The Role of Long Non-Coding RNAs in Endometriosis
Abstract
1. Introduction
2. Aims and Methodology of This Review
3. Evidence for the Role of lncRNAs in Endometriosis
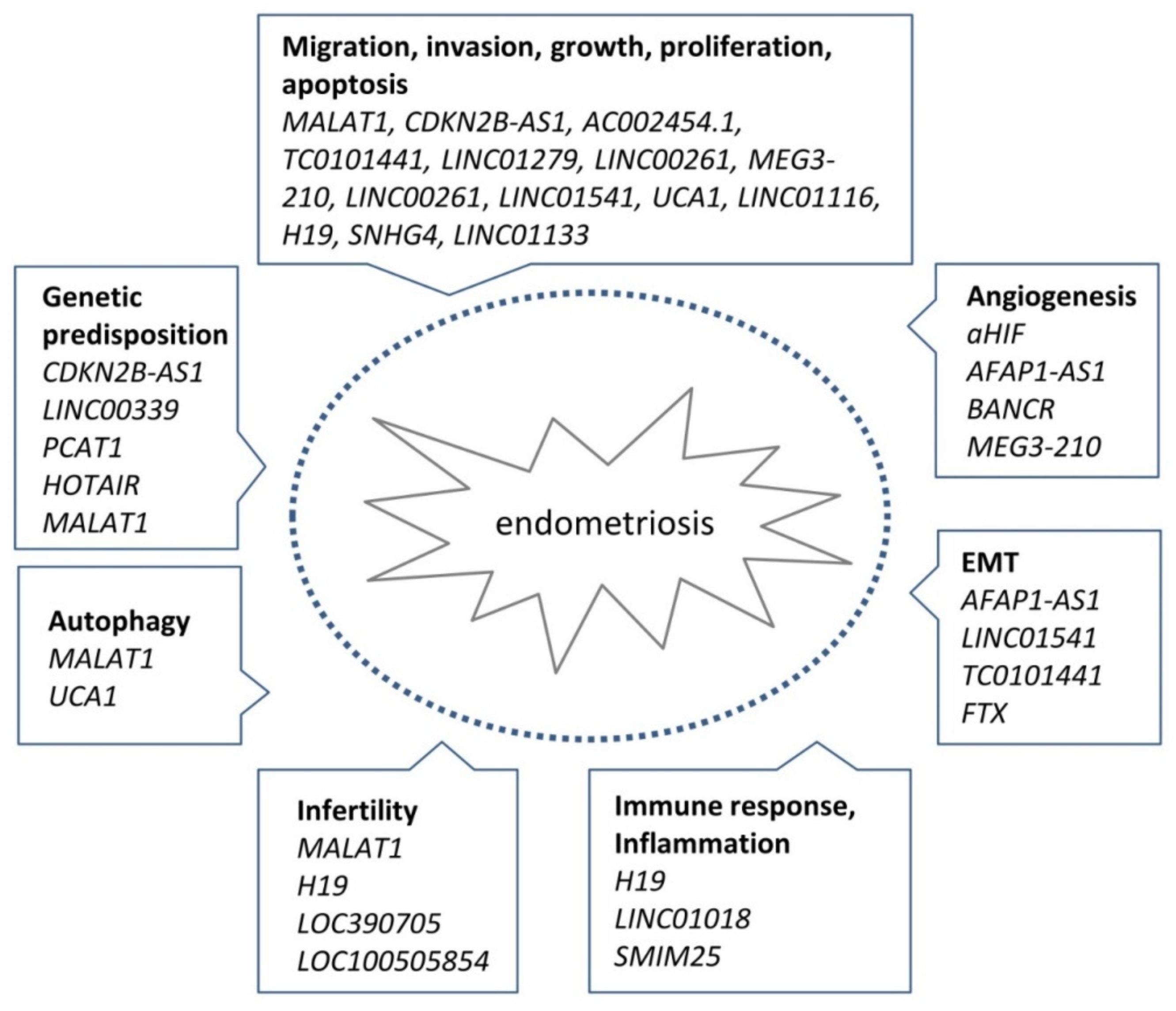
3.1. Genetic Evidence for lncRNA Involvement in Endometriosis
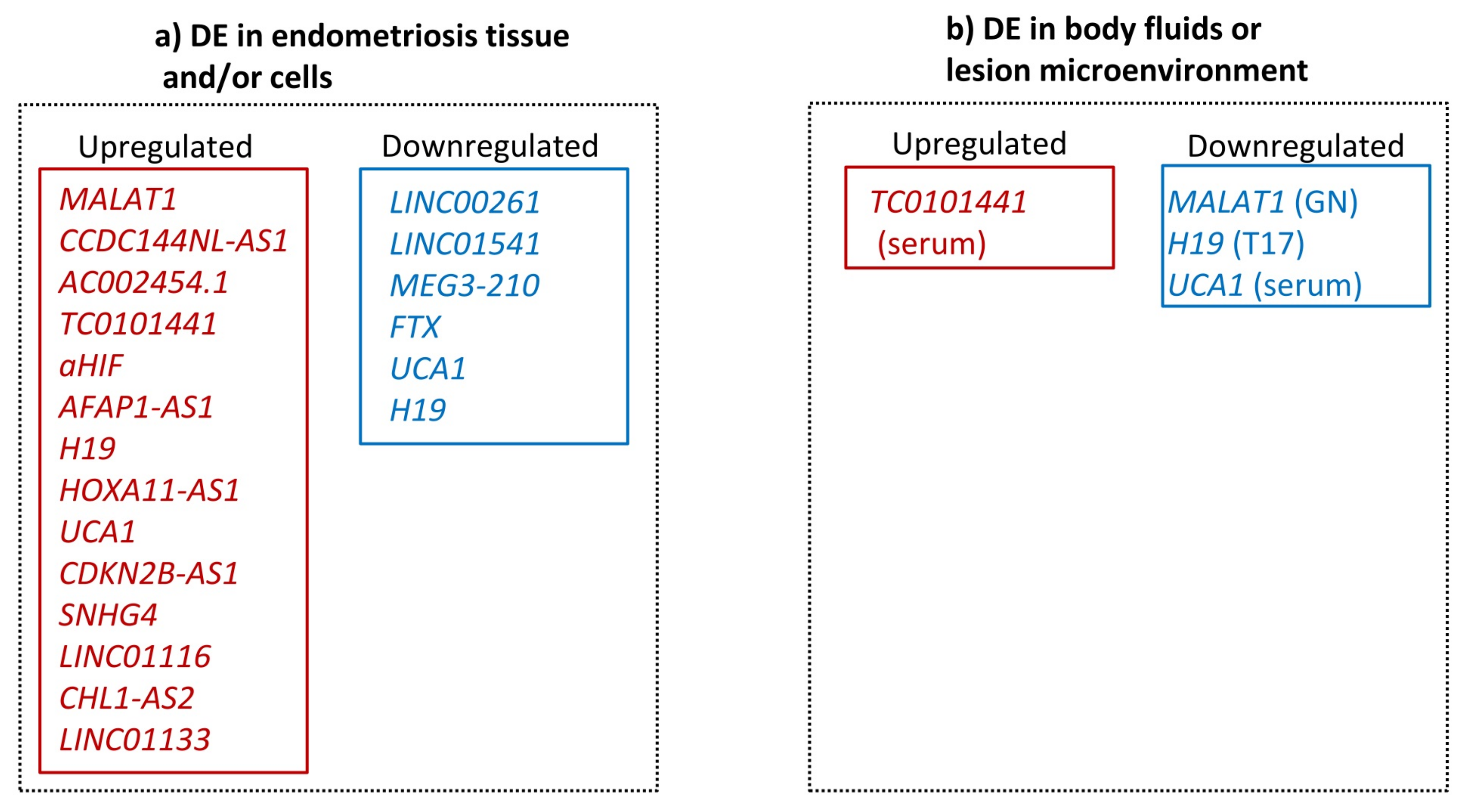
3.2. Transcriptional Evidence for lncRNA Involvement in Endometriosis
| Method (Reference) | Cohort (Tissue Type) | Validation (Type; Cohort; lncRNAs) | Predicted Function in Endometriosis | Limitations |
|---|---|---|---|---|
| Expression Microarray [33] | n = 8 (paired ovarian: 4 eutopic and 4 ectopic tissues) | RT-qPCR n = 42 (paired ovarian: 21 eutopic and 21 ectopic tissues) CHL1-AS2, MGC24103, XLOC_007433, HOXA11-AS, KLP1, LOC100505776, XLOC_012981, LIMS3-LOC4408, LOC389906 | Cis and trans regulation of protein coding genes | 1, 6 |
| Expression Microarray [37] | n = 6 (3 eutopic tissues of women with EM of undefined entity and 3 eutopic tissues of women without EM) | RT-qPCR n = 68 (40 eutopic tissues of women with EM of undefined entity and 28 eutopic tissues of women without EM) RP11-369C8.1, RP11-432J24.5, AC068282.3, GBP1P1, SNHG1, AC002454.1, AC007246.3, FTX | Cell cycle regulation and immune response | 1, 2, 4, 6 |
| Expression Microarray [35] | Serum: n = 20 (10 women with peritoneal and/or ovarian EM and 10 women without EM) Tissue: n = 15 (paired peritoneal and/or ovarian: 5 eutopic and 5 ectopic EM patients and 5 eutopic tissues of women without EM | RT-qPCR Serum: n = 110 (59 women with peritoneal and/or ovarian EM and 51 women without EM) Tissue: qPCR n = 24 (paired peritoneal and/or ovarian: 9 eutopic and 9 ectopic tissues of EM patients and 6 eutopic tissues of women without EM) DE lncRNAs 16 | Combination of NR_038395, NR_038452, ENST00000482343, ENST00000544649, and ENST00000393610 suggested as a non-invasive diagnosis marker | 1, 2 (because of pooling) |
| Expression microarray [38] | n = 8 (paired ovarian: 4 eutopic and 4 ectopic tissues of EM patients) | RT-qPCR n = 87 (paired ovarian: 30 eutopic and 30 ectopic tissues of EM patients and 27 eutopic tissues of women without EM)) CHL1-AS2 | CCDC144NL-AS1 expression was upregulated in ectopic tissues compared to eutopic and control endometrial tissues | 1 |
| Re-analysis of existing microarray data [39] | n = 18 GSE120103 (eutopic tissue from 9 fertile and 9 infertile women with ovarian EM) | Validation of 14 hub mRNAs using GSE26787 (5 fertile and 5 infertile women with unknown EM status) | Identification of putative infertility-associated lncRNAs LOC390705 and LOC100505854 | 1, 3, 6 |
| Re-analysis of existing microarray data [40] | GSE7305 (paired ovarian: 10 eutopic and 10 ectopic tissues of EM patients and 10 eutopic tissues of women without EM) GSE7846 (HEECs of eutopic tissues of 5 EM patients with ovarian EM and HEECs of eutopic tissues of 5 women without EM), GSE29981 (LMD epithelial cells of 20 women without EM) E-MTAB-694 (paired peritoneal: 18 eutopic and 18 ectopic tissues of EM patients and 17 eutopic tissues of women without EM) | No validation | Proposed cell cycle regulatory functions for LINC01279 | 2, 3, 4, 5 |
| RNA-Seq [41] | n = 16 (8 eutopic tissues of women with EM of undefined entity and 8 eutopic tissues of women without EM) | No validation | Predicted oxidative stress and endometrial receptivity regulatory functions | 1, 2, 3, 4, 6 |
