MMP14 Contributes to HDAC Inhibition-Induced Radiosensitization of Glioblastoma
Abstract
:1. Introduction
2. Results
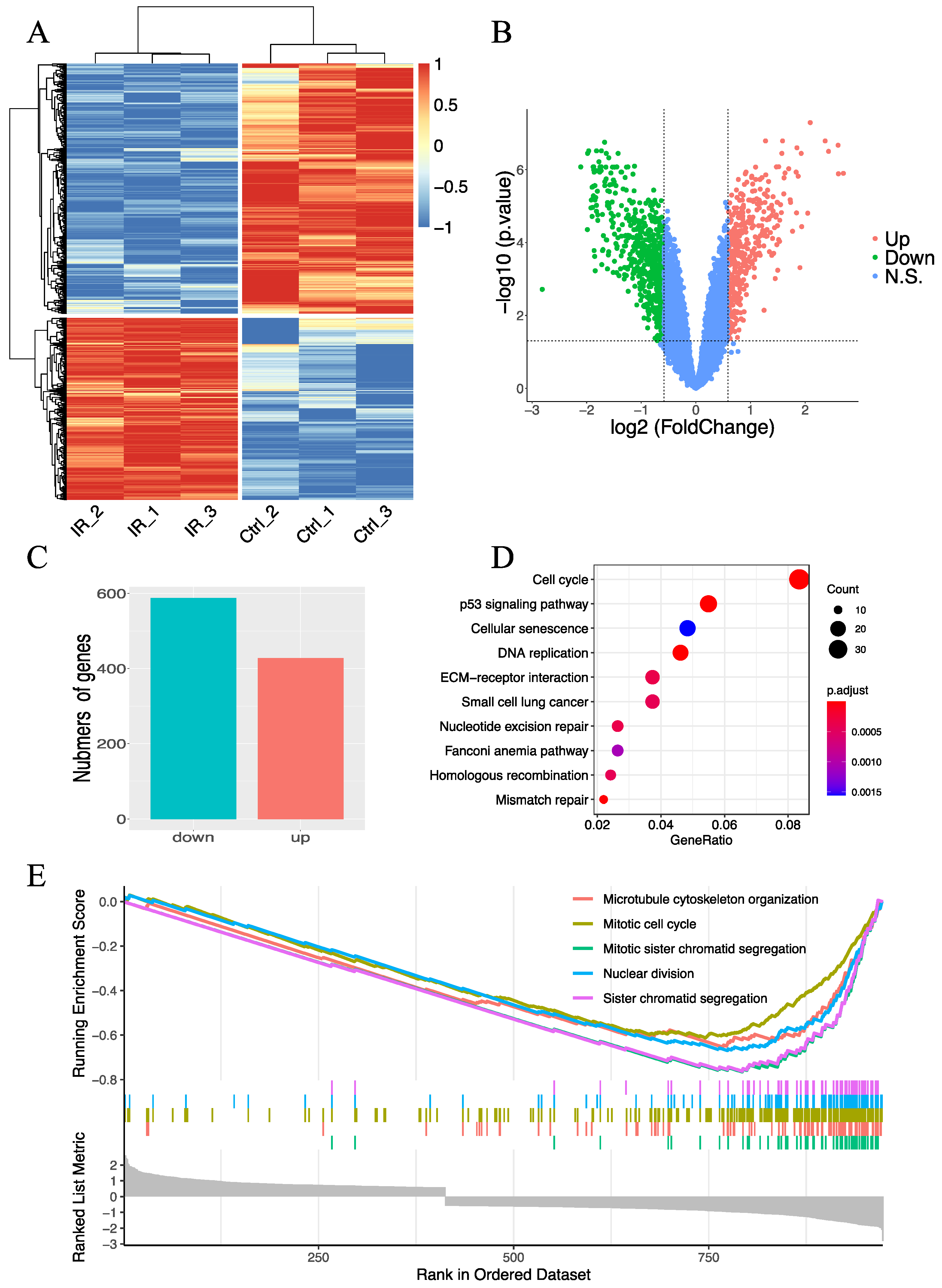
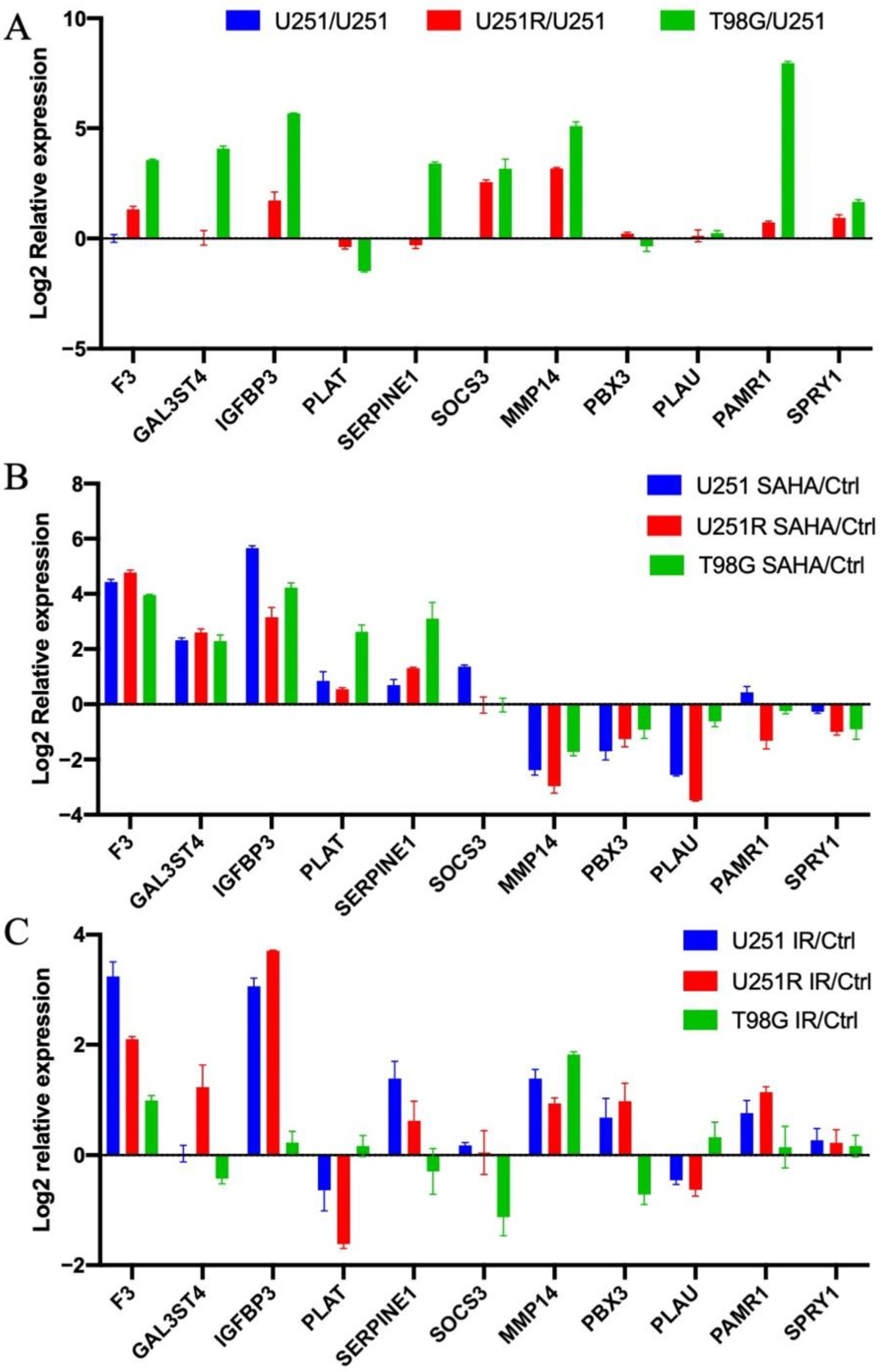
2.1. Evaluation of Radiation-Induced Changes in Gene Expression
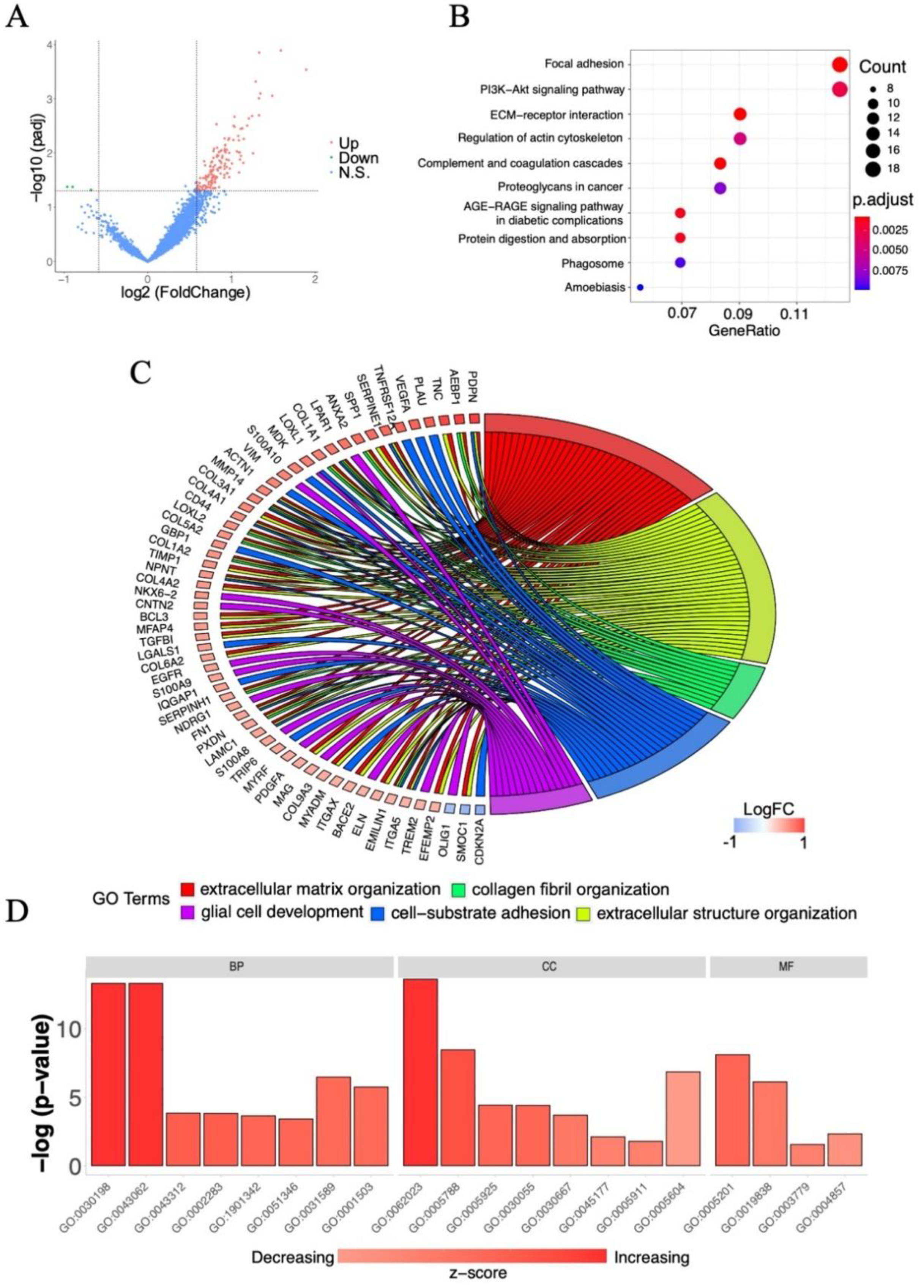
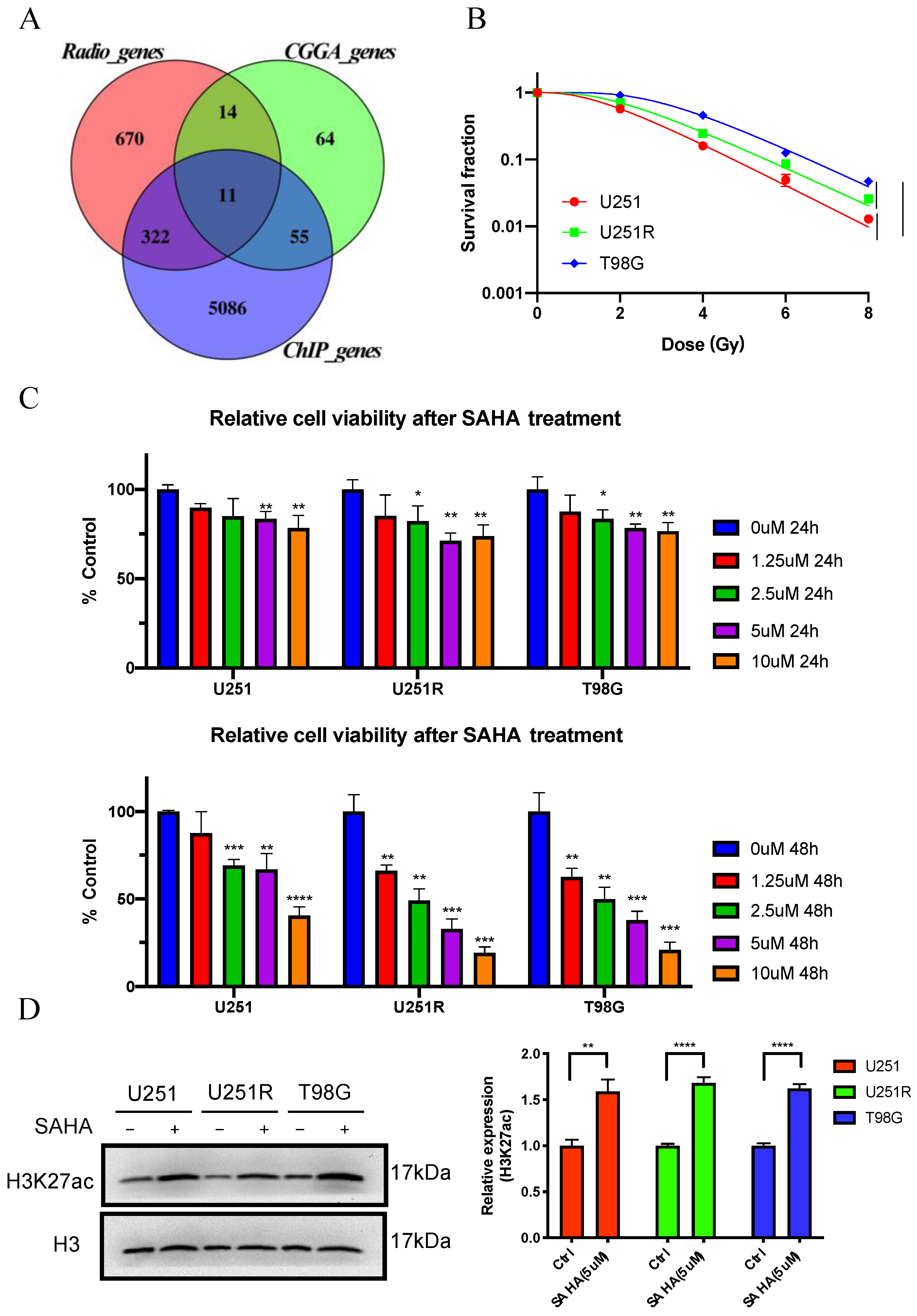
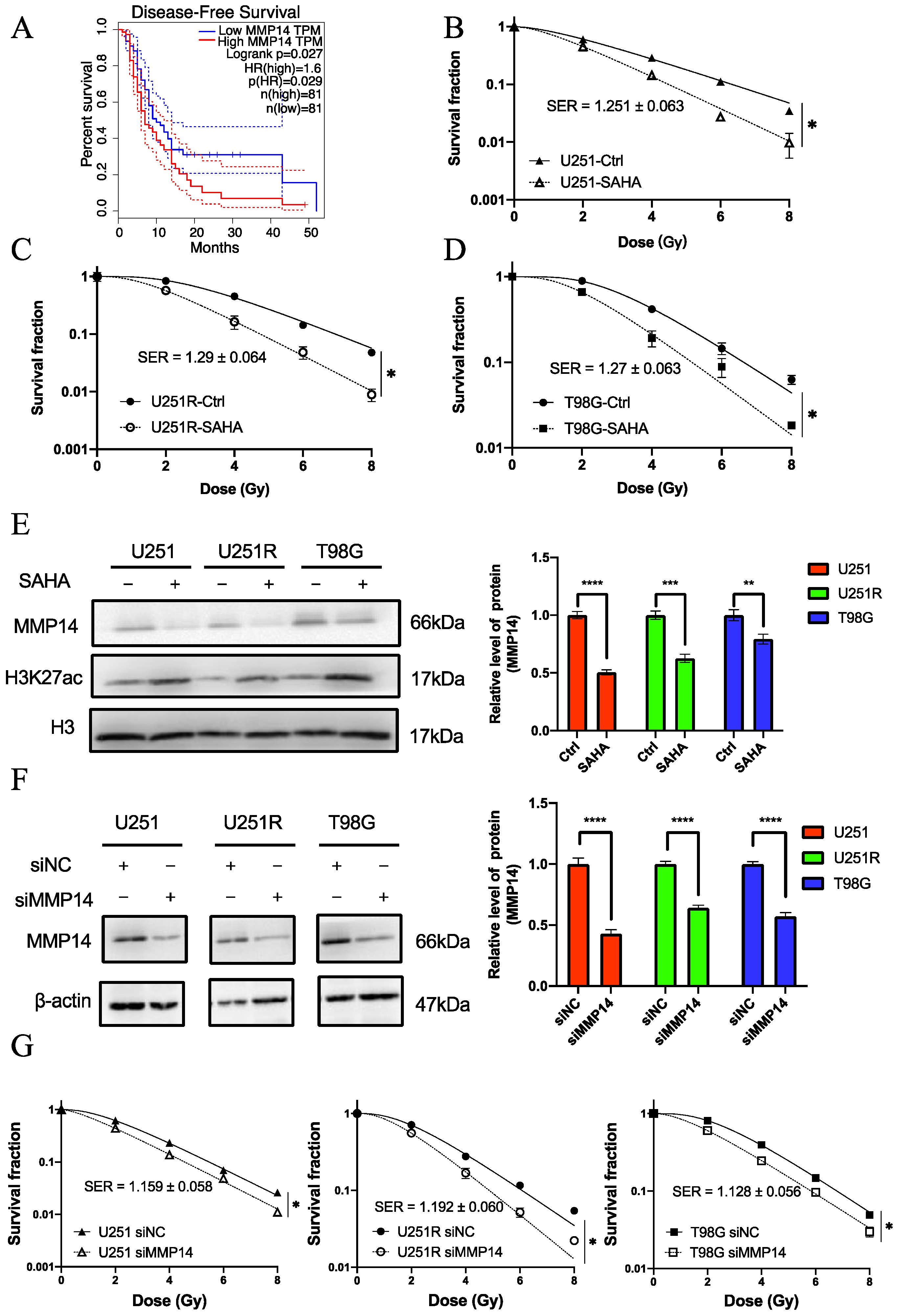
2.2. Screening of Survival-Associated Genes
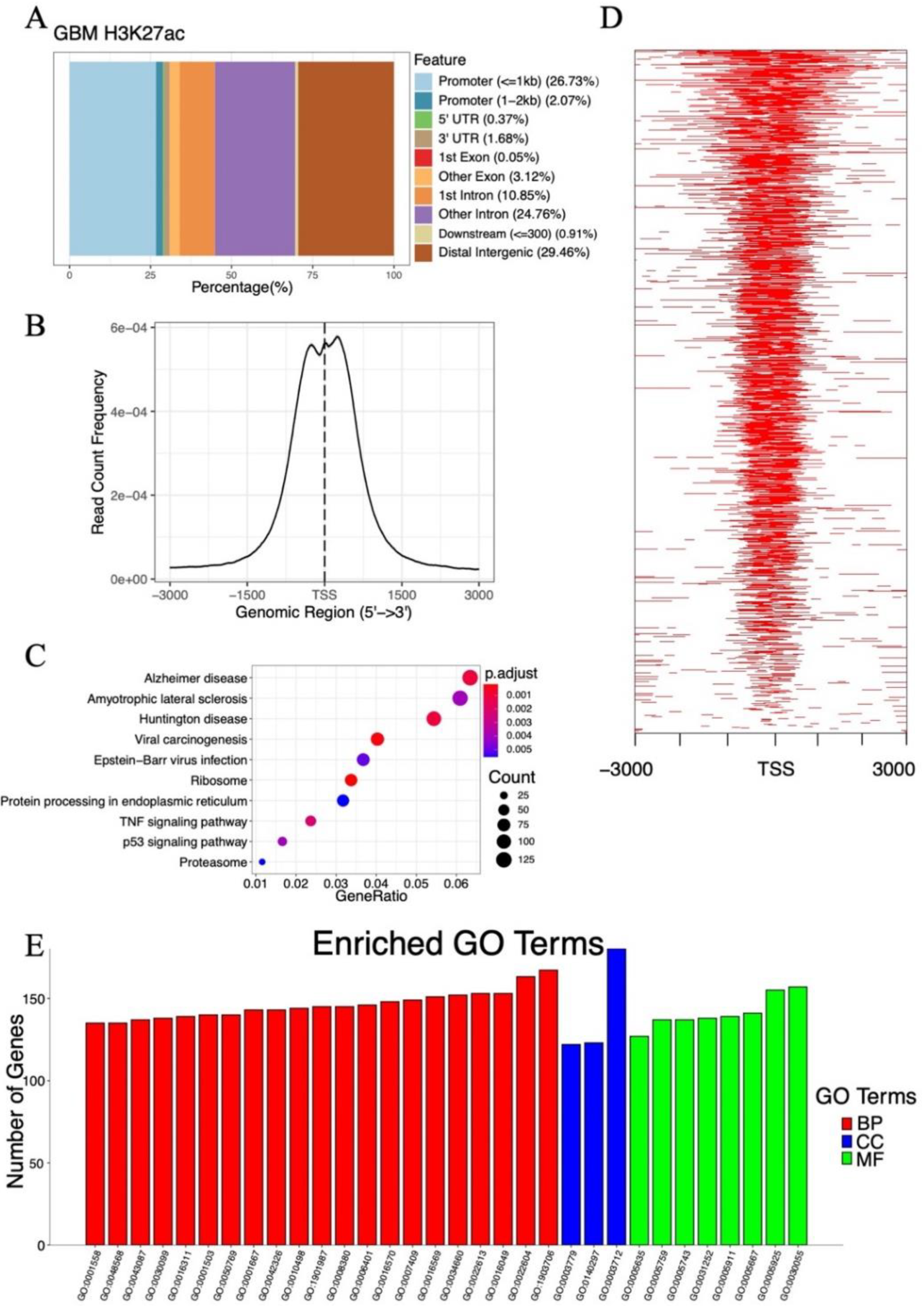
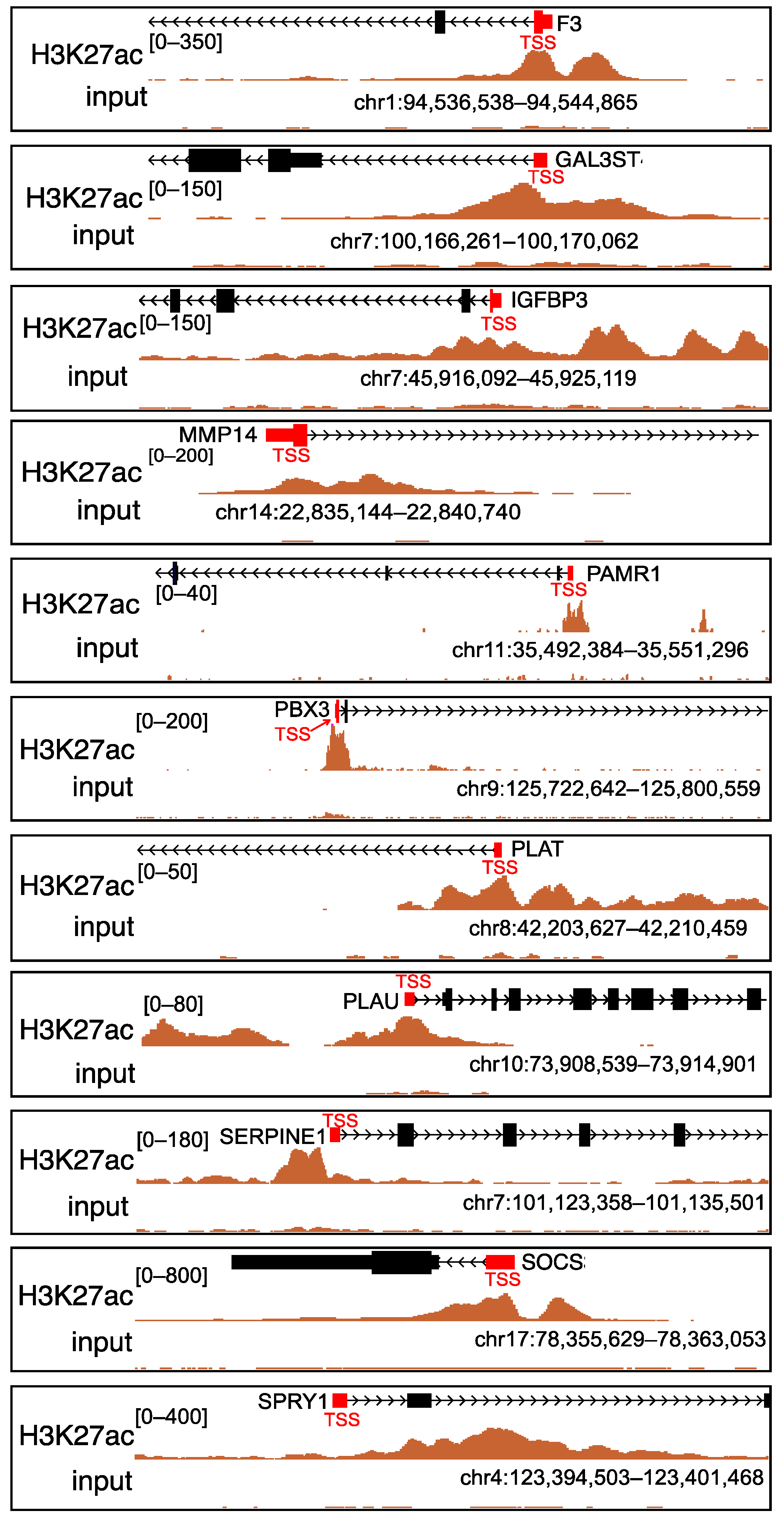
2.3. Analysis of Histone Acetylation-Controlled Genes
2.4. MMP14 Was the Potential Target of SAHA
2.5. The Role of MMP14 in Radiosensitivity
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Irradiation
4.2. SAHA Treatment
4.3. Colony Formation Assay
4.4. RNA Extraction and RT-qPCR Assay
4.5. Western Blot Assay
4.6. Cell Proliferation Assay
4.7. RNA-Seq Data Download, Processing, and Analysis
4.8. ChIP-Seq Data Download, Processing, and Analysis
4.9. Gene Expression Analysis of Clinical Database
4.10. RNA Interference and Drug Treatment
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet. Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Chinnaiyan, P.; Cerna, D.; Burgan, W.E.; Beam, K.; Williams, E.S.; Camphausen, K.; Tofilon, P.J. Postradiation sensitization of the histone deacetylase inhibitor valproic acid. Clin. Cancer Res. 2008, 14, 5410–5415. [Google Scholar] [CrossRef] [Green Version]
