A Novel Regulatory Player in the Innate Immune System: Long Non-Coding RNAs
Abstract
:1. Introduction
2. Long Non-Coding RNAs
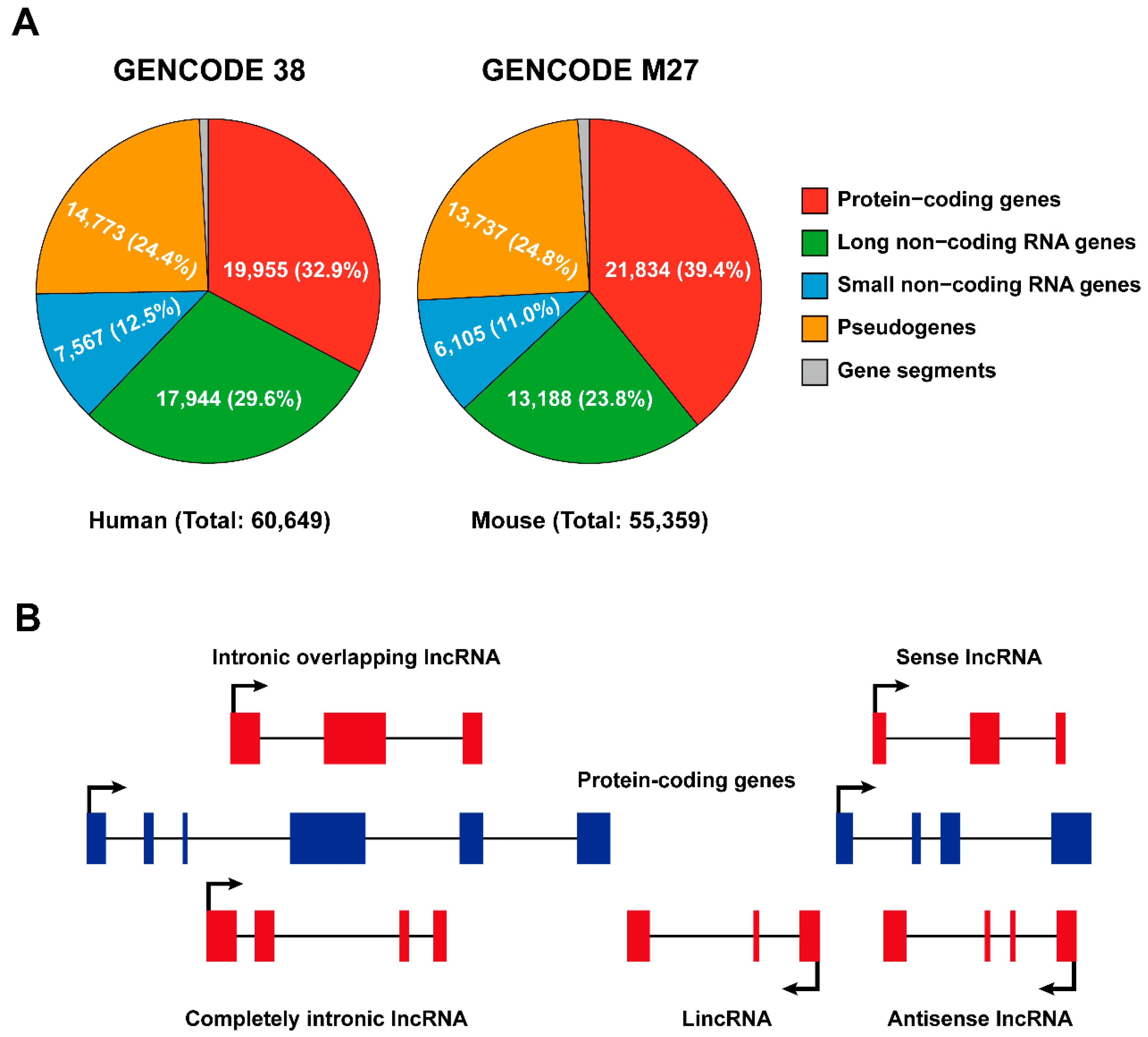
2.1. Classification of Long Non-Coding RNAs
2.2. Transcription and Degradation of Long Non-Coding RNAs
2.3. Alternative Splicing of Long Non-Coding RNAs
2.4. Conservation and Secondary Structure of Long Non-Coding RNAs
2.5. Subcellular Localization of Long Non-Coding RNAs
3. Long Non-Coding RNAs Function as Transcriptional Regulators
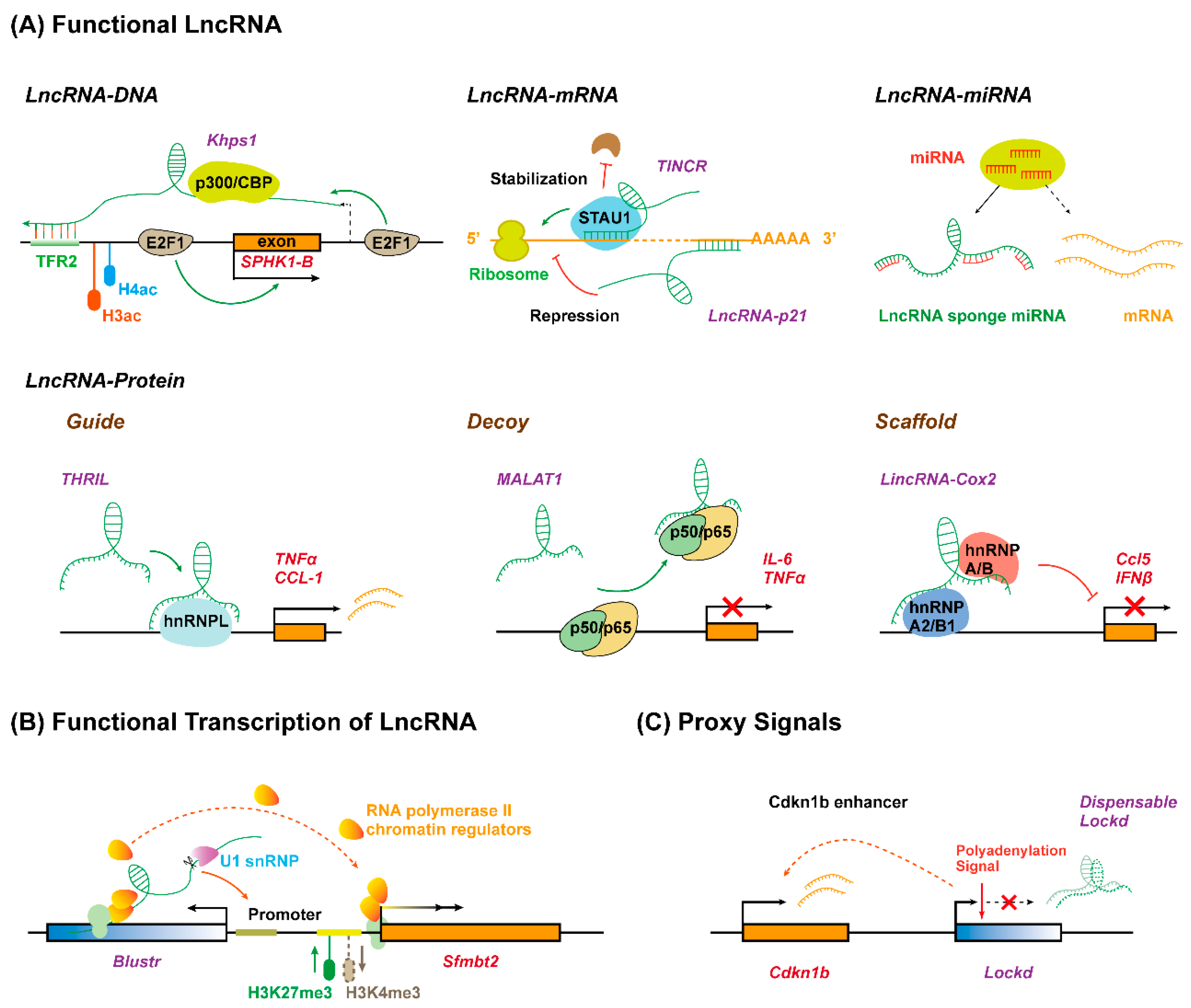
3.1. Functional Long Non-Coding RNA Molecules
3.2. Functional Roles of the Act of Long Non-Coding RNA Transcription
3.3. Long Non-Coding RNAs Act as Proxy Signals for Cis-Regulatory Elements
4. Long Non-Coding RNAs Function in Innate Immunity
4.1. Long Non-Coding RNAs in the Development of Innate Immune Cells
4.1.1. Macrophages
| LncRNA | Target Genes | Functional Consequences | Mechanism | Reference |
|---|---|---|---|---|
| Lnc-MC | miR-199a-5p | Promotes macrophage differentiation | Releases ACVR1B | [88] |
| NTT | PBOV1 | Promotes macrophage differentiation | Recruits hnRNP-U to PBOV1 promoter | [89] |
| LincRNA-Cox2 | NF-κB-mediated cytokines | Inhibits M2 polarization | - | [96] |
| GAS5 | miR-455-5p | Promotes M1 polarization from M2 | Release SOCS3 | [97] |
| MIR-155HG | Proinflammatory cytokines | Induces M1 polarization | - | [98] |
| Mirt2 | TRAF6 | Promotes M2 polarization | Suppresses NF-κB and MAPK pathway | [99] |
