DNA Binding Mode Analysis of a Core-Extended Naphthalene Diimide as a Conformation-Sensitive Fluorescent Probe of G-Quadruplex Structures
Abstract
:1. Introduction
2. Results and Discussion
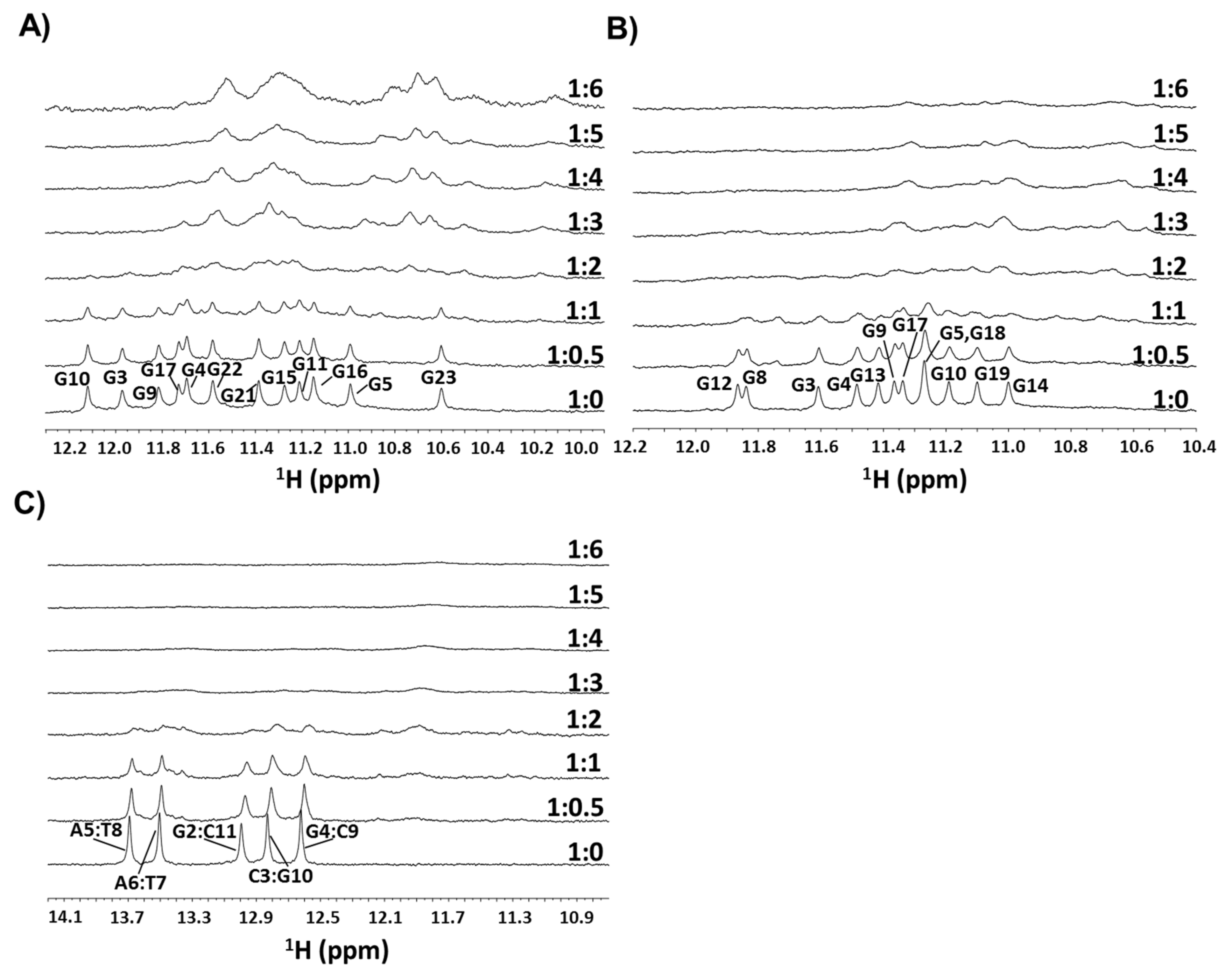
2.1. NMR Experiments
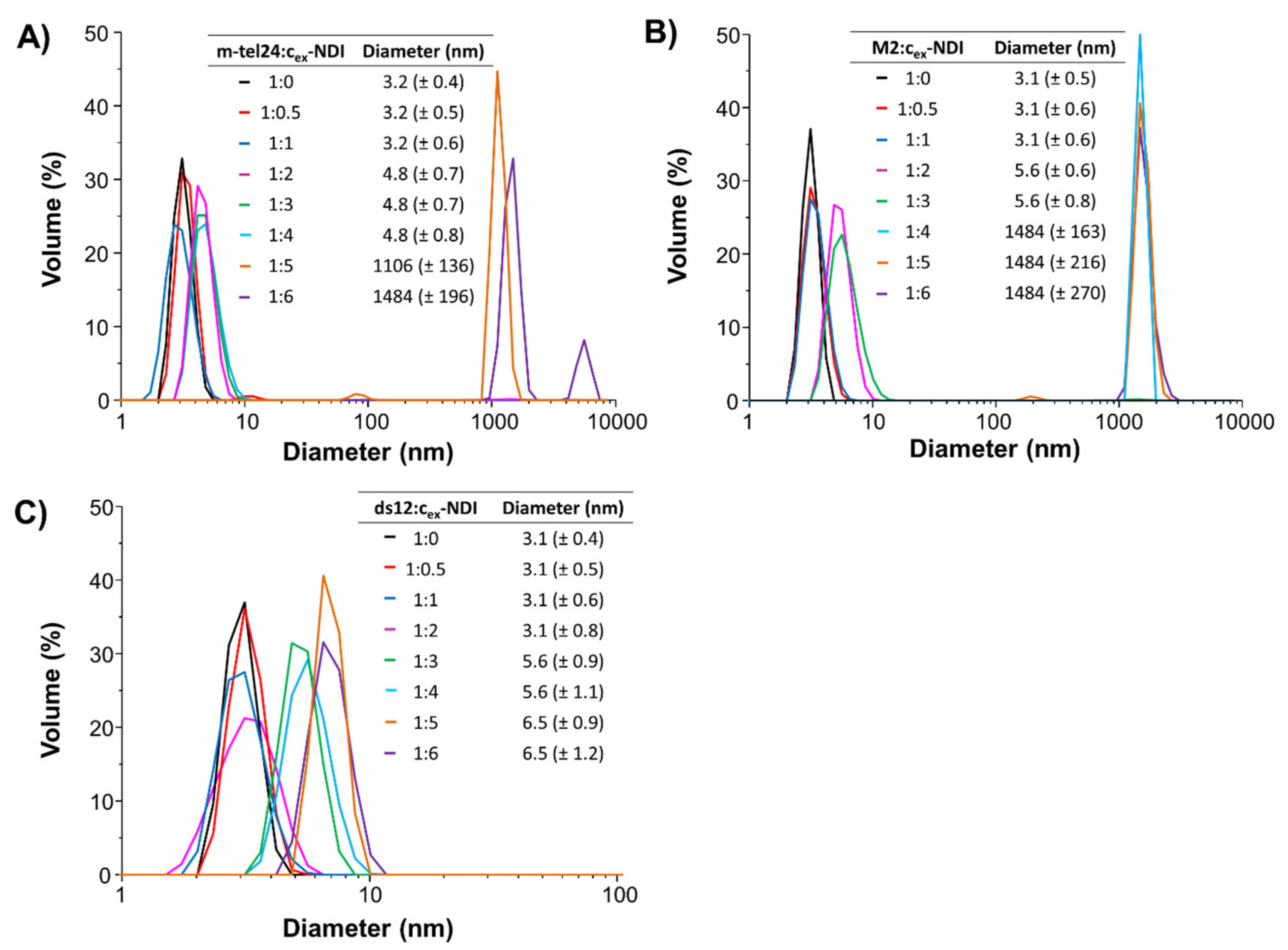
2.2. DLS Experiments
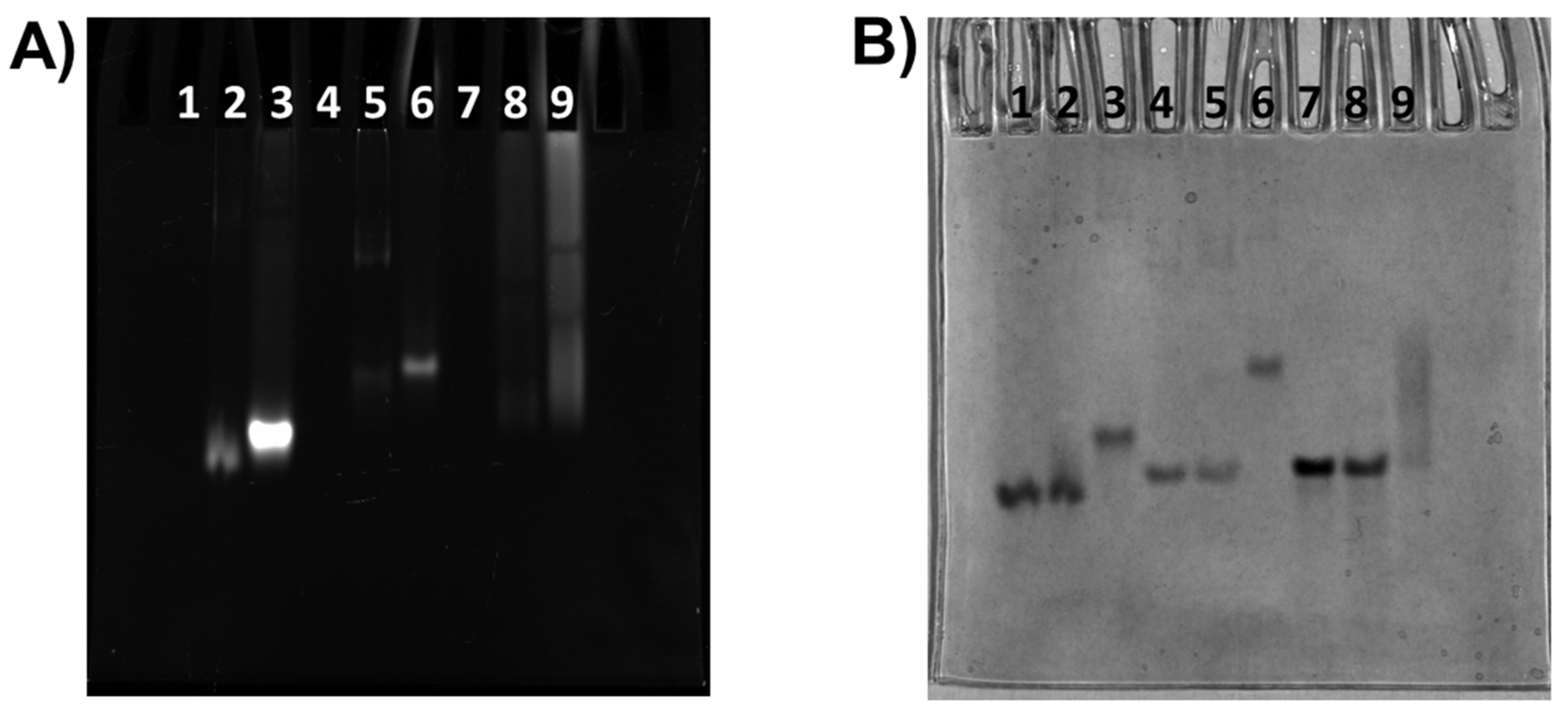
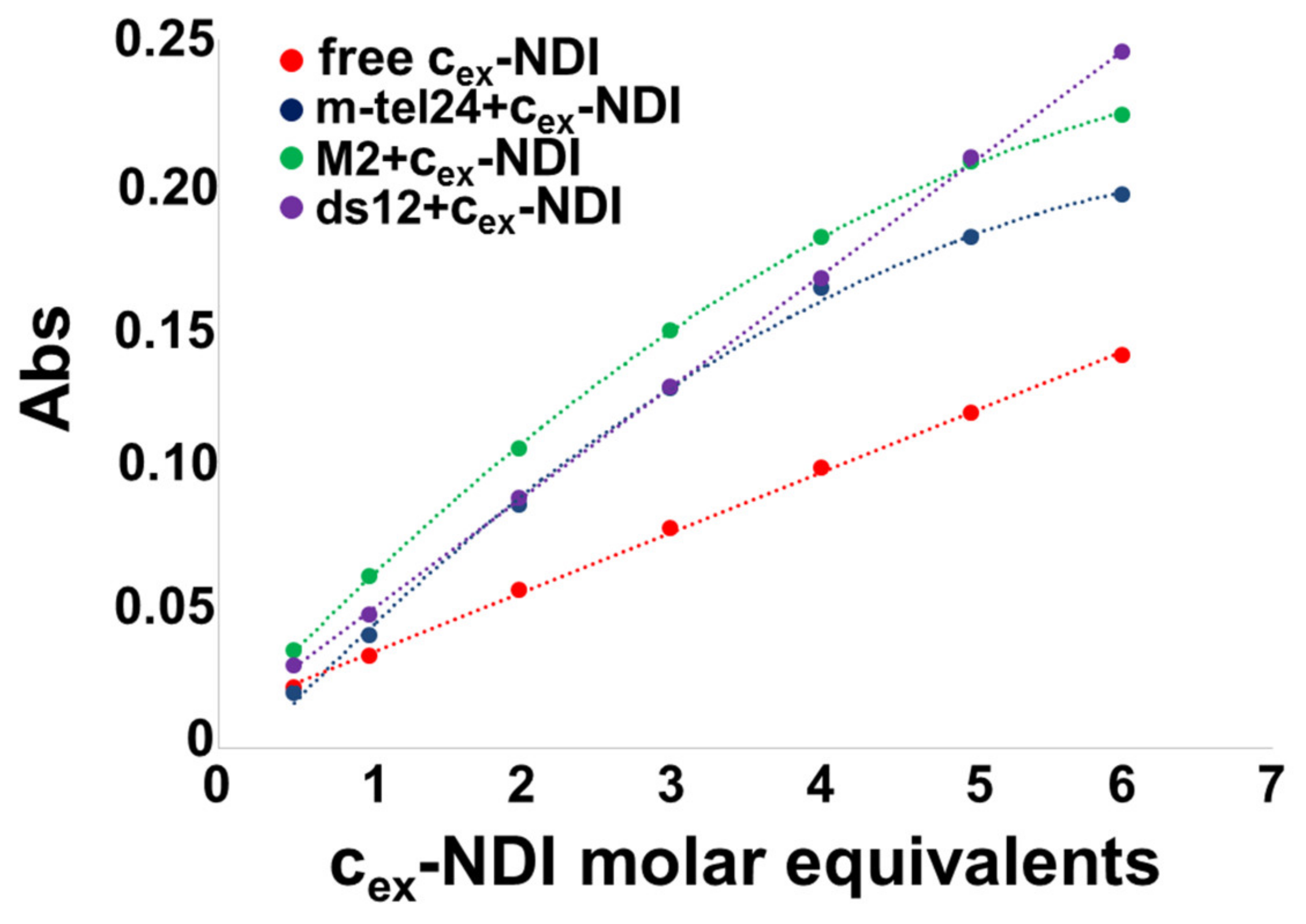
2.3. Gel Electrophoresis Experiments
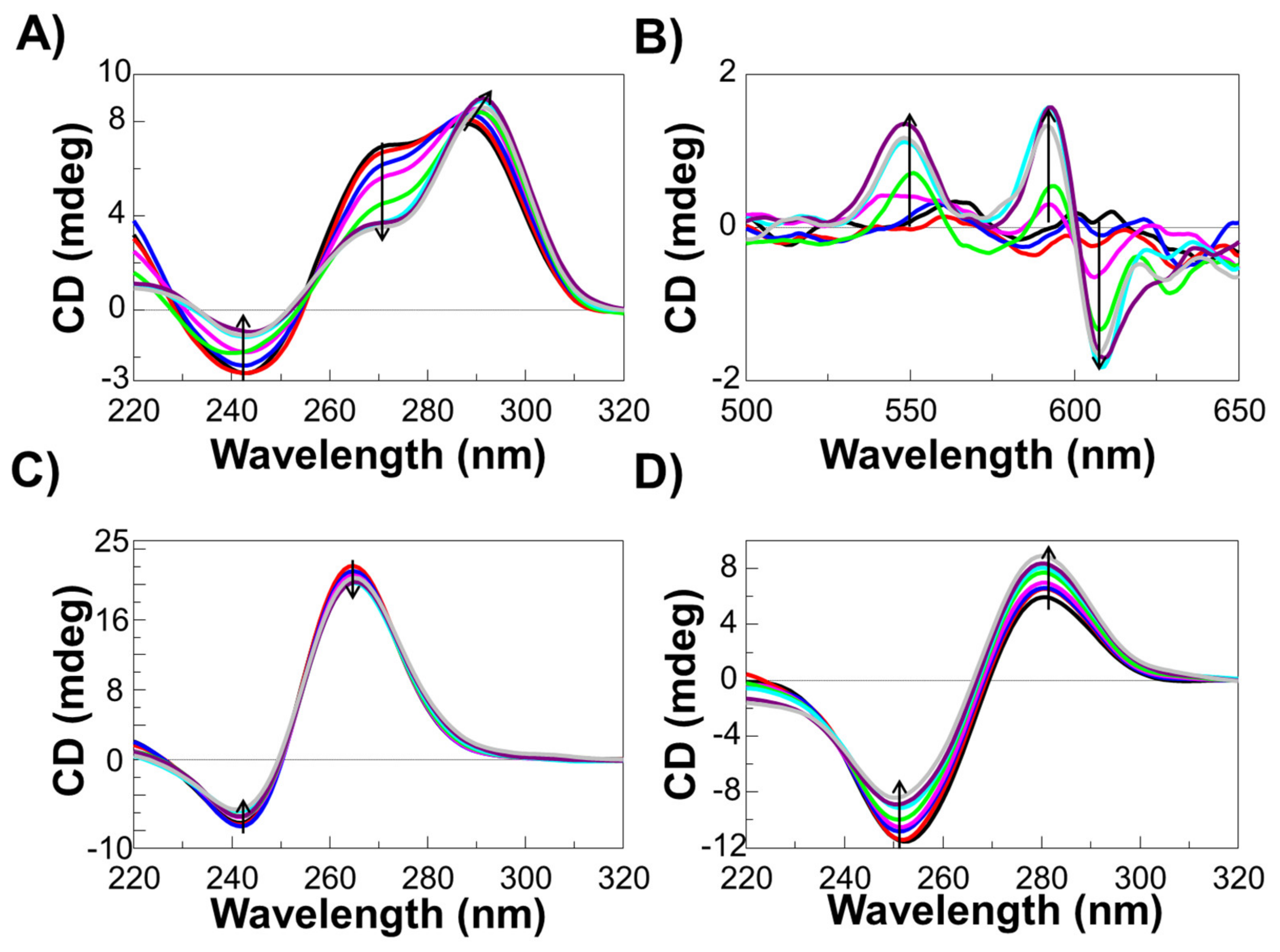
2.4. CD and UV-Vis Experiments
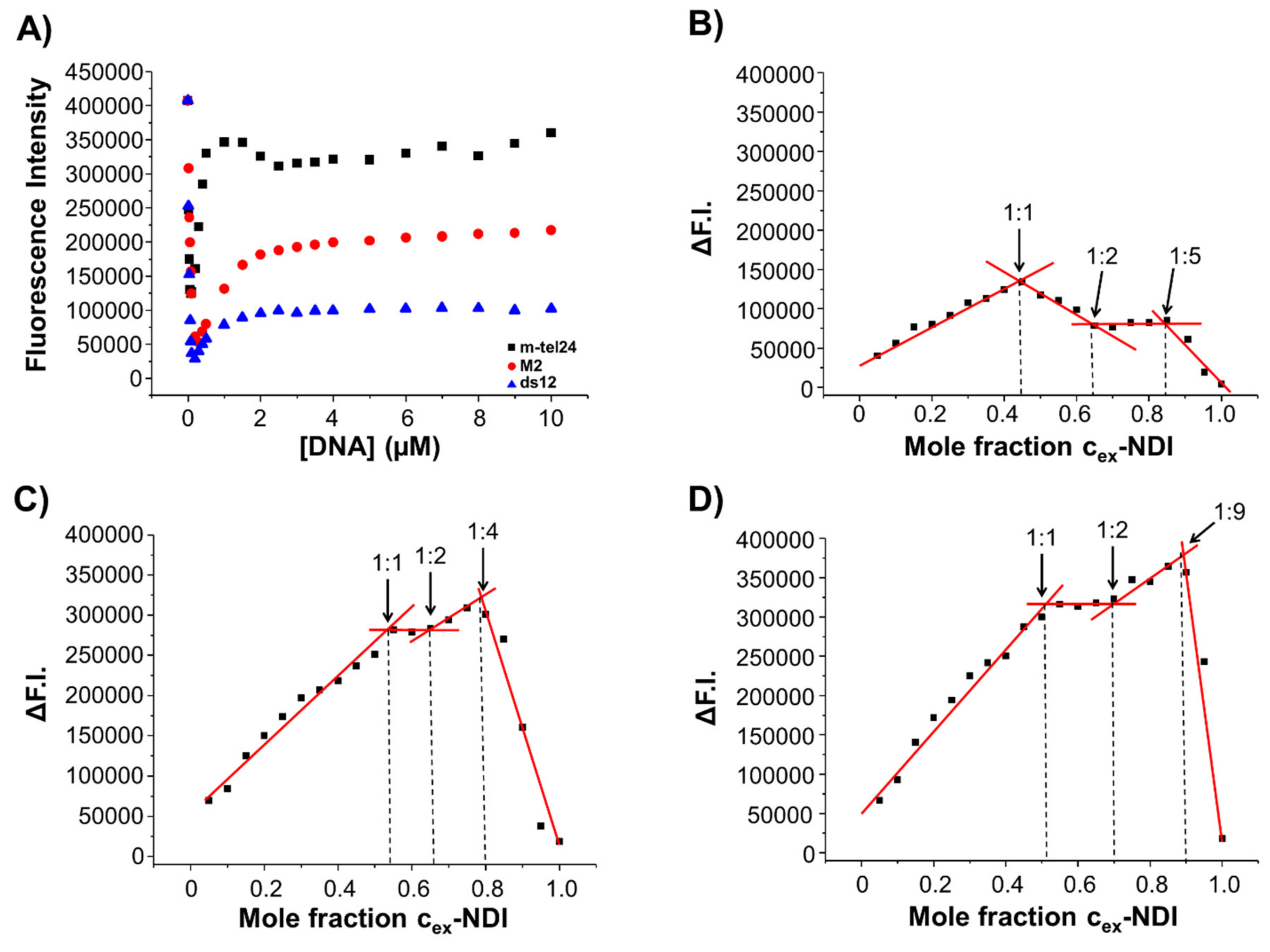
2.5. Fluorescence Experiments
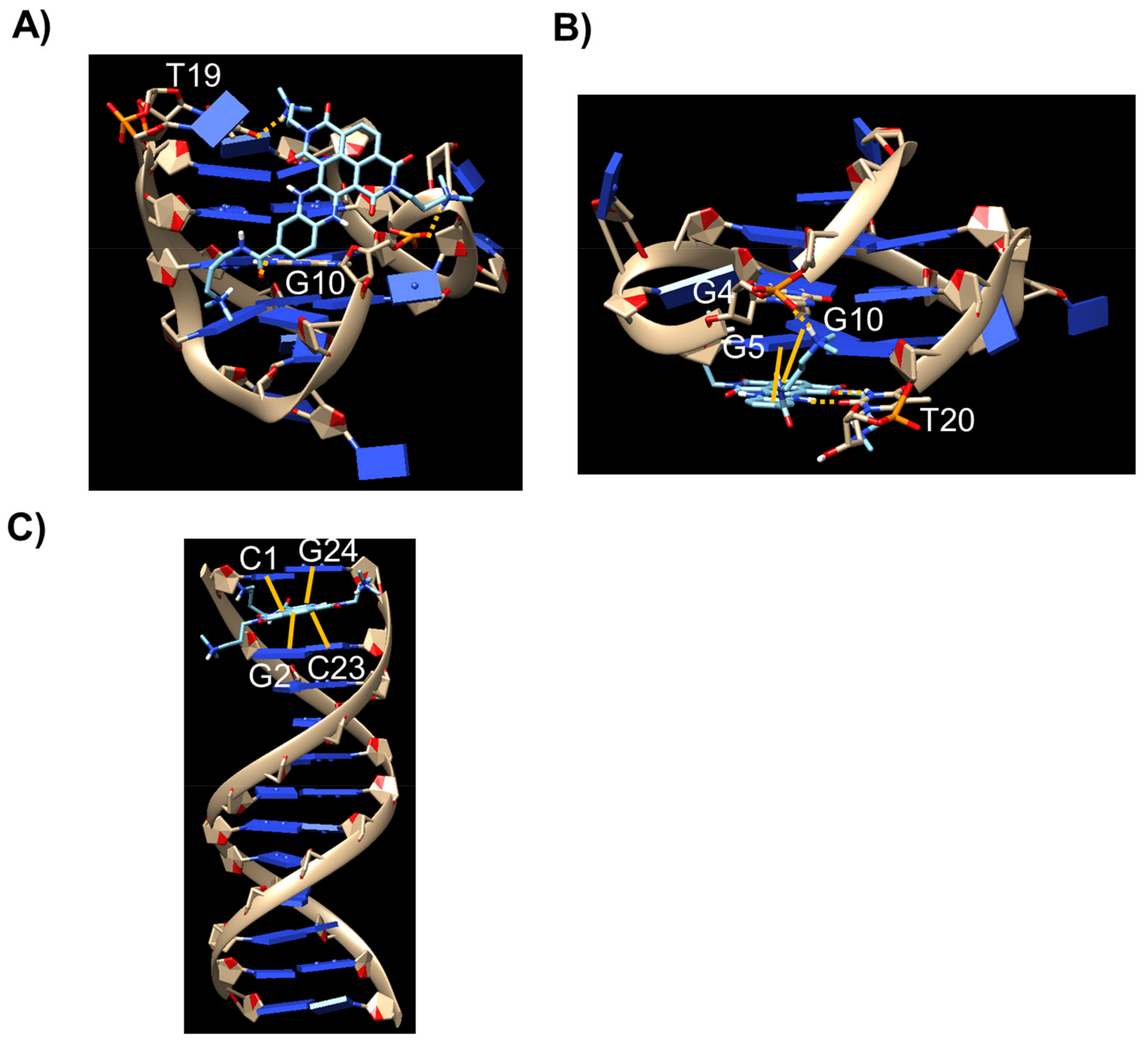
2.6. Docking Studies
2.7. Biological Assays
2.7.1. Analysis of the Effects of cex-NDI on Cancer and Normal Breast Cell Lines
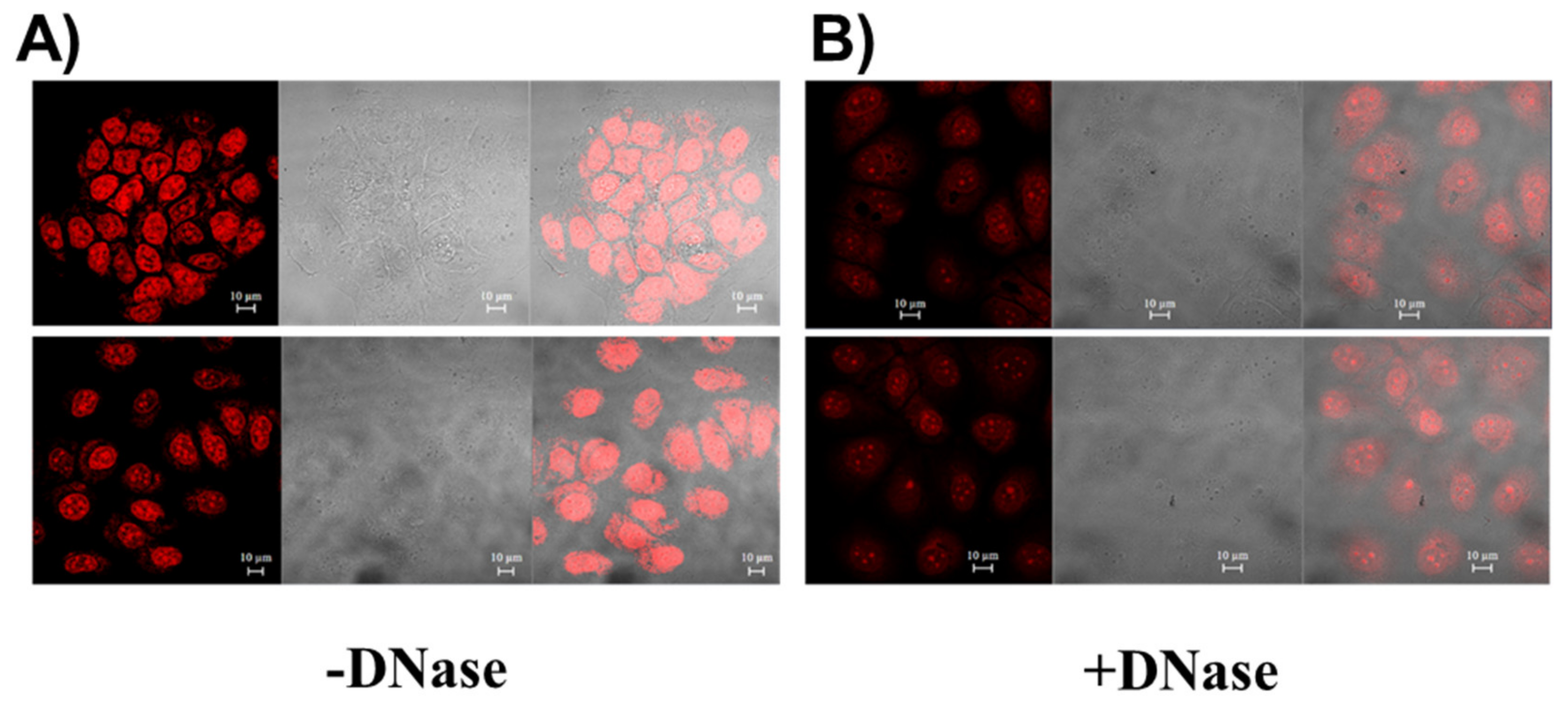
2.7.2. Analysis of cex-NDI Internalisation and Location into Cancer MCF7 and Normal MCF10A Cells by Confocal Laser Scanning Microscopy (CLSM)
3. Materials and Methods
3.1. Sample Preparation
3.2. NMR Experiments
3.3. DLS Experiments
3.4. PAGE Experiments
3.5. CD and UV-Vis Experiments
3.6. Fluorescence Experiments
3.7. Docking Studies
3.8. Biological Assays
3.8.1. Cell Cultures and Cytotoxicity Assays
3.8.2. Internalisation of cex-NDI into MCF7 and MCF10A Cells by CLSM Analyses
3.8.3. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef]
- Xu, Y. Chemistry in human telomere biology: Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011, 40, 2719–2740. [Google Scholar] [CrossRef]
- Murat, P.; Singh, Y.; Defrancq, E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011, 40, 5293–5307. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, C.; Zyner, K.G.; Ohnmacht, S.A.; Robson, M.; Haider, S.M.; Morton, J.P.; Marsico, G.; Vo, T.; Laughlin-Toth, S.; Ahmed, A.A.; et al. Targeting multiple effector pathways in pancreatic ductal adenocarcinoma with a G-quadruplex-binding small molecule. J. Med. Chem. 2018, 61, 2500–2517. [Google Scholar] [CrossRef]