- Ceccacci, E.; Minucci, S. Inhibition of histone deacetylases in cancer therapy: Lessons from leukaemia. Br. J. Cancer 2016, 114, 605–611. [Google Scholar] [CrossRef] [Green Version]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.G.; Allis, C.D.; Chi, P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol. Med. 2007, 13, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Gallinari, P.; Di Marco, S.; Jones, P.; Pallaoro, M.; Steinkühler, C. HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 2007, 17, 195–211. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, A.J.M.; van Gennip, A.H.; Caron, H.N.; Kemp, S.; van Kuilenburg, A.B.P. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, T.; Vanoli, F.; Chiolo, I.; Shubassi, G.; Bernstein, K.A.; Rothstein, R.; Botrugno, O.A.; Parazzoli, D.; Oldani, A.; Minucci, S.; et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011, 471, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Krauze, A.V.; Myrehaug, S.D.; Chang, M.G.; Holdford, D.J.; Smith, S.; Shih, J.; Tofilon, P.J.; Fine, H.A.; Camphausen, K. A Phase 2 Study of Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients With Glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 986–992. [Google Scholar] [CrossRef] [Green Version]
- Galanis, E.; Anderson, S.K.; Miller, C.R.; Sarkaria, J.N.; Jaeckle, K.; Buckner, J.C.; Ligon, K.L.; Ballman, K.V.; Moore, D.F.; Nebozhyn, M.; et al. Phase I/II trial of vorinostat combined with temozolomide and radiation therapy for newly diagnosed glioblastoma: Results of Alliance N0874/ABTC 02. Neuro. Oncol. 2018, 20, 546–556. [Google Scholar] [CrossRef] [Green Version]