| RNA-Seq [36] | n = 10 (5 eutopic tissues of patients with ovarian EM and 5 eutopic tissues of women without EM) | RT-qPCR n = 24 (12 eutopic tissues of patients with ovarian EM and 12 eutopic tissues of women without EM) USP46-AS1, RP11-1143G9.4, RP11-217B1.2, AC004951.6, RP11-182J1.12 | Predicted proliferation, adhesion, migration, invasion, and angiogenesis regulatory functions | 1, 4, 6 |
| RNA-Seq [42] | n = 9 (paired ovarian: 3 eutopic and 3 ectopic tissues of EM patients and 3 eutopic tissues of women without EM) | RT-qPCR n = 45 (paired:15 eutopic and 15 ectopic tissues of EM patients with undefined entity and 15 eutopic tissues of women without EM) PRKAR2B, CLEC2D | Predicted angiogenesis, cell adhesion, cell migration, immune response, inflammatory response, NF-κB signaling, regulatory functions | 1, 2, 4 |
| Re-analysis of existing RNA-Seq and expression array datasets [43] | GSE105764 (paired ovarian: 8 eutopic and 8 ectopic tissues of EM patients) GSE121406 (paired ovarian: FACS sorted stromal cells of 4 eutopic and 4 ectopic tissues of EM patients) GSE105765 (same as GSE105764) | in silico validation by microarray GSE124010 (3 ectopic tissues of patients with EM of undefined entity and 3 eutopic tissues of women without EM) GSE86534 (paired ovarian: 4 eutopic and 4 ectopic tissues of patients with EM) | Predicted function in regulation of inflammation and prediction of sponging LINC01018 and SMIM25 functions for hsa-miR-182-5p in the regulation of CHL1 protein coding gene to promote endometriosis | 1, 4 (validation study), 5, 6 |
| Re-analysis of existing RNA-Seq datasets [44] | n = 28 (paired ovarian: 14 eutopic and 14 ectopic tissues of infertile EM patients) GSE105764 (paired ovarian: 8 eutopic and 8 ectopic tissues of EM patients) GSE105765 (same patients as in GSE105764) GSE25628 (unpaired DIE: 8 eutopic and 8 ectopic tissues of EM patients and 6 eutopic tissues of women without EM) | No validation | Construction of a competitive endogenous (ce) RNA network promoting growth and death of endometrial stroma cells. CDK1 and PCNA proposed as treatment targets for endometriosis- associated infertility | 1, 3, 5 |
| RNA-Seq [45] | n = 12 (paired ovarian: 6 eutopic and 6 ectopic tissues of EM patients) | RT-qPCR n = 60 (paired ovarian: 30 eutopic and 30 ectopic tissues of EM patients) MIR202HG, LINC00261, UCA1, GAGA2-AS1 | Immunity, inflammation | 1, 6 |
| Re-analysis of existing RNA-Seq data [46] | GSE105764 and GSE105765 include same patients n = 16 (paired ovarian: 8 eutopic and 8 ectopic tissues of EM patients) | No validation | LncRNAs:H19, GS1-358P8.4, and RP11-96D1.10 strongly associated with ovarian endometriosis | 1, 3, 6 |
| Animal Studies | ||||
| Expression microarray [34] | Rat n = 35 (EM uterine tissue EM = 13, adipose tissue control = 8, blank = 14) | RT-qPCR; NONRATT003997; gi|672033904|ref|, XR_589853.1|; NONRATT006252; gi|672027621|ref|; XR_592747.1|; gi|672045999|ref|; XR_591544.1| | Regulation of endometrial receptivity | |
3.3. Critical Assessment of the Evidence for the Role of lncRNAs in Endometriosis
4. Functional Evidence for the Mechanism of lncRNA Action in Endometriosis
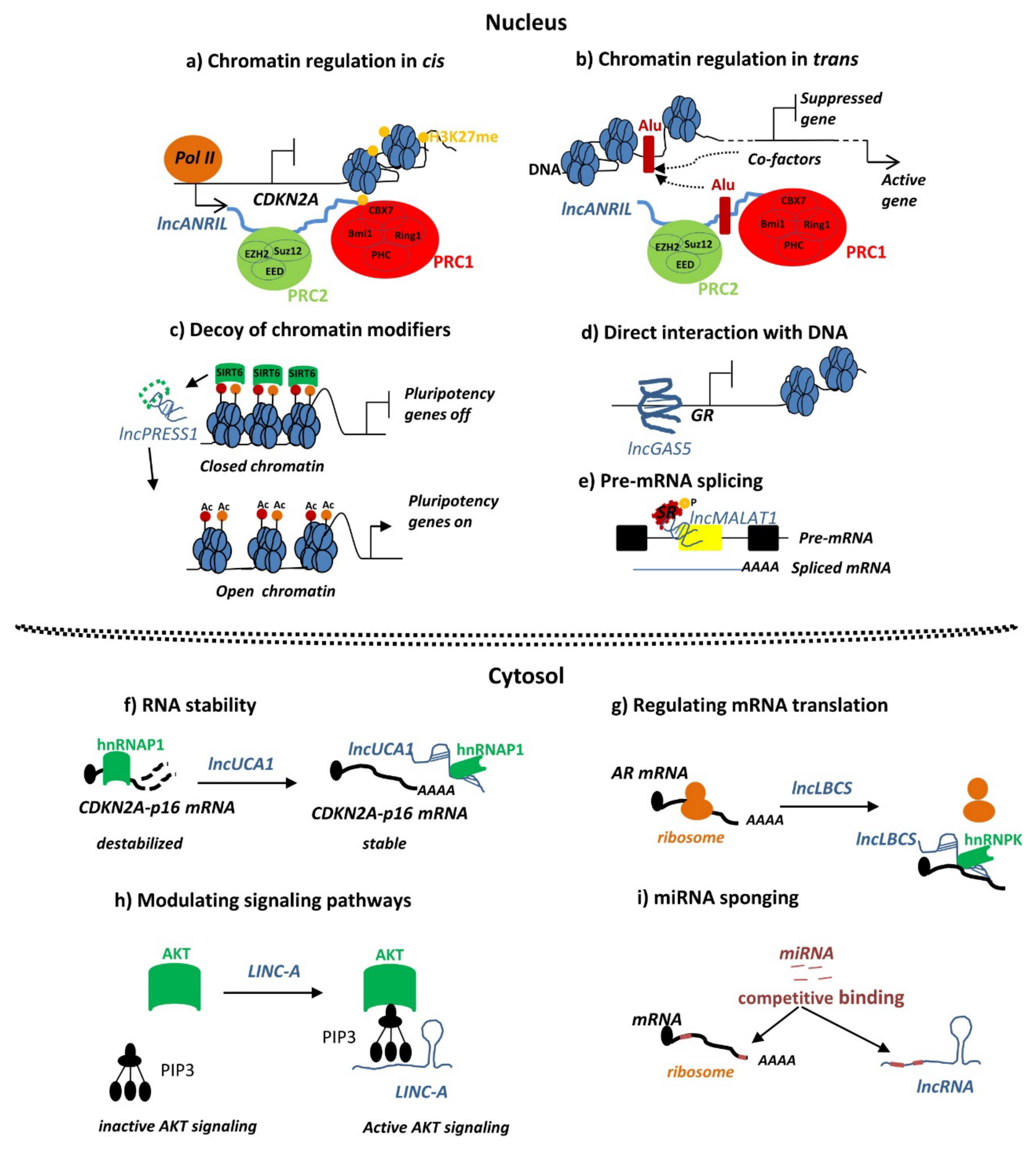
4.1. Chromatin Remodeling and Transcriptional Control by lncRNAs in Endometriosis
4.2. MiRNA Sponging by LncRNAs in Endometriosis
