A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA
Abstract
1. Introduction
2. Results
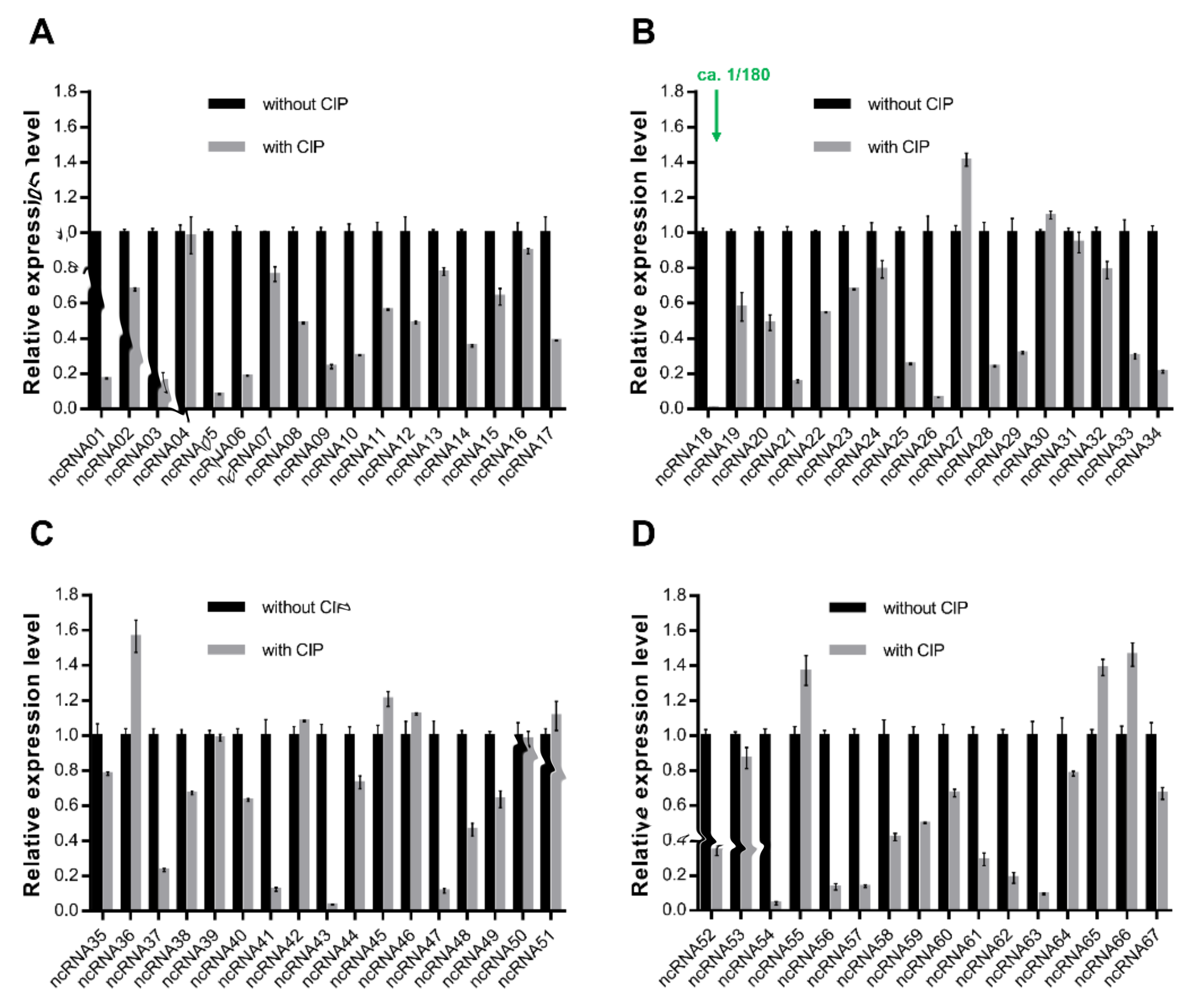
2.1. Response of ncRNA CsiR to Ciprofloxacin
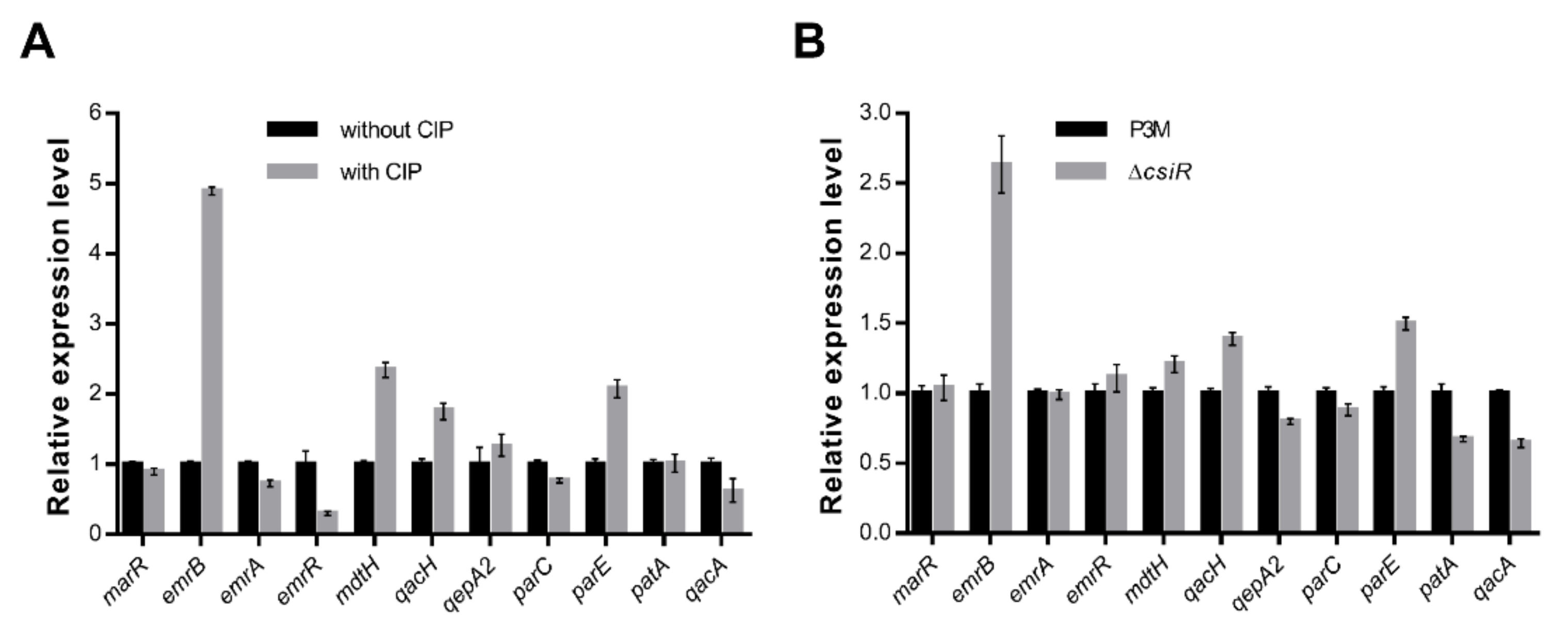
2.2. The Involvement of emrB in the Regulation of Ciprofloxacin Resistance
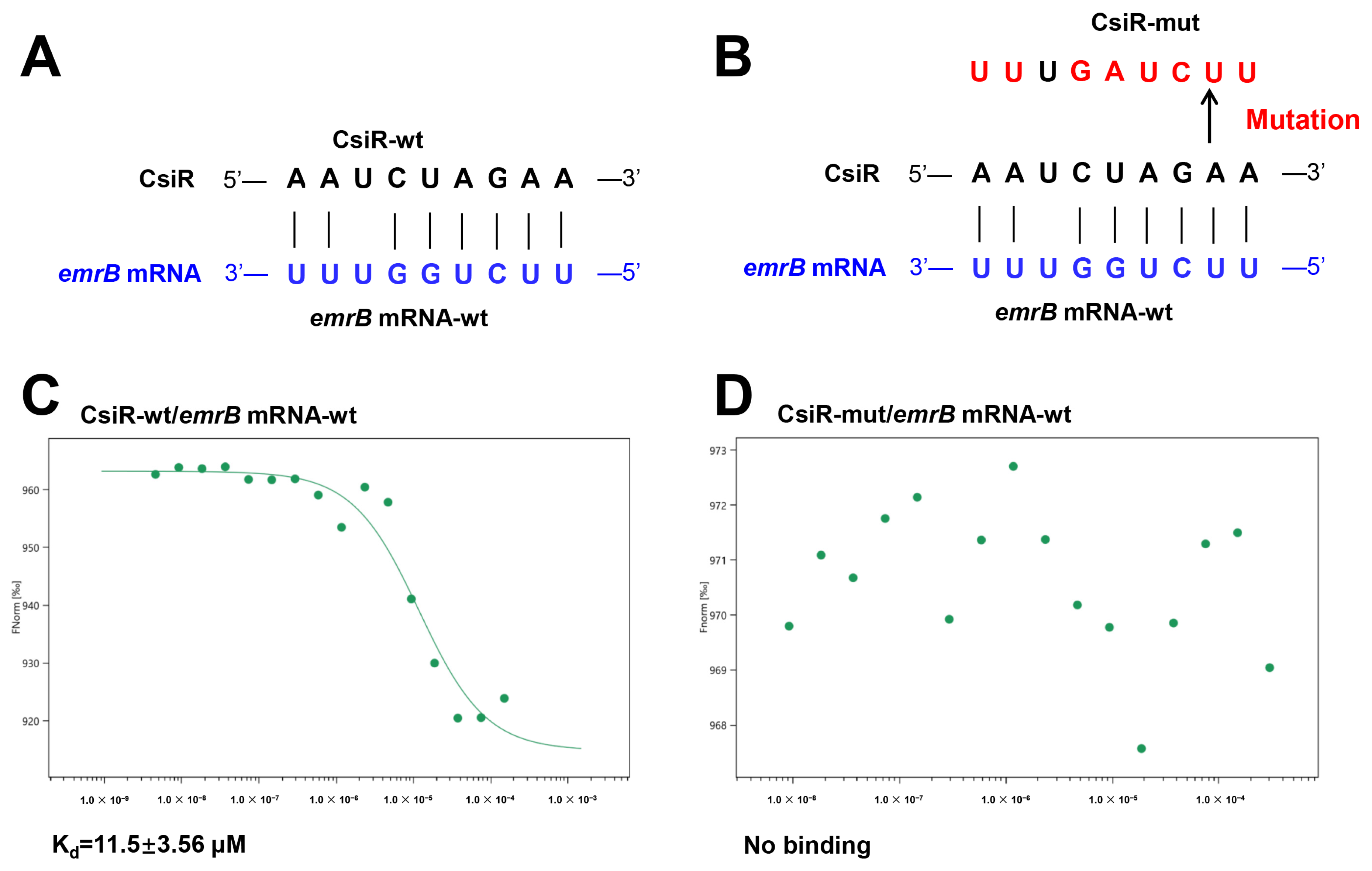
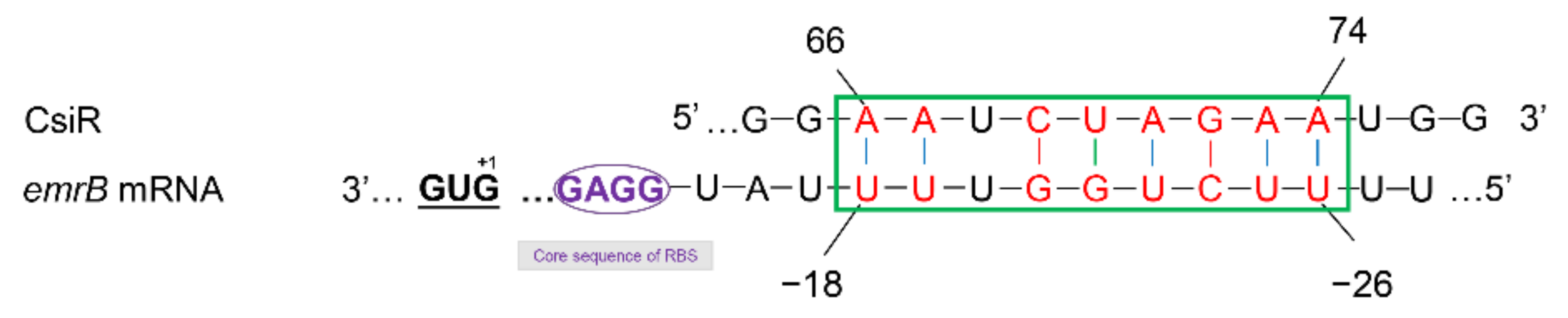
2.3. Interaction Mechanism between CsiR and emrB mRNA
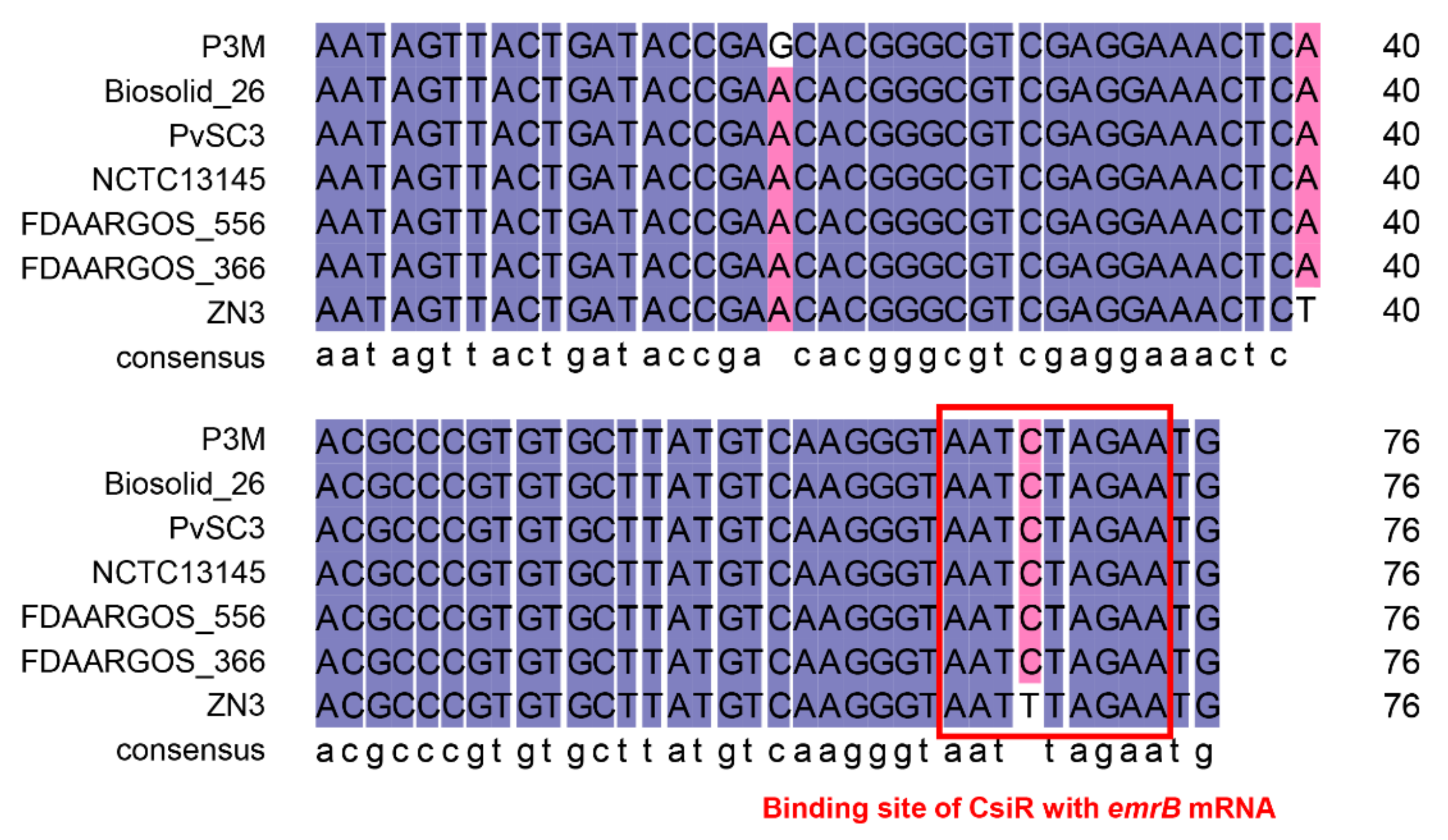
2.4. Species Specificity of CsiR Regulatory Mechanism
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Construction of csiR and emrB Deletion Mutants
4.3. Construction of csiR and emrB Complemented Strains
4.4. Spot Growth Assays
4.5. Quantitative Real-Time PCR
4.6. ncRNA Target Prediction
4.7. RNA Secondary Structure Prediction
4.8. Microscale Thermophoresis Assays
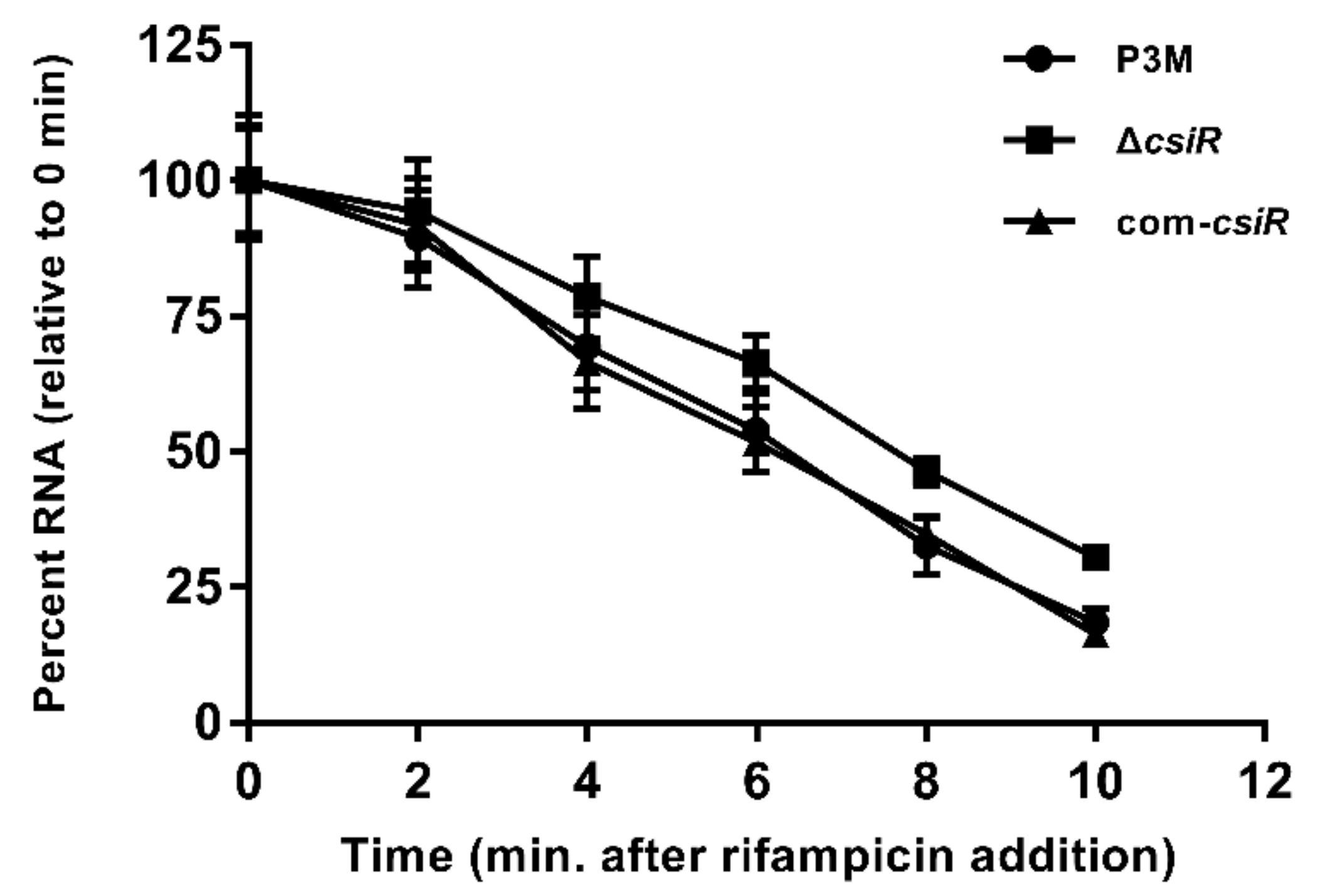
4.9. Half-Life Determination of emrB mRNA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leistra, A.N.; Curtis, N.C.; Contreras, L.M. Regulatory non-coding sRNAs in bacterial metabolic pathway engineering. Metab. Eng. 2019, 52, 190–214. [Google Scholar] [CrossRef]
- Barrandon, C.; Spiluttini, B.; Bensaude, O. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell 2008, 100, 83–95. [Google Scholar] [CrossRef]
- Hör, J.; Gorski, S.A.; Vogel, J. Bacterial RNA biology on a genome scale. Mol. Cell 2018, 70, 785–799. [Google Scholar] [CrossRef]
- Fröhlich, K.S.; Gottesman, S. Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Microbiol. Spectr. 2018, 6, 211–228. [Google Scholar] [CrossRef]