- Karamanou, K.; Franchi, M.; Vynios, D.; Brézillon, S. Epithelial-to-mesenchymal transition and invadopodia markers in breast cancer: Lumican a key regulator. Semin. Cancer Biol. 2020, 62, 125–133. [Google Scholar] [CrossRef]
- Dokmanovic, M.; Clarke, C.; Marks, P.A. Histone deacetylase inhibitors: Overview and perspectives. Mol. Cancer Res. 2007, 5, 981–989. [Google Scholar] [CrossRef] [Green Version]
- Uddin, M.S.; Al Mamun, A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2020, 1044–579X. [Google Scholar]
- Wang, Y.; Wu, K.; Wu, A.; Yang, Z.; Li, J.; Mo, Y.; Xu, M.; Wu, B.; Yang, Z. MMP-14 overexpression correlates with poor prognosis in non-small cell lung cancer. Tumor Biol. 2014, 35, 9815–9821. [Google Scholar] [CrossRef]
- Cui, G.; Cai, F.; Ding, Z.; Gao, L. MMP14 predicts a poor prognosis in patients with colorectal cancer. Hum. Pathol. 2019, 83, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Têtu, B.; Brisson, J.; Wang, C.S.; Lapointe, H.; Beaudry, G.; Blanchette, C.; Trudel, D. The influence of MMP-14, TIMP-2 and MMP-2 expression on breast cancer prognosis. Breast Cancer Res. 2006, 8, R28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasurinen, A.; Tervahartiala, T.; Laitinen, A.; Kokkola, A.; Sorsa, T.; Böckelman, C.; Haglund, C. High serum MMP-14 predicts worse survival in gastric cancer. PLoS ONE 2018, 13, e0208800. [Google Scholar] [CrossRef] [PubMed]