4.3. LncRNAs Modulating Cellular Signaling Pathways in Endometriosis
5. Summary and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sampson, J.A. Peritoneal Endometriosis Due to the Menstrual Dissemination of Endometrial Tissue into the Peritoneal Cavity. Am. J. Obstet. Gynecol. 1927, 14, 442–469. [Google Scholar] [CrossRef]
- Rei, C.; Williams, T.; Feloney, M. Endometriosis in a Man as a Rare Source of Abdominal Pain: A Case Report and Review of the Literature. Case Rep. Obstet. Gynecol. 2018, 2018, 2083121. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Shigesi, N.; Kvaskoff, M.; Kirtley, S.; Feng, Q.; Fang, H.; Knight, J.C.; Missmer, S.A.; Rahmioglu, N.; Zondervan, K.T.; Becker, C.M. The association between endometriosis and autoimmune diseases: A systematic review and meta-analysis. Hum. Reprod. Update 2019, 25, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Delás, M.J.; Hannon, G.J. lncRNAs in development and disease: From functions to mechanisms. Open Biol. 2017, 7, 170121. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.R.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest– and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.P.; Emechebe, U.; Smith, R.; Franklin, S.; Moore, B.; Yandell, M.; Lessnick, S.L.; Moon, A.M. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. Elife 2014, 3, e02805. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Chen, X.; Xie, R.; Xie, W.; Huang, L.; Dong, W.; Han, J.; Liu, X.; Shen, J.; Huang, J.; et al. A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Mol. Cancer 2019, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yao, D.; Huang, B. LncRNA PVT1 promotes cervical cancer progression by sponging miR-503 to upregulate ARL2 expression. Open Life Sci. 2021, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Policarpo, R.; Sierksma, A.; De Strooper, B.; d’Ydewalle, C. From Junk to Function: LncRNAs in CNS Health and Disease. Front. Mol. Neurosci. 2021, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654–3675. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Ou, C.; Xiao, Y.; Han, Q.; Li, H.; Zhou, S. LncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget 2017, 8, 71325–71341. [Google Scholar] [CrossRef] [PubMed]
- Sedano, M.J.; Harrison, A.L.; Zilaie, M.; Das, C.; Choudhari, R.; Ramos, E.; Gadad, S.S. Emerging Roles of Estrogen-Regulated Enhancer and Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 3711. [Google Scholar] [CrossRef] [PubMed]
- Basak, P.; Chatterjee, S.; Bhat, V.; Su, A.; Jin, H.; Lee-Wing, V.; Liu, Q.; Hu, P.; Murphy, L.C.; Raouf, A. Long Non-Coding RNA H19 Acts as an Estrogen Receptor Modulator that is Required for Endocrine Therapy Resistance in ER+ Breast Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 1518–1532. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, Y.; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; Jones, S.; et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, C.S.; Mäkinen, N.; Harris, H.R.; Rahmioglu, N.; Uimari, O.; Cook, J.P.; Shigesi, N.; Ferreira, T.; Velez-Edwards, D.R.; Edwards, T.L.; et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 2019, 10, 4857. [Google Scholar] [CrossRef] [PubMed]