- Guilbaud, G.; Murat, P.; Recolin, B.; Campbell, B.C.; Maiter, A.; Sale, J.E.; Balasubramanian, S. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat. Chem. 2017, 9, 1110–1117. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, S.; Neidle, S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009, 13, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasiri, H.R.; Bell, N.M.; McLuckie, K.I.E.; Husby, J.; Abell, C.; Neidle, S.; Balasubramanian, S. Targeting a c-MYC G-quadruplex DNA with a fragment library. Chem. Commun. 2014, 50, 1704–1707. [Google Scholar] [CrossRef] [PubMed]
- Mulliri, S.; Laaksonen, A.; Spanu, P.; Farris, R.; Farci, M.; Mingoia, F.; Roviello, G.N.; Mocci, F. Spectroscopic and in silico studies on the interaction of substituted pyrazolo[1,2-a]benzo[1,2,3,4]tetrazine-3-one derivatives with c-myc G4-DNA. Int. J. Mol. Sci. 2021, 22, 6028. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Napolitano, E.; Riccardi, C.; Musumeci, D.; Montesarchio, D. Disentangling the structure–activity relationships of naphthalene diimides as anticancer G-quadruplex-targeting drugs. J. Med. Chem. 2021, 64, 3578–3603. [Google Scholar] [CrossRef]
- Platella, C.; Raucci, U.; Rega, N.; D’Atri, S.; Levati, L.; Roviello, G.N.; Fuggetta, M.P.; Musumeci, D.; Montesarchio, D. Shedding light on the interaction of polydatin and resveratrol with G-quadruplex and duplex DNA: A biophysical, computational and biological approach. Int. J. Biol. Macromol. 