- Zarrabi, K.; Dufour, A.; Li, J.; Kuscu, C.; Pulkoski-Gross, A.; Zhi, J.; Hu, Y.; Sampson, N.S.; Zucker, S.; Cao, J. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J. Biol. Chem. 2011, 286, 33167–33177. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.-H.; Du, L.; Li, N.; Zhang, Y.; Xiang, Y.; Tang, J.-H.; Xia, S.; Zhang, E.E.; Lv, S.-Q. Methylation-mediated miR-155-FAM133A axis contributes to the attenuated invasion and migration of IDH mutant gliomas. Cancer Lett. 2018, 432, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Matsuoka, S.; Carpenter, P.B.; Elledge, S.J. 53BP1, a mediator of the DNA damage checkpoint. Science 2002, 298, 1435–1438. [Google Scholar] [CrossRef]
- Wesseling, P.; Capper, D. WHO 2016 Classification of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef] [Green Version]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Turner, K.M.; Nguyen, N.; Raviram, R.; Erb, M.; Santini, J.; Luebeck, J.; Rajkumar, U.; Diao, Y.; Li, B.; et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019, 575, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Bergström, T.; Jiang, Y.; Johansson, P.; Marinescu, V.D.; Lindberg, N.; Segerman, A.; Wicher, G.; Niklasson, M.; Baskaran, S.; et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2015, 2, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Chen, Q.; Liu, H.; Hu, S.; Zhou, Y.; Bai, Y.; Zhang, J.; Pan, Y.; Shao, C. CD81 Enhances Radioresistance of Glioblastoma by Promoting Nuclear Translocation of Rad51. Cancers 2021, 13, 1998. [Google Scholar] [CrossRef]