- Zhang, H.; Zhan, Y.; Yan, Y.; Liu, Y.; Hu, G.; Wang, S.; Yang, H.; Qiu, X.; Liu, Y.; Li, J.; et al. The Pseudomonas stutzeri-specific regulatory noncoding RNA NfiS targets katB mRNA encoding a catalase essential for optimal oxidative resistance and nitrogenase activity. J. Bacteriol. 2019, 201, e00334-19. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, R.; Tang, H.; Osei-Adjei, G.; Xu, S.; Zhang, Y.; Huang, X. A 3′ UTR-derived non-coding RNA RibS increases expression of cfa and promotes biofilm formation of Salmonella enterica serovar Typhi. Res. Microbiol. 2018, 169, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhu, W.; Chen, D.; Zhou, Y.; Lin, H. Small noncoding RNA sRNA0426 is involved in regulating biofilm formation in Streptococcus mutans. Microbiologyopen 2020, 9, e1096. [Google Scholar] [CrossRef]
- Orell, A.; Tripp, V.; Aliaga-Tobar, V.; Albers, S.V.; Maracaja-Coutinho, V.; Randau, L. A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius. Nucleic Acids Res. 2018, 46, 4794–4806. [Google Scholar] [CrossRef]
- Hücker, S.M.; Simon, S.; Scherer, S.; Neuhaus, K. Transcriptional and translational regulation by RNA thermometers, riboswitches and the sRNA DsrA in Escherichia coli O157:H7 Sakai under combined cold and osmotic stress adaptation. FEMS Microbiol. Lett. 2017, 364, fnw262. [Google Scholar] [CrossRef] [PubMed]
- Bojanovič, K.; D’Arrigo, I.; Long, K.S. Global transcriptional responses to osmotic, oxidative, and imipenem stress conditions in Pseudomonas putida. Appl. Environ. Microbiol. 2017, 83, e03236-16. [Google Scholar] [CrossRef] [PubMed]