2020, 151, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Pirota, V.; Musumeci, D.; Rizzi, F.; Iachettini, S.; Zizza, P.; Biroccio, A.; Freccero, M.; Montesarchio, D.; Doria, F. Trifunctionalized naphthalene diimides and dimeric analogues as G-quadruplex-targeting anticancer agents selected by affinity chromatography. Int. J. Mol. Sci. 2020, 21, 1964. [Google Scholar] [CrossRef] [Green Version]
- Pirota, V.; Platella, C.; Musumeci, D.; Benassi, A.; Amato, J.; Pagano, B.; Colombo, G.; Freccero, M.; Doria, F.; Montesarchio, D. On the binding of naphthalene diimides to a human telomeric G-quadruplex multimer model. Int. J. Biol.Macromol. 2020, 166, 1320–1334. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; Miller, J.; Howat, W.J.; Balasubramanian, S. Elevated levels of G-quadruplex formation in human stomach and liver cancer tissues. PLoS ONE 2014, 9, e102711. [Google Scholar] [CrossRef]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G- quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.L.; Zhang, Z.; Wang, M.; Lu, L.; Zhong, H.J.; Leung, C.H. Recent developments in G-quadruplex probes. Chem. Biol. 2015, 22, 812–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summers, P.A.; Lewis, B.W.; Gonzalez-Garcia, J.; Porreca, R.M.; Lim, A.H.M.; Cadinu, P.; Martin-Pintado, N.; Mann, D.J.; Edel, J.B.; Vannier, J.B.; et al. Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T. Single-molecule visualisation of DNA G-quadruplex formation in live cells. Nat. Chem. 2021, 12, 832–837. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Chen, H.; Li, Q.; Guan, A.; Wang, L. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1101–1106. [Google Scholar] [CrossRef]