- He, L.; Bhat, K.; Ioannidis, A.; Zhang, L.; Nguyen, N.T.; Allen, J.E.; Nghiemphu, P.L.; Cloughesy, T.F.; Liau, L.M.; Kornblum, H.I.; et al. Effects of the DRD2/3 antagonist ONC201 and radiation in glioblastoma. Radiother. Oncol. 2021, 161, 140–147. [Google Scholar] [CrossRef]
- Gonzalez-Molina, J.; Gramolelli, S.; Liao, Z.; Carlson, J.W.; Ojala, P.M.; Lehti, K. MMP14 in Sarcoma: A Regulator of Tumor Microenvironment Communication in Connective Tissues. Cells 2019, 8, 991. [Google Scholar] [CrossRef] [Green Version]
- Torrisi, F.; Vicario, N.; Spitale, F.M.; Cammarata, F.P.; Minafra, L.; Salvatorelli, L.; Russo, G.; Cuttone, G.; Valable, S.; Gulino, R.; et al. The Role of Hypoxia and SRC Tyrosine Kinase in Glioblastoma Invasiveness and Radioresistance. Cancers 2020, 12, 2860. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, M.; Zhou, Y.; Guo, W.; Yi, M.; Zhang, Z.; Ding, Y.; Wang, Y. The application of histone deacetylases inhibitors in glioblastoma. J. Exp. Clin. Cancer Res. 2020, 39, 138. [Google Scholar] [CrossRef] [PubMed]
- Kunadis, E.; Lakiotaki, E.; Korkolopoulou, P.; Piperi, C. Targeting post-translational histone modifying enzymes in glioblastoma. Pharmacol. Ther. 2021, 220, 107721. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Zhang, Y.; Shang, E.; Shu, C.; Torrini, C.; Zhao, J.; Bianchetti, E.; Mela, A.; Humala, N.; Mahajan, A.; et al. HDAC inhibitors elicit metabolic reprogramming by targeting super-enhancers in glioblastoma models. J. Clin. Investg. 2020, 130, 3699–3716. [Google Scholar] [CrossRef]