- Giral, H.; Landmesser, U.; Kratzer, A. Into the Wild: GWAS Exploration of Non-coding RNAs. Front. Cardiovasc. Med. 2018, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.H.; Kaur, S.; Brorsson, C.A.; Pociot, F. Effects of GWAS-associated genetic variants on lncRNAs within IBD and T1D candidate loci. PLoS ONE 2014, 9, e105723. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Choi, Y.M.; Hong, M.A.; Yoon, S.H.; Kim, J.J.; Hwang, K.; Chae, S.J. Association of CDKN2B-AS and WNT4 genetic polymorphisms in Korean patients with endometriosis. Fertil. Steril. 2014, 102, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Fung, J.N.; Shakhbazov, K.; Sapkota, Y.; Cloonan, N.; Hemani, G.; Hillman, K.M.; Kaufmann, S.; Luong, H.T.; Bowdler, L.; et al. Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum. Mol. Genet. 2016, 25, 5046–5058. [Google Scholar] [PubMed]
- Chang, C.Y.; Tseng, C.C.; Lai, M.T.; Chiang, A.J.; Lo, L.C.; Chen, C.M.; Yen, M.J.; Sun, L.; Yang, L.; Hwang, T.; et al. Genetic impacts on thermostability of onco-lncRNA HOTAIR during the development and progression of endometriosis. PLoS ONE 2021, 16, e0248168. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, M.; Liang, Z.; Chen, S.; Chen, F.; Zhu, J.; Zhao, M.; Xu, C.; He, J.; Hua, W.; et al. Association of polymorphisms in MALAT1 with the risk of endometriosis in Southern Chinese women. Biol. Reprod. 2019, 102, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xing, Q.; Feng, T.; He, M.; Yu, W.; Chen, H. SNP rs710886 A>G in long noncoding RNA PCAT1 is associated with the risk of endometriosis by modulating expression of multiple stemness-related genes via microRNA-145 signaling pathway. J. Cell Biochem. 2020, 121, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.R.; Jia, S.Z.; Lin, H.; Leng, J.H.; Lang, J.H. Genome-wide profiling of long noncoding ribonucleic acid expression patterns in ovarian endometriosis by microarray. Fertil. Steril. 2014, 101, 1038–1346. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhu, X.; Li, Z.; Zhu, Y.; Lang, J. lncRNA/mRNA profiling of endometriosis rat uterine tissues during the implantation window. Int. J. Mol. Med. 2019, 44, 2145–2160. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; Sun, Y.M.; Huang, W.; He, B.; Zhao, Y.N.; Chen, Y.Q. Genome-wide Long Non-coding RNA Analysis Identified Circulating LncRNAs as Novel Non-invasive Diagnostic Biomarkers for Gynecological Disease. Sci. Rep. 2016, 6, 23343. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Ma, J.; Liu, Y.; Lin, K.; Jiang, X.; Qu, Y.; Lin, J.; Xu, K. Analysis of long non-coding RNA expression profiles using RNA sequencing in ovarian endometriosis. Gene 2018, 673, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yang, Z.; Liu, K.; Wang, D. Genome-Wide Microarray Analysis of Long Non-Coding RNAs in Eutopic Secretory Endometrium with Endometriosis. Cell Physiol. Biochem. 2015, 37, 2231–2245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, W.; Ye, X.; Ma, R.; Luo, J.; Zhu, H.; Chang, X. Aberrant expression of CHL1 gene and long non-coding RNA CHL1-AS1, CHL1-AS2 in ovarian endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 236, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xia, X.; Hu, Y.; Fang, X.; Orsulic, S. Identification of Infertility-Associated Topologically Important Genes Using Weighted Co-expression Network Analysis. Front. Genet. 2021, 12, 580190. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Q.; Zhang, R.; Zhang, C.; Lin, J.; Huang, X. Identification of LINC01279 as a cell cycle-associated long non-coding RNA in endometriosis with GBA analysis. Mol. Med. Rep. 2018, 18, 3850–3858. