-Coding RNAs. High. Throughput 2017, 6, 12. [Google Scholar]
- Carrara, M.; Fuschi, P.; Ivan, C.; Martelli, F. Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J. Cell Mol. Med. 2018, 22, 5176–5187. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, Y.; Zhou, W.; Peng, Z.; Hong, X.; Zhang, Y. Circular RNA expression in ovarian endometriosis. Epigenomics 2018, 10, 559–572. [Google Scholar] [CrossRef]
- Bi, J.; Wang, D.; Cui, L.; Yang, Q. RNA sequencing-based long non-coding RNA analysis and immunoassay in ovarian endometriosis. Am. J. Reprod. Immunol. 2021, 85, e13359. [Google Scholar] [CrossRef]
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2019, 20, 1420–1433. [Google Scholar] [CrossRef]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef]
- Wu, J.; Huang, H.; Huang, W.; Wang, L.; Xia, X.; Fang, X. Analysis of exosomal lncRNA, miRNA and mRNA expression profiles and ceRNA network construction in endometriosis. Epigenomics 2020, 12, 1193–1213. [Google Scholar] [CrossRef]
- Wang, D.; Luo, Y.; Wang, G.; Yang, Q. Circular RNA expression profiles and bioinformatics analysis in ovarian endometriosis. Mol. Genet. Genom. Med. 2019, 7, e00756. [Google Scholar] [CrossRef]
- Xu, D.; Cai, Y.; Tang, L.; Han, X.; Gao, F.; Cao, H.; Qi, F.; Kapranov, P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020, 10, 1794. [Google Scholar] [CrossRef]
- Holdsworth-Carson, S.J.; Churchill, M.; Donoghue, J.F.; Mortlock, S.; Fung, J.N.; Sloggett, C.; Chung, J.; Cann, L.; Teh, W.T.; Campbell, K.R.; et al. Elucidating the role of long intergenic non-coding RNA 339 in human endometrium and endometriosis. Mol. Hum. Reprod. 2021, 27, gaab010. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef]
- Wang, H.; Sha, L.; Huang, L.; Yang, S.; Zhou, Q.; Luo, X.; Shi, B. LINC00261 functions as a competing endogenous RNA to regulate BCL2L11 expression by sponging miR-132-3p in endometriosis. Am. J. Transl. Res. 2019, 11, 2269–2279. [Google Scholar] [PubMed]
- Brosens, I.A.; Brosens, J.J. Is laparoscopy the gold standard for the diagnosis of endometriosis? Eur J. Obs. Gynecol. Reprod. Biol. 2000, 88, 117–119. [Google Scholar] [CrossRef]
- Tammaa, A.; Fritzer, N.; Strunk, G.; Krell, A.; Salzer, H.; Hudelist, G. Learning curve for the detection of pouch of Douglas obliteration and deep infiltrating endometriosis of the rectum. Hum. Reprod. 2014, 29, 1199–1204. [Google Scholar] [CrossRef]
- Canis, M.; Mage, G.; Wattiez, A.; Pouly, J.L.; Bruhat, M.A. The ovarian endometrioma: Why is it so poorly managed? Laparoscopic treatment of large ovarian endometrioma: Why such a long learning curve? Hum. Reprod. 2003, 18, 5–7. [Google Scholar] [CrossRef][Green Version]
- Balasch, J.; Creus, M.; Fabregues, F.; Carmona, F.; Ordi, J.; Martinez-Roman, S.; Vanrell, J.A. Visible and non-visible endometriosis at laparoscopy in fertile and infertile women and in patients with chronic pelvic pain: A prospective study. Hum. Reprod. 1996, 11, 387–391. [Google Scholar] [CrossRef]
- Parasar, P.; Ozcan, P.; Terry, K.L. Endometriosis: Epidemiology, Diagnosis and Clinical Management. Curr. Obs. Gynecol. Rep. 2017, 6, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Savelli, L. Transvaginal sonography for the assessment of ovarian and pelvic endometriosis: How deep is our understanding? Ultrasound Obs. Gynecol. 2009, 33, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; Sun, Y.M.; Huang, W.; He, B.; Zhao, Y.N.; Chen, Y.Q. Genome-wide Long Non-coding RNA Analysis Identified Circulating LncRNAs as Novel Non-invasive Diagnostic Biomarkers for Gynecological Disease. Sci. Rep. 2016, 6, 23343. [Google Scholar] [CrossRef]