- Heim, C.E.; Bosch, M.E.; Yamada, K.J.; Aldrich, A.L.; Chaudhari, S.S.; Klinkebiel, D.; Gries, C.M.; Alqarzaee, A.A.; Li, Y.; Thomas, V.C.; et al. Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 2020, 5, 1271–1284. [Google Scholar] [CrossRef]
- Thakur, V.; Zhang, K.; Savadelis, A.; Zmina, P.; Aguila, B.; Welford, S.M.; Abdul-Karim, F.; Bonk, K.W.; Keri, R.A.; Bedogni, B. The membrane tethered matrix metalloproteinase MT1-MMP triggers an outside-in DNA damage response that impacts chemo- and radiotherapy responses of breast cancer. Cancer Lett. 2019, 443, 115–124. [Google Scholar] [CrossRef]
- Zhou, H.; Miki, R.; Eeva, M.; Fike, F.M.; Seligson, D.; Yang, L.; Yoshimura, A.; Teitell, M.A.; Jamieson, C.A.M.; Cacalano, N.A. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin. Cancer Res. 2007, 13, 2344–2353. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, K.; Motoyama, S.; Koyota, S.; Shibuya, K.; Usami, S.; Maruyama, K.; Saito, H.; Minamiya, Y.; Sugiyama, T.; Ogawa, J. IGFBP3 and BAG1 enhance radiation-induced apoptosis in squamous esophageal cancer cells. Biochem. Biophys. Res. Commun. 2011, 404, 1070–1075. [Google Scholar] [CrossRef]
- Kawamata, N.; Chen, J.; Koeffler, H.P. Suberoylanilide hydroxamic acid (SAHA; vorinostat) suppresses translation of cyclin D1 in mantle cell lymphoma cells. Blood 2007, 110, 2667–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Cheng, H.; Shao, Q.; Dong, Z.; Xie, Q.; Zhao, L.; Wang, Q.; Kong, B.; Qu, X. Leptin-promoted human extravillous trophoblast invasion is MMP14 dependent and requires the cross talk between Notch1 and PI3K/Akt signaling. Biol. Reprod. 