| LncRNA-MM2P | STAT6 | Promotes M2 polarization | Increases phosphorylation of STAT6 | [100] |
| PTPRE-AS1 | PTPRE | Inhibits M2 activation | Recruits WDR5 to PTPRE promoter | [95] |
| Lnc-DC | STAT3 | Promotes DCs differentiation | Prevents Y705 dephosphorylation of STAT3 by SHP1 | [101] |
| MALAT1 | miR-155 | Induces tolerogenic DCs | Releases DC-SIGH and IL-10 | [102] |
| HOTAIRM1 | HOXA cluster, CD11b and CD18 | Promotes granulocyte differentiation and maturation | - | [103] |
| Lnc-CD56 | CD56 | Promotes CD56 NK cell development | - | [104] |
| GAS5 | miR-544 | Enhances CD107a+ NK cells and its cytotoxicity | Upregulates RUNX3 as a sponge | [105] |
| Linc-EPHA6-1 | has-miR-4885-5p | Promotes cytotoxicity of NK cells | Upregulates NKp46 expression as a sponge | [106] |
4.1.2. Dendritic Cells
4.1.3. Granulocytes
4.1.4. Natural Killer Cells
4.2. Long Non-Coding RNAs Function in Host Inflammatory Response Triggered by the Innate Immune System
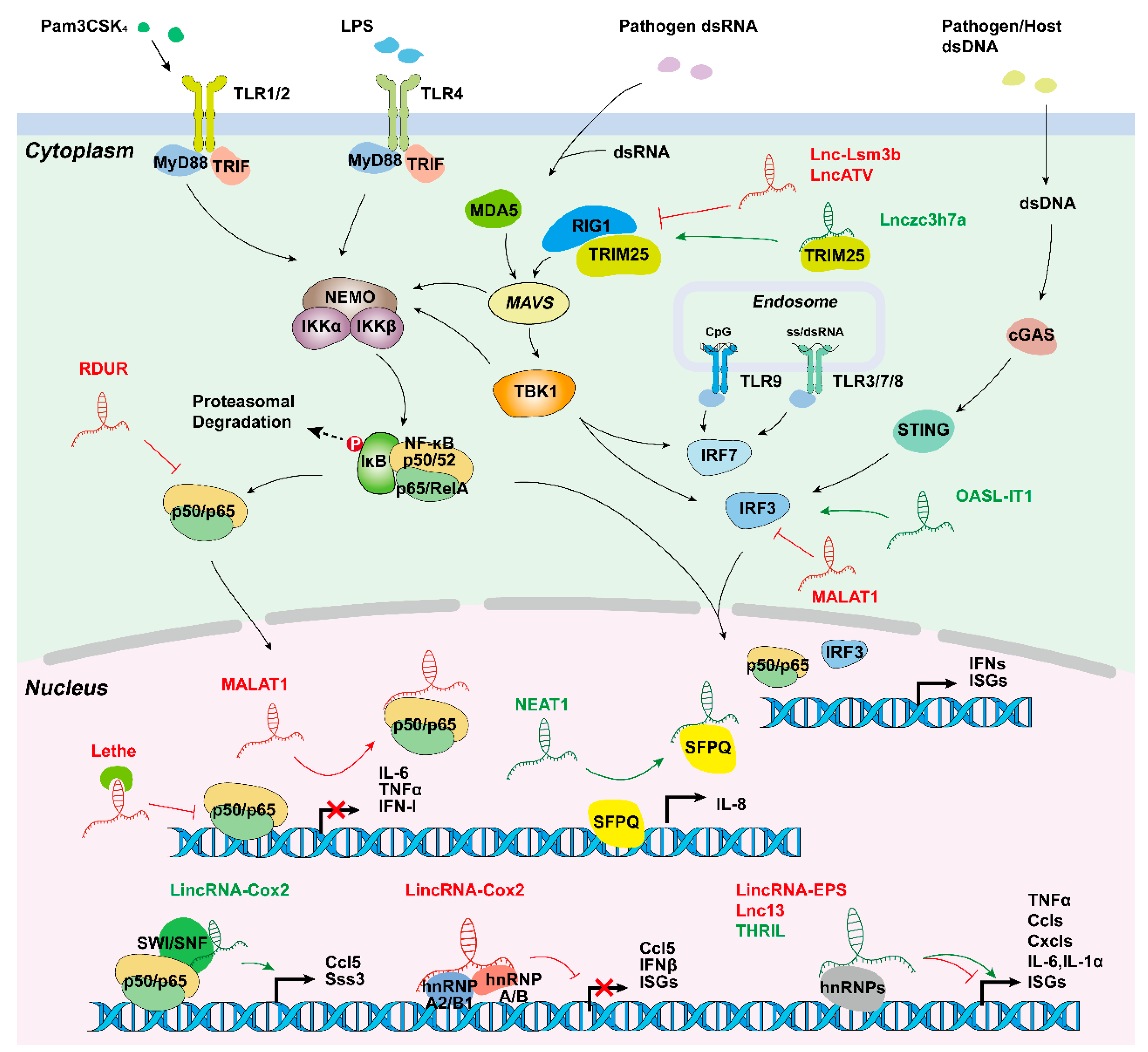
4.2.1. Inflammatory Signaling Triggered by PAMPs and DAMPs
4.2.2. Long Non-Coding RNAs Promote the Inflammatory Response
4.2.3. Long Non-Coding RNAs Inhibit the Inflammatory Response
4.3. Long Non-Coding RNAs Function in Antiviral Innate Immune Response
4.3.1. Antiviral Signaling
4.3.2. Long Non-Coding RNAs Promote Antiviral Innate Immune Response
4.3.3. Long Non-Coding RNAs Inhibit Antiviral Innate Immune Response
5. Innate Immune Long Non-Coding RNAs in Non-Infectious Diseases
5.1. Hematological Diseases