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, Q. Endometriosis-related ceRNA network to identify predictive biomarkers of endometrial receptivity. Epigenomics 2019, 11, 147–167. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-P.; Tian, X.; Cui, H.-Y.; Zhang, Q.; Hua, K.-Q. The messenger RNA and long non-coding RNA expression profiles in ectopic and eutopic endometrium provide novel insights into endometriosis. Reprod. Dev. Med. 2019, 3, 11. [Google Scholar]
- Jiang, L.; Zhang, M.; Wang, S.; Xiao, Y.; Wu, J.; Zhou, Y.; Fang, X. LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction. Int. J. Immunopathol. Pharmacol. 2020, 34, 2058738420976309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, J.; Duan, S.; Fang, Z.; Tian, J.; Yin, H.; Zhai, Q.; Wang, X.; Zhang, L. Comprehensive characterization of endometrial competing endogenous RNA network in infertile women of childbearing age. Aging 2020, 12, 4204–4221. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Wang, D.; Cui, L.; Yang, Q. RNA sequencing-based long non-coding RNA analysis and immunoassay in ovarian endometriosis. Am. J. Reprod. Immunol. 2021, 85, e13359. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wang, B.; Wang, T.; Ren, W. Identification of Functional lncRNAs Associated With Ovarian Endometriosis Based on a ceRNA Network. Front. Genet. 2021, 12, 534054. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Huang, Q.; Wu, R.; Dai, S.; Huang, Z.; Ren, L.; Huang, S.; Chen, Q. Long non-coding RNA AFAP1-AS1 promoting epithelial-mesenchymal transition of endometriosis is correlated with transcription factor ZEB1. Am. J. Reprod. Immunol. 2019, 81, e13074. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xin, W.; Tang, X.; Qiu, J.; Zhang, Y.; Hua, K. LncRNA H19 Overexpression in Endometriosis and its Utility as a Novel Biomarker for Predicting Recurrence. Reprod. Sci. 2020, 27, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Taylor, H.S. The Role of Hox Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harb. Perspect. Med. 2015, 6, a023002. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Liu, Z.L.; Sun, M.; Liu, J.; Wang, Z.X.; De, W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013, 13, 464. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.F.; Chen, Z.X.; Zhou, W.D.; Li, Y.Z.; Huang, Z.X.; Lin, D.C.; Ren, L.L.; Chen, Q.X.; Chen, Q.H. High expression of ZEB1 in endometriosis and its role in 17beta-estradiol-induced epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 2018, 11, 4744–4758. [Google Scholar] [PubMed]
- Ghazal, S.; McKinnon, B.; Zhou, J.; Mueller, M.; Men, Y.; Yang, L.; Mueller, M.; Flannery, C.; Huang, Y.; Taylor, H.S. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol. Med. 2015, 7, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qiu, J.; Tang, X.; Cui, H.; Zhang, Q.; Yang, Q. LncRNA-H19 regulates cell proliferation and invasion of ectopic endometrium by targeting ITGB3 via modulating miR-124-3p. Exp. Cell Res. 2019, 381, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, L.; Zhong, Y.; Cai, M.; Gao, J.; Tan, C.; Han, X.; Guo, R.; Han, L. LncRNA H19 over-expression inhibited Th17 cell differentiation to relieve endometriosis through miR-342-3p/IER3 pathway. Cell Biosci. 