- Desgranges, E.; Marzi, S.; Moreau, K.; Romby, P.; Caldelari, I. Noncoding RNA. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Zapf, R.L.; Wiemels, R.E.; Keogh, R.A.; Holzschu, D.L.; Howell, K.M.; Trzeciak, E.; Caillet, A.R.; King, K.A.; Selhorst, S.A.; Naldrett, M.J.; et al. The small RNA Teg41 regulates expression of the alpha phenol-soluble modulins and is required for virulence in Staphylococcus aureus. mBio 2019, 10, e02484-18. [Google Scholar] [CrossRef]
- Dar, D.; Sorek, R. Regulation of antibiotic-resistance by non-coding RNAs in bacteria. Curr. Opin. Microbiol. 2017, 36, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Mediati, D.G.; Wu, S.; Wu, W.; Tree, J.J. Networks of resistance: Small RNA control of antibiotic resistance. Trends Genet. 2021, 37, 35–45. [Google Scholar] [CrossRef]
- Dersch, P.; Khan, M.A.; Mühlen, S.; Görke, B. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 2017, 8, 803. [Google Scholar] [CrossRef] [PubMed]
- Villa, J.K.; Su, Y.; Contreras, L.M.; Hammond, M.C. Synthetic biology of small RNAs and riboswitches. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Nshogozabahizi, J.C.; Aubrey, K.L.; Ross, J.A.; Thakor, N. Applications and limitations of regulatory RNA elements in synthetic biology and biotechnology. J. Appl. Microbiol. 2019, 127, 968–984. [Google Scholar] [CrossRef]