5.2. Rheumatoid Arthritis
5.3. Cardiovascular Diseases
5.4. Intestinal Diseases
5.5. Diabetes Mellitus
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, S. Long noncoding RNA: Novel links between gene expression and innate immunity. Virus Res. 2016, 212, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinf. 2016, 14, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Deb, A.; Parida, S.; Mondal, S.; Khatua, S.; Ghosh, Z. LncRBase V. 2: An updated resource for multispecies lncRNAs and ClinicLSNP hosting genetic variants in lncRNAs for cancer patients. RNA Biol. 2021, 18, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.P.; Leonardi, T.; Han, N.; Viré, E.; Gascoigne, D.K.; Arias-Carrasco, R.; Büscher, M.; Pandolfini, L.; Zhang, A.; Pluchino, S. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018, 19, 32. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Atianand, M.K.; Hu, W.; Satpathy, A.T.; Shen, Y.; Ricci, E.P.; Alvarez-Dominguez, J.R.; Bhatta, A.; Schattgen, S.A.; McGowan, J.D.; Blin, J. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 2016, 165, 1672–1685. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef]
- Natoli, G.; Andrau, J. Noncoding transcription at enhancers: General principles and functional models. Annu. Rev. Genet. 2012, 46, 1–19. [Google Scholar] [CrossRef]