- Cui, D.; Ma, J.; Liu, Y.; Lin, K.; Jiang, X.; Qu, Y.; Lin, J.; Xu, K. Analysis of long non-coding RNA expression profiles using RNA sequencing in ovarian endometriosis. Gene 2018, 673, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhu, Z.; Song, Y. Downregulation of lncrna uca1 as a diagnostic and prognostic biomarker for ovarian endometriosis. Rev. Assoc. Med. Bras. 2019, 65, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Jia, S.Z.; Dai, Y.; Zhang, J.J.; Li, X.Y.; Shi, J.H.; Leng, J.H.; Lang, J.H. Identification of Circular RNAs as a Novel Biomarker for Ovarian Endometriosis. Chin. Med. J. 2018, 131, 559–566. [Google Scholar] [CrossRef]
- Xu, X.; Jia, S.Z.; Dai, Y.; Zhang, J.J.; Li, X.; Shi, J.; Leng, J.; Lang, J. The Relationship of Circular RNAs With Ovarian Endometriosis. Reprod. Sci. 2018, 25, 1292–1300. [Google Scholar] [CrossRef]
- Khalaj, K.; Miller, J.E.; Lingegowda, H.; Fazleabas, A.T.; Young, S.L.; Lessey, B.A.; Koti, M.; Tayade, C. Extracellular vesicles from endometriosis patients are characterized by a unique miRNA-lncRNA signature. JCI Insight 2019, 4, e128846. [Google Scholar] [CrossRef]
- Qiu, J.J.; Lin, Y.Y.; Tang, X.Y.; Ding, Y.; Yi, X.F.; Hua, K.Q. Extracellular vesicle-mediated transfer of the lncRNA-TC0101441 promotes endometriosis migration/invasion. Exp. Cell Res. 2020, 388, 111815. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xin, W.; Tang, X.; Qiu, J.; Zhang, Y.; Hua, K. LncRNA H19 Overexpression in Endometriosis and its Utility as a Novel Biomarker for Predicting Recurrence. Reprod. Sci. 2020, 27, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef]
- Sukowati, C.H.C.; Cabral, L.K.D.; Tiribelli, C.; Pascut, D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines 2021, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Xu, Z.Y.; Ruan, S.M.; Mo, S.; Qin, J.J.; Cheng, X.D. Long non-coding RNAs towards precision medicine in gastric cancer: Early diagnosis, treatment, and drug resistance. Mol. Cancer 2020, 19, 96. [Google Scholar] [CrossRef]
- Lessey, B.A.; Kim, J.J. Endometrial receptivity in the eutopic endometrium of women with endometriosis: It is affected, and let me show you why. Fertil. Steril. 2017, 108, 19–27. [Google Scholar] [CrossRef]
- Xu, X.; Li, Z.; Liu, J.; Yu, S.; Wei, Z. MicroRNA expression profiling in endometriosis-associated infertility and its relationship with endometrial receptivity evaluated by ultrasound. J. Xray Sci. Technol. 2017, 25, 523–532. [Google Scholar] [CrossRef]
- Mari-Alexandre, J.; Barcelo-Molina, M.; Belmonte-Lopez, E.; Garcia-Oms, J.; Estelles, A.; Braza-Boils, A.; Gilabert-Estelles, J. Micro-RNA profile and proteins in peritoneal fluid from women with endometriosis: Their relationship with sterility. Fertil. Steril. 2018, 109, 675–684.e2. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, W.; Fu, J.; Xu, Y.; Gu, R.; Qu, R.; Li, L.; Sun, Y.; Sun, X. MicroRNA-451 is downregulated in the follicular fluid of women with endometriosis and influences mouse and human embryonic potential. Reprod. Biol. Endocrinol. 2019, 17, 96. [Google Scholar] [CrossRef]
- Pearce, C.L.; Templeman, C.; Rossing, M.A.; Lee, A.; Near, A.M.; Webb, P.M.; Nagle, C.M.; Doherty, J.A.; Cushing-Haugen, K.L.; Wicklund, K.G.; et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case-control studies. Lancet Oncol. 2012, 13, 385–394. [Google Scholar] [CrossRef]
- Nakamura, N.; Terai, Y.; Nunode, M.; Kokunai, K.; Konishi, H.; Taga, S.; Nakamura, M.; Yoo, M.; Hayashi, M.; Yamashita, Y.; et al. The differential expression of miRNAs between ovarian endometrioma and endometriosis-associated ovarian cancer. J. Ovarian Res. 2020, 13, 51. [Google Scholar] [CrossRef]