- Patel, S.; Panchasara, H.; Braddick, D.; Gohil, N.; Singh, V. Synthetic small RNAs: Current status, challenges, and opportunities. J. Cell. Biochem. 2018, 119, 9619–9639. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Deng, Z.; Yan, Y.; Zhang, H.; Lu, C.; Yang, Z.; Shang, L.; Huang, Y.; Lv, F.; Liu, Y.; et al. NfiR, a new regulatory noncoding RNA (ncRNA), is required in concert with the NfiS ncRNA for optimal expression of nitrogenase genes in Pseudomonas stutzeri A1501. Appl. Environ. Microbiol. 2019, 85, e00762-19. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Zhang, G.F.; Liu, X.; Zhang, S.; Pan, B.; Liu, M.L. Ciprofloxacin derivatives and their antibacterial activities. Eur. J. Med. Chem. 2018, 146, 599–612. [Google Scholar] [CrossRef]
- Castro, W.; Navarro, M.; Biot, C. Medicinal potential of ciprofloxacin and its derivatives. Future Med. Chem. 2013, 5, 81–96. [Google Scholar] [CrossRef]
- Thomson, C.J. The global epidemiology of resistance to ciprofloxacin and the changing nature of antibiotic resistance: A 10 year perspective. J. Antimicrob. Chemother. 1999, 43, 31–40. [Google Scholar] [CrossRef]
- Fantin, B.; Duval, X.; Massias, L.; Alavoine, L.; Chau, F.; Retout, S.; Andremont, A.; Mentré, F. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J. Infect. Dis. 2009, 200, 390–398. [Google Scholar] [CrossRef]
- Hamilton, A.L.; Kamm, M.A.; Ng, S.C.; Morrison, M. Proteus spp. as putative gastrointestinal pathogens. Clin. Microbiol. Rev. 2018, 31, e00085-17. [Google Scholar] [CrossRef] [PubMed]
- Dominika, D. Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar]