2014, 90, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nebbioso, A.; Carafa, V.; Conte, M.; Tambaro, F.P.; Abbondanza, C.; Martens, J.; Nees, M.; Benedetti, R.; Pallavicini, I.; Minucci, S.; et al. c-Myc Modulation and Acetylation Is a Key HDAC Inhibitor Target in Cancer. Clin. Cancer Res. 2017, 23, 2542–2555. [Google Scholar] [CrossRef] [Green Version]
- Tasset, C.; Singh Yadav, A.; Sureshkumar, S.; Singh, R.; van der Woude, L.; Nekrasov, M.; Tremethick, D.; van Zanten, M.; Balasubramanian, S. POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007280. [Google Scholar] [CrossRef] [Green Version]
- Haldar, S.; Dru, C.; Mishra, R.; Tripathi, M.; Duong, F.; Angara, B.; Fernandez, A.; Arditi, M.; Bhowmick, N.A. Histone deacetylase inhibitors mediate DNA damage repair in ameliorating hemorrhagic cystitis. Sci. Rep. 2016, 6, 39257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [Green Version]
| CGGA_genes | Radio_genes | ||||
|---|---|---|---|---|---|
| GeneID | Log2FC | p_Value | GeneID | Log2FC | p_Value |
| PLAU | 1.18857781 | 0.00313738 | PLAT | 1.6308572 | 5.23E−09 |
| SERPINE1 | 1.09935193 | 0.00977508 | PLAU | 1.26191627 | 0.00000183 |
| SOCS3 | 1.03888471 | 0.00329858 | SERPINE1 | 1.24394928 | 0.00000518 |
| F3 | 0.92412421 | 0.00724404 | MMP14 | 1.09278289 | 2.4E−09 |
| MMP14 | 0.91836114 | 0.00541851 | PAMR1 | 0.95922717 | 2.13E−07 |
| IGFBP3 | 0.91328995 | 0.02870288 | F3 | 0.80955228 | 0.00000134 |