- Werner, A.; Carlile, M.; Swan, D. What do natural antisense transcripts regulate? RNA Biol. 2009, 6, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A.; Valenstein, M.L.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA 2012, 109, 19202–19207. [Google Scholar] [CrossRef] [Green Version]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly (A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [Green Version]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [Green Version]
- Vinogradov, A.E. Dualism of gene GC content and CpG pattern in regard to expression in the human genome: Magnitude versus breadth. Trends Genet. 2005, 21, 639–643. [Google Scholar] [CrossRef]
- Cawley, S.; Bekiranov, S.; Ng, H.H.; Kapranov, P.; Sekinger, E.A.; Kampa, D.; Piccolboni, A.; Sementchenko, V.; Cheng, J.; Williams, A.J. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 2004, 116, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Uesaka, M.; Nishimura, O.; Go, Y.; Nakashima, K.; Agata, K.; Imamura, T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genom. 2014, 15, 35. [Google Scholar] [CrossRef] [Green Version]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deveson, I.W.; Brunck, M.E.; Blackburn, J.; Tseng, E.; Hon, T.; Clark, T.A.; Clark, M.B.; Crawford, J.; Dinger, M.E.; Nielsen, L.K. Universal alternative splicing of noncoding exons. Cell Syst. 2018, 6, 245–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuckerman, B.; Ulitsky, I. Predictive models of subcellular localization of long RNAs. RNA 2019, 25, 557–572. [Google Scholar] [CrossRef]