- Ishida, H.; Fuziwara, H.; Kaibori, Y.; Horiuchi, T.; Sato, K.; Osada, Y. Cloning of multidrug resistance gene pqrA from Proteus vulgaris. Antimicrob. Agents Chemother. 1995, 39, 453–457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, J.H.; Sheng, W.H.; Hsueh, P.R.; SMART Program. Antimicrobial susceptibility and distribution of extended-spectrum β-lactamases, AmpC β-lactamases and carbapenemases among Proteus, Providencia and Morganella isolated from global hospitalised patients with intra-abdominal and urinary tract infections: Results of the study for monitoring antimicrobial resistance trends (SMART), 2008-2011. J. Glob. Antimicrob. Resist. 2020, 22, 398–407. [Google Scholar] [PubMed]
- Bilal, S.; Anam, S.; Mahmood, T.; Abdullah, R.M.; Nisar, S.; Kalsoom, F.; Luqman, M.; Anjum, F.R. Antimicrobial profiling and molecular characterization of antibiotic resistant genes of Proteus vulgaris isolated from tertiary care hospital, Islamabad, Pakistan. Pak. J. Pharm. Sci. 2019, 32, 2887–2891. [Google Scholar]
- Zhang, H.; Wang, H.; Ma, Z.; Liu, Y.; Wu, Z.; Xu, H.; Qiao, M. Characterization of Proteus vulgaris Strain P3M, a foodborne multidrug-resistant bacterium isolated from Penaeus vannamei in China. Microb. Drug. Resist. 2021. [Google Scholar] [CrossRef]
- Sabrin, A.; Gioe, B.W.; Gupta, A.; Grove, A. An EmrB multidrug efflux pump in Burkholderia thailandensis with unexpected roles in antibiotic resistance. J. Biol. Chem. 2019, 294, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, H.; Tsay, J.T.; Jackowski, S.; Takamura, Y.; Rock, C.O. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J. Bacteriol. 1993, 175, 3723–3729. [Google Scholar] [CrossRef]