- Łęczkowska, A.; Gonzalez-Garcia, J.; Perez-Arnaiz, C.; Garcia, B.; White, A.J.P.; Vilar, R. Binding studies of metal–salphen and metal–bipyridine complexes towards G-quadruplex DNA. Chem. Eur. J. 2018, 24, 11785–11794. [Google Scholar] [CrossRef]
- Shivalingam, A.; Vyšniauskas, A.; Albrecht, T.; White, A.J.P.; Kuimova, M.K.; Vilar, R. Trianguleniums as optical probes for G-quadruplexes: A photophysical, electrochemical, and computational study. Chem. Eur. J. 2016, 22, 4129–4139. [Google Scholar] [CrossRef] [Green Version]
- Berrones Reyes, J.; Kuimova, M.K.; Vilar, R. Metal complexes as optical probes for DNA sensing and imaging. Curr. Opin. Chem. Biol. 2021, 61, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, V.; Harikrishna, S.; Bhasikuttan, A.C.; Pradeepkumar, P.I. Topology specific stabilization of promoter over telomeric g-quadruplex dnas by bisbenzimidazole carboxamide derivatives. ACS Chem. Biol. 2015, 10, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, R.; Li, Q.; Li, S.; Xiang, J.; Guo, X.; Wang, S.; Zeng, Y.; Li, Y.; Yang, G. Visualization of parallel G-quadruplexes in cells with a series of new developed bis(4-aminobenzylidene)acetone derivatives. ACS Omega 2018, 3, 10487–10492. [Google Scholar] [CrossRef]
- Laguerre, A.; Wong, J.M.Y.; Monchaud, D. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zuffo, M.; Guedin, A.; Leriche, E.; Doria, F.; Pirota, V.; Gabelica, V.; Mergny, J.L.; Freccero, M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef] [PubMed]
- Grande, V.; Doria, F.; Freccero, M.; Würthner, F. An aggregating amphiphilic squaraine: A light-up probe that discriminates parallel G-quadruplexes. Angew. Chem. Int. Ed. 2017, 56, 7520–7524. [Google Scholar] [CrossRef]