2019, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yi, M.; Zhang, X.; Zhang, T.; Jiang, L.; Cao, L.; Zhou, Y.; Fang, X. Effects of CDKN2B-AS1 on cellular proliferation, invasion and AKT3 expression are attenuated by miR-424-5p in a model of ovarian endometriosis. Reprod. Biomed. Online 2021, 42, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Chen, S.; Wang, D.; Yang, Q. LINC01116 promotes proliferation and migration of endometrial stromal cells by targeting FOXP1 via sponging miR-9-5p in endometriosis. J. Cell Mol. Med. 2021, 25, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, Y.; Zhao, Y.; Xu, C.; Zhang, A.; Zhang, Q.; Wang, D.; He, J.; Hua, W.; Duan, P. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell Res. Ther. 2017, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhang, Z.; Xiong, W.; Li, N.; Liu, H.; He, H.; Li, Q.; Liu, Y.; Zhang, L. Estradiol promotes EMT in endometriosis via MALAT1/miR200s sponge function. Reproduction 2019, 157, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, L.; Yu, Q.; Zhang, Y.; Yan, L.; Chen, Z.J. The estrogen-regulated lncRNA H19/miR-216a-5p axis alters stromal cell invasion and migration via ACTA2 in endometriosis. Mol. Hum. Reprod. 2019, 25, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Xu, H.; Lin, H.; Wei, Y.; Yin, Y.; Huang, Y.; Huang, S.; Liao, Y. LINC01541 Functions as a ceRNA to Modulate the Wnt/beta-Catenin Pathway by Decoying miR-506-5p in Endometriosis. Reprod. Sci. 2021, 28, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, X.; Lu, D.; Feng, Y.; Xu, R.; Li, X.; Yin, C.; Xue, B.; Zhao, H.; Wang, S.; et al. LncRNA SNHG4 promotes the increased growth of endometrial tissue outside the uterine cavity via regulating c-Met mediated by miR-148a-3p. Mol. Cell Endocrinol. 2020, 514, 110887. [Google Scholar] [CrossRef] [PubMed]
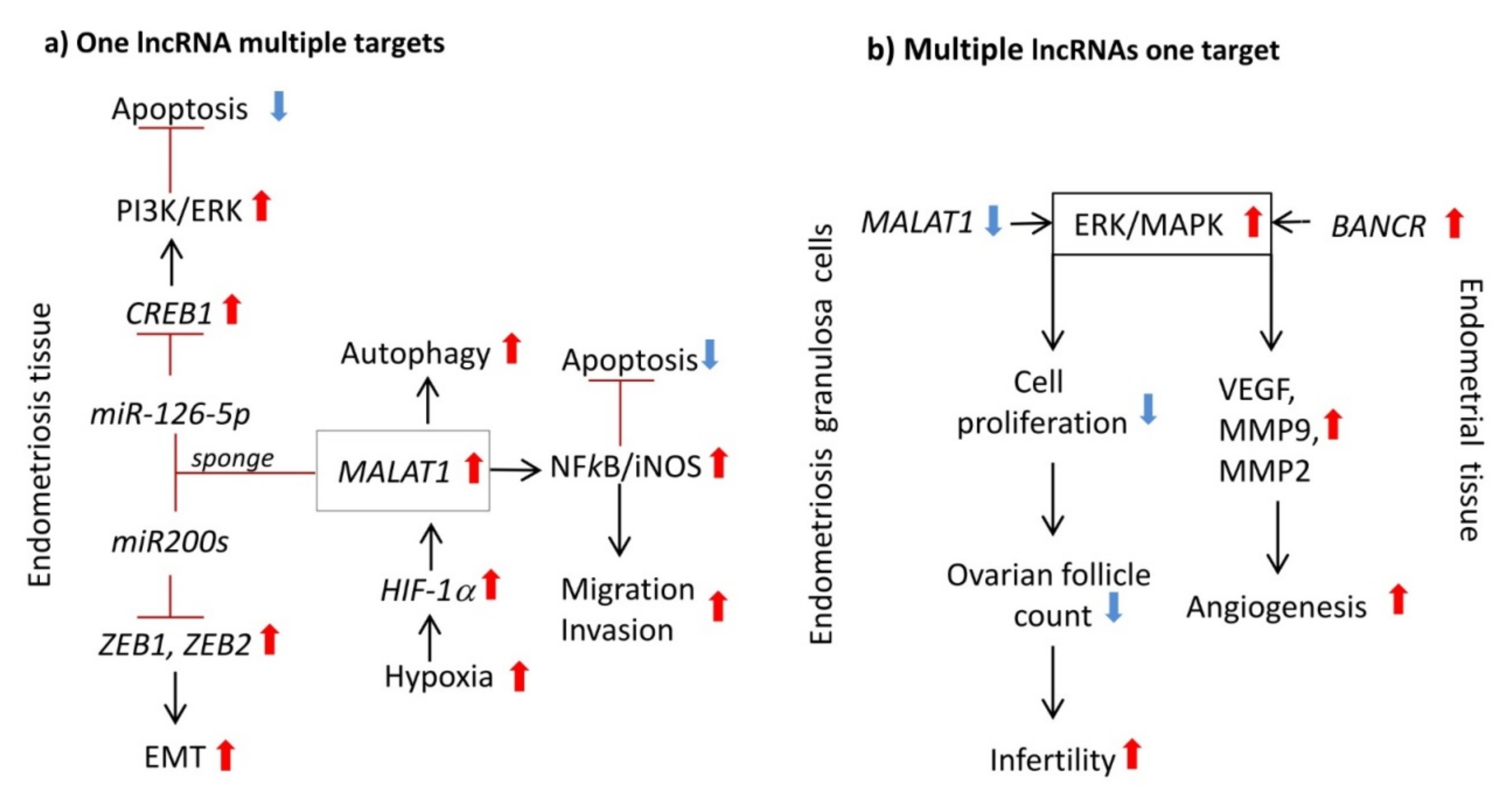
- Feng, Y.; Tan, B.Z. LncRNA MALAT1 inhibits apoptosis of endometrial stromal cells through miR-126-5p-CREB1 axis by activating PI3K-AKT pathway. Mol. Cell Biochem. 2020, 475, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sha, L.; Huang, L.; Yang, S.; Zhou, Q.; Luo, X.; Shi, B. LINC00261 functions as a competing endogenous RNA to regulate BCL2L11 expression by sponging miR-132-3p in endometriosis. Am. J. Transl. Res. 2019, 11, 2269–2279. [Google Scholar] [PubMed]
- Liu, Y.; Ma, J.; Cui, D.; Fei, X.; Lv, Y.; Lin, J. LncRNA MEG3-210 regulates endometrial stromal cells migration, invasion and apoptosis through p38 MAPK and PKA/SERCA2 signalling via interaction with Galectin-1 in endometriosis. Mol. Cell Endocrinol. 2020, 513, 110870. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, L.H.; Zhang, B.; Zheng, Q.M. The modulation of endometriosis by lncRNA MALAT1 via NF-kappaB/iNOS. Eur Rev. Med. Pharmacol. Sci. 2019, 23, 4073–4080. [Google Scholar] [PubMed]
- Li, Y.; Liu, Y.D.; Chen, S.L.; Chen, X.; Ye, D.S.; Zhou, X.Y.; Zhe, J.; Zhang, J. Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway. Mol. Hum. Reprod. 