| PAMR1 | 0.77039761 | 0.01443068 | GAL3ST4 | 0.76133027 | 0.000011 |
| PLAT | 0.76908966 | 0.02181263 | IGFBP3 | 0.75038818 | 0.00000156 |
| SPRY1 | 0.72918236 | 0.03779377 | SPRY1 | −0.6228777 | 0.00073747 |
| PBX3 | 0.72507171 | 0.02756671 | SOCS3 | −0.7459958 | 0.0000168 |
| GAL3ST4 | 0.61764614 | 0.04685508 | PBX3 | −1.0973208 | 0.00000114 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Liu, H.; Zheng, W.; Chen, Q.; Hu, S.; Pan, Y.; Bai, Y.; Zhang, J.; Shao, C. MMP14 Contributes to HDAC Inhibition-Induced Radiosensitization of Glioblastoma. Int. J. Mol. Sci. 2021, 22, 10403. https://doi.org/10.3390/ijms221910403
Zhou Y, Liu H, Zheng W, Chen Q, Hu S, Pan Y, Bai Y, Zhang J, Shao C. MMP14 Contributes to HDAC Inhibition-Induced Radiosensitization of Glioblastoma. International Journal of Molecular Sciences. 2021; 22(19):10403. https://doi.org/10.3390/ijms221910403
Chicago/Turabian StyleZhou, Yuchuan, Hongxia Liu, Wang Zheng, Qianping Chen, Songling Hu, Yan Pan, Yang Bai, Jianghong Zhang, and Chunlin Shao. 2021. "MMP14 Contributes to HDAC Inhibition-Induced Radiosensitization of Glioblastoma" International Journal of Molecular Sciences 22, no. 19: 10403. https://doi.org/10.3390/ijms221910403
APA StyleZhou, Y., Liu, H., Zheng, W., Chen, Q., Hu, S., Pan, Y., Bai, Y., Zhang, J., & Shao, C. (2021). MMP14 Contributes to HDAC Inhibition-Induced Radiosensitization of Glioblastoma. International Journal of Molecular Sciences, 22(19), 10403. https://doi.org/10.3390/ijms221910403