- Doria, F.; Nadai, M.; Zuffo, M.; Perrone, R.; Freccero, M.; Richter, S.N. A red-NIR fluorescent dye detecting nuclear DNA G-quadruplexes: In vitro analysis and cell imaging. Chem. Commun. 2017, 53, 2268–2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef]
- Shivalingam, A.; Izquierdo, M.A.; Le Marois, A.; Vyšniauskas, A.; Suhling, K.; Kuimova, M.K.; Vilar, R. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015, 6, 8178. [Google Scholar] [CrossRef] [Green Version]
- Zuffo, M.; Doria, F.; Botti, S.; Bergamaschi, G.; Freccero, M. G-quadruplex fluorescence sensing by core-extended naphthalene diimides. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1303–1311. [Google Scholar] [CrossRef]
- Platella, C.; Musumeci, D.; Arciello, A.; Doria, F.; Freccero, M.; Randazzo, A.; Amato, J.; Pagano, B.; Montesarchio, D. Controlled Pore Glass-based oligonucleotide affinity support: Towards High Throughput Screening methods for the identification of conformation-selective G-quadruplex ligands. Anal. Chim. Acta 2018, 1030, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.N.; Tu, A.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovski, M.; Endoh, T.; Tateishi-karimata, H.; Ohyama, T.; Tanaka, S. Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations. Nucleic Acids Res. 2018, 46, 4301–4315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Trajkovski, M.; Doria, F.; Freccero, M.; Montesarchio, D.; Plavec, J. On the interaction of an anticancer trisubstituted naphthalene diimide with G-quadruplexes of different topologies: A structural insight. Nucleic Acids Res. 2020, 48, 12380–12393. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Dickerhoff, J.; Yang, D. NMR studies of G-quadruplex structures and G-quadruplex-interactive compounds. In G-Quadruplex Nucleic Acids: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2019; Volume 2035, pp. 157–175. ISBN 9784431538592. [Google Scholar]