2019, 25, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Z.; Xiong, W.; Zhang, L.; Du, Y.; Liu, Y.; Xiong, X. Long non-coding RNA MALAT1 mediates hypoxia-induced pro-survival autophagy of endometrial stromal cells in endometriosis. J. Cell Mol. Med. 2019, 23, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Wang, W.; Zhang, Y.; Song, E.; Fan, Y.; Wei, B. Hypoxia-inducible factor-1alpha: A promising therapeutic target in endometriosis. Biochimie 2016, 123, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Wei, Y.; Yin, Y.; Huang, S.; Lin, H.; Liao, Y.; Liu, X.; Chen, X.; Shi, H.; Liu, C.; et al. LINC01541 overexpression attenuates the 17beta-Estradiol-induced migration and invasion capabilities of endometrial stromal cells. Syst. Biol. Reprod. Med. 2019, 65, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.J.; Lin, X.J.; Zheng, T.T.; Tang, X.Y.; Zhang, Y.; Hua, K.Q. The Exosomal Long Noncoding RNA aHIF is Upregulated in Serum From Patients With Endometriosis and Promotes Angiogenesis in Endometriosis. Reprod. Sci. 2019, 26, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Yotova, I.; Hudson, Q.J.; Pauler, F.M.; Proestling, K.; Haslinger, I.; Kuessel, L.; Perricos, A.; Husslein, H.; Wenzl, R. LINC01133 Inhibits Invasion and Promotes Proliferation in an Endometriosis Epithelial Cell Line. Int. J. Mol. Sci. 2021, 22, 8385. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ni, C.; Xiao, W.; Wang, S. Role of lncRNA FTX in invasion, metastasis, and epithelial-mesenchymal transition of endometrial stromal cells caused by endometriosis by regulating the PI3K/Akt signaling pathway. Ann. Transl. Med. 2020, 8, 1504. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.B.; Chen, L.P.; Hu, M.; Shi, Z.; Liu, Y.N. Effects of lncRNA BANCR on endometriosis through ERK/MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6806–6812. [Google Scholar] [PubMed]
- Jiang, L.; Wan, Y.; Feng, Z.; Liu, D.; Ouyang, L.; Li, Y.; Liu, K. Long Noncoding RNA UCA1 Is Related to Autophagy and Apoptosis in Endometrial Stromal Cells. Front. Oncol. 2020, 10, 618472. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Chen, P.; Ma, Y.; Wang, S.; Tian, Y.; Wang, A.; Wang, D. AC002454.1 and CDK6 synergistically promote endometrial cell migration and invasion in endometriosis. Reproduction 2019, 157, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, W.; Zhu, H.; Yu, X.; Zhang, Y.; Ye, X.; Cheng, H.; Ma, R.; Cui, H.; Luo, J.; et al. Knockdown of long noncoding RNA CCDC144NL-AS1 attenuates migration and invasion phenotypes in endometrial stromal cells from endometriosis†. Biol. Reprod. 2019, 100, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.J.; Lin, Y.Y.; Tang, X.Y.; Ding, Y.; Yi, X.F.; Hua, K.Q. Extracellular vesicle-mediated transfer of the lncRNA-TC0101441 promotes endometriosis migration/invasion. Exp. Cell Res. 2020, 388, 111815. [Google Scholar] [CrossRef] [PubMed]
- Korucuoglu, U.; Biri, A.A.; Konac, E.; Alp, E.; Onen, I.H.; Ilhan, M.N.; Turkyilmaz, E.; Erdem, A.; Erdem, M.; Menevse, S. Expression of the imprinted IGF2 and H19 genes in the endometrium of cases with unexplained infertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 2010, 149, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Lottin, S.; Dugimont, T.; Fauquette, W.; Coll, J.; Dupouy, J.P.; Boilly, B.; Curgy, J.J. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene 1999, 18, 4460–4473. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, K.; Zhan, H.; Ma, J.; Xu, K.; Wu, R.; Zhou, C.; Lin, J. Silencing of SRA1 Regulates ER Expression and Attenuates the Growth of Stromal Cells in Ovarian Endometriosis. Reprod. Sci. 2017, 24, 836–843. [Google Scholar] [CrossRef] [PubMed]
| lncRNA (Reference) | Sponged miRNA | Target mRNA (Pathway) in Endometriosis |
|---|---|---|