- Suryawanshi, S.; Vlad, A.M.; Lin, H.M.; Mantia-Smaldone, G.; Laskey, R.; Lee, M.; Lin, Y.; Donnellan, N.; Klein-Patel, M.; Lee, T.; et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin. Cancer Res. 2013, 19, 1213–1224. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Ghazal, S.; McKinnon, B.; Zhou, J.; Mueller, M.; Men, Y.; Yang, L.; Mueller, M.; Flannery, C.; Huang, Y.; Taylor, H.S. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol. Med. 2015, 7, 996–1003. [Google Scholar] [CrossRef]
- Gu, Z.Y.; Jia, S.Z.; Leng, J.H. Establishment of endometriotic models: The past and future. Chin. Med. J. 2020, 133, 1703–1710. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Xanthoulea, S.; Giacomini, E.; Delvoux, B.; Alleva, E.; Vigano, P. Endometriotic cell culture contamination and authenticity: A source of bias in in vitro research? Hum. Reprod. 2020, 35, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Huan, Q.; Cheng, S.C.; Du, Z.H.; Ma, H.F.; Li, C. LncRNA AFAP1-AS1 regulates proliferation and apoptosis of endometriosis through activating STAT3/TGF-beta/Smad signaling via miR-424-5p. J. Obs. Gynaecol. Res. 2021, 47, 2394–2405. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Huang, Q.; Wu, R.; Dai, S.; Huang, Z.; Ren, L.; Huang, S.; Chen, Q. Long non-coding RNA AFAP1-AS1 promoting epithelial-mesenchymal transition of endometriosis is correlated with transcription factor ZEB1. Am. J. Reprod Immunol. 2019, 81, e13074. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, X.; Lu, D.; Feng, Y.; Xu, R.; Li, X.; Yin, C.; Xue, B.; Zhao, H.; Wang, S.; et al. LncRNA SNHG4 promotes the increased growth of endometrial tissue outside the uterine cavity via regulating c-Met mediated by miR-148a-3p. Mol. Cell Endocrinol. 2020, 514, 110887. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, J.; Cui, D.; Fei, X.; Lv, Y.; Lin, J. LncRNA MEG3-210 regulates endometrial stromal cells migration, invasion and apoptosis through p38 MAPK and PKA/SERCA2 signalling via interaction with Galectin-1 in endometriosis. Mol. Cell Endocrinol. 2020, 513, 110870. [Google Scholar] [CrossRef]
- Cakmak, H.; Seval-Celik, Y.; Arlier, S.; Guzeloglu-Kayisli, O.; Schatz, F.; Arici, A.; Kayisli, U.A. p38 Mitogen-Activated Protein Kinase is Involved in the Pathogenesis of Endometriosis by Modulating Inflammation, but not Cell Survival. Reprod. Sci. 2018, 25, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Chetry, M.; Thapa, S.; Hu, X.; Song, Y.; Zhang, J.; Zhu, H.; Zhu, X. The Role of Galectins in Tumor Progression, Treatment and Prognosis of Gynecological Cancers. J. Cancer 2018, 9, 4742–4755. [Google Scholar]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. Author Correction: MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2019, 10, 5290. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, S.M.; Creighton, C.J.; Han, D.Y.; Zariff, A.; Anderson, M.L.; Gunaratne, P.H.; Matzuk, M.M. Functional microRNA involved in endometriosis. Mol. Endocrinol. 2011, 25, 821–832. [Google Scholar] [CrossRef]
- Dong, L.; Zhang, L.; Liu, H.; Xie, M.; Gao, J.; Zhou, X.; Zhao, Q.; Zhang, S.; Yang, J. Circ_0007331 knock-down suppresses the progression of endometriosis via miR-200c-3p/HiF-1alpha axis. J. Cell Mol. Med. 2020, 24, 12656–12666. [Google Scholar] [CrossRef]
- He, X.; Liu, N.; Mu, T.; Lu, D.; Jia, C.; Wang, S.; Yin, C.; Liu, L.; Zhou, L.; Huang, X.; et al. Oestrogen induces epithelial-mesenchymal transition in endometriosis via circ_0004712/miR-148a-3p sponge function. J. Cell. Mol. Med. 2020, 24, 9658–9666. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Chang, C.Y.; Tseng, C.C.; Lai, M.T.; Chiang, A.J.; Lo, L.C.; Chen, C.M.; Yen, M.J.; Sun, L.; Yang, L.; Hwang, T.; et al. Genetic impacts on thermostability of onco-lncRNA HOTAIR during the development and progression of endometriosis. PLoS ONE 2021, 16, e0248168. [Google Scholar] [CrossRef]