- Musumeci, D.; Amato, J.; Zizza, P.; Platella, C.; Cosconati, S.; Cingolani, C.; Biroccio, A.; Novellino, E.; Randazzo, A.; Giancola, C.; et al. Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1341–1352. [Google Scholar] [CrossRef]
- Amato, J.; Platella, C.; Iachettini, S.; Zizza, P.; Musumeci, D.; Cosconati, S.; Pagano, A.; Novellino, E.; Biroccio, A.; Randazzo, A.; et al. Tailoring a lead-like compound targeting multiple G-quadruplex structures. Eur. J. Med. Chem. 2019, 163, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Mazzini, S.; Napolitano, E.; Mattio, L.M.; Beretta, G.L.; Zaffaroni, N.; Pinto, A.; Montesarchio, D.; Dallavalle, S. Plant-derived stilbenoids as DNA-binding agents: From monomers to dimers. Chem. Eur. J. 2021, 27, 8832–8845. [Google Scholar] [CrossRef]
- Šket, P.; Plavec, J. Tetramolecular DNA quadruplexes in solution: Insights into structural diversity and cation movement. J. Am. Chem. Soc. 2010, 132, 12724–12732. [Google Scholar] [CrossRef] [PubMed]
- Elsharawy, K.A.; Althobiti, M.; Mohammed, O.J.; Aljohani, A.I.; Toss, M.S.; Green, A.R.; Rakha, E.A. Nucleolar protein 10 (NOP10) predicts poor prognosis in invasive breast cancer. Breast Cancer Res. Treat. 2021, 185, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, R.L.; Ziegler, T.D.; Supakorndej, T.; Terns, R.M.; Terns, M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 2006, 17, 955–965. [Google Scholar] [CrossRef] [Green Version]