- Kawamoto, H.; Koide, Y.; Morita, T.; Aiba, H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006, 61, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Papenfort, K.; Reinhardt, R.; Sharma, C.M.; Vogel, J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012, 31, 4005–4019. [Google Scholar] [CrossRef] [PubMed]
- Hüttenhofer, A.; Noller, H.F. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994, 13, 3892–3901. [Google Scholar] [CrossRef]
- Udekwu, K.I.; Darfeuille, F.; Vogel, J.; Reimegård, J.; Holmqvist, E.; Wagner, E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005, 19, 2355–2366. [Google Scholar] [CrossRef]
- Zhan, Y.; Yan, Y.; Deng, Z.; Chen, M.; Lu, W.; Lu, C.; Shang, L.; Yang, Z.; Zhang, W.; Wang, W.; et al. The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 2016, 113, E4348–E4356. [Google Scholar] [CrossRef] [PubMed]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Marques, C.N.H.; Nelson, S.M. Pharmacodynamics of ciprofloxacin against Pseudomonas aeruginosa planktonic and biofilm-derived cells. Lett. Appl. Microbiol. 2019, 68, 350–359. [Google Scholar] [CrossRef]
- Jakobsen, L.; Lundberg, C.V.; Frimodt-Møller, N. Ciprofloxacin pharmacokinetics/pharmacodynamics against susceptible and low-level resistant Escherichia coli isolates in an experimental ascending urinary tract infection model in mice. Antimicrob. Agents Chemother. 2020, 65, e01804-20. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; De Vecchi, E.; Mombelli, B.; Nicola, L.; Valli, M.; Gismondo, M.R. Activity of levofloxacin and ciprofloxacin against urinary pathogens. J. Antimicrob. Chemother. 