- Du, H.; Taylor, H.S. The Role of Hox Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harb. Perspect. Med. 2015, 6, a023002. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Ozes, A.R.; Miller, D.F.; Ozes, O.N.; Fang, F.; Liu, Y.; Matei, D.; Huang, T.; Nephew, K.P. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35, 5350–5361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, W.; Zhu, H.; Yu, X.; Zhang, Y.; Ye, X.; Cheng, H.; Ma, R.; Cui, H.; Luo, J.; et al. Knockdown of long noncoding RNA CCDC144NL-AS1 attenuates migration and invasion phenotypes in endometrial stromal cells from endometriosisdagger. Biol. Reprod. 2019, 100, 939–949. [Google Scholar] [CrossRef]
- Kennedy, S.; Bergqvist, A.; Chapron, C.; D’Hooghe, T.; Dunselman, G.; Greb, R.; Hummelshoj, L.; Prentice, A.; Saridogan, E.; Soriano, D.; et al. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum. Reprod. 2005, 20, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Krakowsky, R.H.; Tollefsbol, T.O. Impact of Nutrition on Non-Coding RNA Epigenetics in Breast and Gynecological Cancer. Front. Nutr. 2015, 2, 16. [Google Scholar] [CrossRef]
- Agrawal, S.; Tapmeier, T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.; Becker, C. The miRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci 2018, 19, 599. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.M.; Laufer, M.R.; Stratton, P.; Hummelshoj, L.; Missmer, S.A.; Zondervan, K.T.; Adamson, G.D.; Group, W.E.W. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: I. Surgical phenotype data collection in endometriosis research. Fertil. Steril. 2014, 102, 1213–1222. [Google Scholar] [CrossRef]
- Ciebiera, M.; Esfandyari, S.; Siblini, H.; Prince, L.; Elkafas, H.; Wojtyla, C.; Al-Hendy, A.; Ali, M. Nutrition in Gynecological Diseases: Current Perspectives. Nutrients 2021, 13, 1178. [Google Scholar] [CrossRef]
- Kalra, E.K. Nutraceutical—Definition and introduction. AAPS PharmSci. 2003, 5, E25. [Google Scholar] [CrossRef]
- Machairiotis, N.; Vasilakaki, S.; Kouroutou, P. Natural products: Potential lead compounds for the treatment of endometriosis. Eur. J. Obs. Gynecol. Reprod. Biol. 2020, 245, 7–12. [Google Scholar] [CrossRef]
| Technique | Description | Ref |
|---|---|---|
| Microarray | Identify lncRNAs | [39] |
| RNA-sequencing | Identify lncRNAs | [40,41] |
| Tiling arrays | Identify and characterize lncRNAs | [42] |
| CAGE (Cap analysis of gene expression) | Identify lncRNAs | [43] |
| ChIRP-seq (chromatin isolation by RNA purification sequencing) | Identify lncRNA-chromatin interactions | [44,45] |
| CHART-seq (capture hybridization analysis of RNA targets) | Identify lncRNA-chromatin interactions | [46] |
| 3C (chromosome confirmation capture) | Characterize lncRNA-genome binding site | [47] |
| RAP (RNA antisense purification) | Characterize lncRNA-genome binding site | [48] |
| RIP-seq (RNA immunoprecipitation) | Identify lncRNA-protein interactions | [49,50] |
| PAR-CLIP (Photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation) | Identify lncRNA-protein interactions | [51] |
| RNA pull-downs | Identify lncRNA-protein interactions | [52] |
| EMSA (Electrophoretic mobility shift assay) | Characterize lncRNA-protein complexes | [53] |
| RT-qPCR (real-time quantitative polymerase chain reactions) | Cellular localization and expression | [54] |
| RNA-FISH (RNA-fluorescent in situ hybridization) | Cellular localization | [55] |
| RNA-ISH (RNA- in situ hybridization) | Cellular localization | [54] |
| RNAi (RNA interference) | Knockdown lncRNAs | [56,57] |
| CRISPR-Cas9 | Knockdown lncRNAs | [58,59,60,61] |
| ASO (Antisense oligonucleotides) | Knockdown lncRNAs | [62] |
| Clinical Application | ncRNA | Methods | Number of Patients | Tissue Types | Cycle Phase | Diagnostic Value | Study Type | Ref. |
|---|---|---|---|---|---|---|---|---|