- Khurts, S.; Masutomi, K.; Delgermaa, L.; Arai, K.; Oishi, N.; Mizuno, H.; Hayashi, N.; Hahn, W.C.; Murakami, S. Nucleolin interacts with telomerase. J. Biol. Chem. 2004, 279, 51508–51515. [Google Scholar] [CrossRef] [Green Version]
- Stewart, S.A. Multiple levels of telomerase regulation. Mol. Interv. 2002, 2, 481–483. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Di Girolamo, R.; Tartaglione, L.; Pizzo, E.; Arciello, A. Cryptides identified in human apolipoprotein b as new weapons to fight antibiotic resistance in cystic fibrosis disease. Int. J. Mol. Sci. 2020, 21, 2049. [Google Scholar] [CrossRef] [Green Version]
- Gaglione, R.; Dell’Olmo, E.; Bosso, A.; Chino, M.; Pane, K.; Ascione, F.; Itri, F.; Caserta, S.; Amoresano, A.; Lombardi, A.; et al. Novel human bioactive peptides identified in Apolipoprotein B: Evaluation of their therapeutic potential. Biochem. Pharm. 2017, 130, 34–50. [Google Scholar] [CrossRef]
- Dell’Olmo, E.; Gaglione, R.; Cesaro, A.; Cafaro, V.; Teertstra, W.R.; de Cock, H.; Notomista, E.; Haagsman, H.P.; Veldhuizen, E.J.A.; Arciello, A. Host defence peptides identified in human apolipoprotein B as promising antifungal agents. Appl. Microbiol. Biotechnol. 2021, 105, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Pirone, L.; Farina, B.; Fusco, S.; Smaldone, G.; Aulitto, M.; Dell’Olmo, E.; Roscetto, E.; Del Gatto, A.; Fattorusso, R.; et al. Insights into the anticancer properties of the first antimicrobial peptide from Archaea. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2155–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lampitella, E.; Landi, N.; Oliva, R.; Gaglione, R.; Bosso, A.; De Lise, F.; Ragucci, S.; Arciello, A.; Petraccone, L.; Pizzo, E.; et al. Toxicity and membrane perturbation properties of the ribotoxin-like protein Ageritin. J. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]