| H19 [53] | Let-7; miR-125-3p; miR-342-3p; miR-216a-5p | IGF1R; ITGB3; IER3; ACTA2 |
| CDKN2B-AS1 [56] | miR-424-5p | AKT3 |
| LINC01541 [60] | miR-506-3p | WIF1 (Wnt/β-catenin) |
| LINC01116 [57] | miR-9-5p | FOXP1 |
| SNHG4 [61] | miR-148a-3p | c-Met |
| LINC01018 [43] | miR-182-5p | CHLI (inflammatory) |
| SMIM25 [43] | miR-182-5p | CHLI (inflammatory) |
| MALAT1 [62] | miR-126-5p; miR200s; miR200c | CREB1 (PI3K/AKT); ZEB1, ZEB2, VIM (EMT); ZEB1, ZEB2; CDH2 (EMT) |
| LINC00261 [63] | miR-132-3p | BCL2L11 |
| PCAT1 [32] | miR-145 | FASCIN1, SOX2, MSI2, SERPIN |
| lncRNA (Reference) | Model System | Signaling Molecules | Signaling Pathways | Type of Regulation | Function in Endometriosis |
|---|---|---|---|---|---|
| MEG3-210 [64] | Primary HESC, HEEC EM mouse model | Galectin-1 | P38 MAPK, PKA/SERCA2 | direct | Regulation of migration, invasion and apoptosis, lesion growth and vascularization |
| MALAT1 [65] | EMs cells | Caspase-3, MMP-9 | NFkB/iNOS | Indirect | Regulation of apoptosis, migration, invasion |
| MALAT1 [66] | Granulosa cells (KGN cell line) | p21, p53, CDK1 | ERK/MAPK | Indirect | Regulation of cell proliferation, ovarian follicle count, infertility |
| MALAT1 [67] | HESC | HIF-1α, LC3-II, beclin1 | - | Indirect | Regulation of hypoxia-induced pro-survival and autophagy |
| LINC01541 [68] | HESC | β-Catenin, VEGFA, BCL2, caspase-3 | WNT/β-Catenin | Indirect | Regulates EMT, migration, invasion, survival, and angiogenesis |
| LINC01541 [69] | 12Z epithelial endometriosis cell line | p21, cyclin A | TESK1/Cofilin | Indirect | Regulation of cell proliferation and invasion |
| FTX [70] | EESC (ectopic endometrial stromal cells), HESC (normal) | E-Cadherin, N-cadherin, ZEB1, vimentin | PI3K/AKT | Indirect | Regulates EMT and cell cycle |
| BANCR [71] | Rat model of EM, ectopic tissue, and serum | VEGF, MMP-9, MMP-2 | MAPK/ERK | Indirect | Regulation of angiogenesis |
| UCA1 [72] | HESC | IC3, VMP1 | - | Indirect | Regulation of autophagy and apoptosis |
| AC002454.1 [73] | EESC | CDK6 | - | Indirect | Regulation of cell migration and invasion |
| CCDC144NL-AS1 [74] | HESC | Vimentin, MMP-9 | - | Indirect | Regulation of cell migration and invasion |
| TC0101441 [75] | ECSC | N-Cadherin, SNAIL, SLUG, TCF8/ZEB1 | - | Indirect | Promotes endometriosis cyst stromal cell (ECSC) migration and invasion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudson, Q.J.; Proestling, K.; Perricos, A.; Kuessel, L.; Husslein, H.; Wenzl, R.; Yotova, I. The Role of Long Non-Coding RNAs in Endometriosis. Int. J. Mol. Sci. 2021, 22, 11425. https://doi.org/10.3390/ijms222111425
Hudson QJ, Proestling K, Perricos A, Kuessel L, Husslein H, Wenzl R, Yotova I. The Role of Long Non-Coding RNAs in Endometriosis. International Journal of Molecular Sciences. 2021; 22(21):11425. https://doi.org/10.3390/ijms222111425
Chicago/Turabian StyleHudson, Quanah J., Katharina Proestling, Alexandra Perricos, Lorenz Kuessel, Heinrich Husslein, René Wenzl, and Iveta Yotova. 2021. "The Role of Long Non-Coding RNAs in Endometriosis" International Journal of Molecular Sciences 22, no. 21: 11425. https://doi.org/10.3390/ijms222111425
APA StyleHudson, Q. J., Proestling, K., Perricos, A., Kuessel, L., Husslein, H., Wenzl, R., & Yotova, I. (2021). The Role of Long Non-Coding RNAs in Endometriosis. International Journal of Molecular Sciences, 22(21), 11425. https://doi.org/10.3390/ijms222111425