| Noninvasive diagnostic biomarkers | NR_038395, NR_038452, ENST00000482343, ENST00000544649, ENST00000393610 | Genome-wide transcriptome array | 59 endometriosis 51 controls | Case: Endometriosis tissue (eutopic and ectopic endometrium) and blood samples Control: Eutopic endometrium, blood sample | Follicular, 50; luteal, 9 of 59 endometriosis patients Follicular, 44; luteal, 7 of 51 control patients | ENST0000048234 alone: 72.41% sensitivity and 71.74% specificity Panel of NR_038395, NR_038452, ENST00000482343, ENST00000544649 ENST00000393610: 89.66% sensitivity and 73.17% specificity | Case-Control | [83] |
| Biomarker discovery | 86 total differently expressed SNORD3A TCONS_00006582 ABO TCONS_08347373 | RNA Sequencing | 17 endometriosis 17 controls | Case: Ectopic endometrium Control: eutopic endometrium | Proliferative | Not evaluated | Case-control | [84] |
| Noninvasive diagnostic biomarkers | UCA1 | qRT PCR | 98 endometriosis 28 controls | Case: Serum, eutopic-ectopic endometrium Controls: serum | Not evaluated | Stage I: specificity of 80.1% and sensitivity of 76.7% Stage II: specificity of 85.6% and sensitivity of 81.1% Stage III: specificity of 89.1% and sensitivity of 88.1 Stage IV: specificity of 90.5 % and sensitivity of 89.0% | Case-control | [85] |
| Biomarker discovery | circ_0004712 circ_0002198 | CircRNA array | 41 endometriosis 22 controls | Case: Ectopic, eutopic endometrium Control: Eutopic endometrium | Proliferative, 28; secretive, 13 of 41 endometriosis patients Proliferative, 16; secretive, 6 of 22 control patients | Not evaluated | Case-control | [86] |
| Biomarker discovery Therapeutic target discovery | circ_0004712, circ_0002198, circ_0003570, circ_0008951, circ_0017248 | CircRNA array | 41 endometriosis | Case: Ectopic, eutopic endometrium Control: Eutopic endometrium | Proliferative, 30; secretory, 11 of 41 endometriosis patients | Not evaluated | Discovery | [87] |
| Noninvasive diagnostic biomarkers | MEG8, SNHG25, LINC00293, LINC00929, RP5-898J17.1, NEAT1, H19 | Small RNA sequencing | 6 endometriosis Controls none | Case Eutopic ectopic endometrium, plasma, peritoneal fluid (PF) | Not evaluated | Not evaluated | Discovery | [88] |
| Noninvasive diagnostic biomarkers | TC0101441 | FISH qRT-PCR | 10 endometriosis 10 control | Case: Ectopic, eutopic endometrium, serum Controls: Eutopic endometrium, serum | Not evaluated | Not evaluated | Discovery | [89] |
| Noninvasive diagnostic biomarkers | TC0101441 | qRT-PCR | 29 endometriosis 16 controls | Case: Serum Controls: Serum | Not evaluated | Not evaluated | Discovery | [89] |
| Noninvasive diagnostic biomarker Recurrence | H19 | qRT-PCR | 104 endometriosis 50 controls | Case: Ectopic, eutopic endometrium Controls: Eutopic endometrium | Proliferative | sensitivity 90.9% and specificity 61.0%, for predicting recurrence | Case-control | [90] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Parodi, L.; Hawkins, S.M. Translational Applications of Linear and Circular Long Noncoding RNAs in Endometriosis. Int. J. Mol. Sci. 2021, 22, 10626. https://doi.org/10.3390/ijms221910626
Wang X, Parodi L, Hawkins SM. Translational Applications of Linear and Circular Long Noncoding RNAs in Endometriosis. International Journal of Molecular Sciences. 2021; 22(19):10626. https://doi.org/10.3390/ijms221910626
Chicago/Turabian StyleWang, Xiyin, Luca Parodi, and Shannon M. Hawkins. 2021. "Translational Applications of Linear and Circular Long Noncoding RNAs in Endometriosis" International Journal of Molecular Sciences 22, no. 19: 10626. https://doi.org/10.3390/ijms221910626
APA StyleWang, X., Parodi, L., & Hawkins, S. M. (2021). Translational Applications of Linear and Circular Long Noncoding RNAs in Endometriosis. International Journal of Molecular Sciences, 22(19), 10626. https://doi.org/10.3390/ijms221910626