| IC50 (µM) | |||||
|---|---|---|---|---|---|
| MCF7 | MCF10A | ||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h |
| N/A | 12 | 9 | N/A | 16 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Platella, C.; Gaglione, R.; Napolitano, E.; Arciello, A.; Pirota, V.; Doria, F.; Musumeci, D.; Montesarchio, D. DNA Binding Mode Analysis of a Core-Extended Naphthalene Diimide as a Conformation-Sensitive Fluorescent Probe of G-Quadruplex Structures. Int. J. Mol. Sci. 2021, 22, 10624. https://doi.org/10.3390/ijms221910624
Platella C, Gaglione R, Napolitano E, Arciello A, Pirota V, Doria F, Musumeci D, Montesarchio D. DNA Binding Mode Analysis of a Core-Extended Naphthalene Diimide as a Conformation-Sensitive Fluorescent Probe of G-Quadruplex Structures. International Journal of Molecular Sciences. 2021; 22(19):10624. https://doi.org/10.3390/ijms221910624
Chicago/Turabian StylePlatella, Chiara, Rosa Gaglione, Ettore Napolitano, Angela Arciello, Valentina Pirota, Filippo Doria, Domenica Musumeci, and Daniela Montesarchio. 2021. "DNA Binding Mode Analysis of a Core-Extended Naphthalene Diimide as a Conformation-Sensitive Fluorescent Probe of G-Quadruplex Structures" International Journal of Molecular Sciences 22, no. 19: 10624. https://doi.org/10.3390/ijms221910624
APA StylePlatella, C., Gaglione, R., Napolitano, E., Arciello, A., Pirota, V., Doria, F., Musumeci, D., & Montesarchio, D. (2021). DNA Binding Mode Analysis of a Core-Extended Naphthalene Diimide as a Conformation-Sensitive Fluorescent Probe of G-Quadruplex Structures. International Journal of Molecular Sciences, 22(19), 10624. https://doi.org/10.3390/ijms221910624