2001, 48, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhang, J.; Sun, W.; He, W.; Jiang, H.; Chen, D.; Murchie, A.I.H. Riboswitch control of aminoglycoside antibiotic resistance. Cell 2013, 152, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Planson, A.G.; Sauveplane, V.; Dervyn, E.; Jules, M. Bacterial growth physiology and RNA metabolism. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194502. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C. Metabolic monitoring by bacterial mRNAs. Arch. Microbiol. 2005, 183, 151–159. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, V.; Pérez-Martín, J. Regulatory noise in prokaryotic promoters: How bacteria learn to respond to novel environmental signals. Mol. Microbiol. 1996, 19, 1177–1184. [Google Scholar] [CrossRef]
- Huang, W.; Wilks, A. A rapid seamless method for gene knockout in Pseudomonas aeruginosa. BMC Microbiol. 2017, 17, 199. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.B.; Puhler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Nunn, D.; Bergman, S.; Lory, S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 1990, 172, 2911–2919. [Google Scholar] [CrossRef]
- Zhang, H.; Chang, M.; Zhang, X.; Cai, P.; Dai, Y.; Song, T.; Wu, Z.; Xu, H.; Qiao, M. Functional identification and evolutionary analysis of two novel plasmids mediating quinolone resistance in Proteus vulgaris. Microorganisms 2020, 8, 1074. [Google Scholar] [CrossRef]
- Ying, X.; Cao, Y.; Wu, J.; Liu, Q.; Cha, L.; Li, W. sTarPicker: A method for efficient prediction of bacterial sRNA targets based on a two-step model for hybridization. PLoS ONE 2011, 6, e22705. [Google Scholar] [CrossRef] [PubMed]
- Eggenhofer, F.; Tafer, H.; Stadler, P.F.; Hofacker, I.L. RNApredator: Fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011, 39, W149–W154. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Seidel, S.A.I.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Lippok, S.; Seidel, S.A.; Duhr, S.; Uhland, K.; Holthoff, H.P.; Jenne, D.; Braun, D. Direct detection of antibody concentration and affinity in human serum using microscale thermophoresis. Anal. Chem. 2012, 84, 3523–3530. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Song, T.; Qin, C.; Xu, H.; Qiao, M. A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA. Int. J. Mol. Sci. 2021, 22, 10627. https://doi.org/10.3390/ijms221910627
Zhang H, Song T, Qin C, Xu H, Qiao M. A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA. International Journal of Molecular Sciences. 2021; 22(19):10627. https://doi.org/10.3390/ijms221910627
Chicago/Turabian StyleZhang, Hongyang, Tongzhen Song, Chuhan Qin, Haijin Xu, and Mingqiang Qiao. 2021. "A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA" International Journal of Molecular Sciences 22, no. 19: 10627. https://doi.org/10.3390/ijms221910627
APA StyleZhang, H., Song, T., Qin, C., Xu, H., & Qiao, M. (2021). A Novel Non-Coding RNA CsiR Regulates the Ciprofloxacin Resistance in Proteus vulgaris by Interacting with emrB mRNA. International Journal of Molecular Sciences, 22(19), 10627. https://doi.org/10.3390/ijms221910627