- Yoon, J.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Kornblihtt, A.R.; Schor, I.E.; Alló, M.; Dujardin, G.; Petrillo, E.; Muñoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Ares, M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.; Bartel, D.P. Long noncoding RNAs in C. elegans. Genome Res. 2012, 22, 2529–2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs; Springer: Singapore, 2017; pp. 1–46. [Google Scholar]
- Nitsche, A.; Rose, D.; Fasold, M.; Reiche, K.; Stadler, P.F. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 2015, 21, 801–812. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010, 151, 939–947. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, J. Human long noncoding RNAs are substantially less folded than messenger RNAs. Mol. Biol. Evol. 2015, 32, 970–977. [Google Scholar] [CrossRef] [Green Version]
- Kaewsapsak, P.; Shechner, D.M.; Mallard, W.; Rinn, J.L.; Ting, A.Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife 2017, 6, e29224. [Google Scholar] [CrossRef]
- Rinn, J.; Guttman, M. RNA and dynamic nuclear organization. Science 2014, 345, 1240–1241. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Sunwoo, H.; Colognori, D.; Froberg, J.E.; Jeon, Y.; Lee, J.T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. USA 2017, 114, 10654–10659. [Google Scholar] [CrossRef] [Green Version]
- Bresson, S.M.; Hunter, O.V.; Hunter, A.C.; Conrad, N.K. Canonical poly (A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet. 2015, 11, e1005610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levsky, J.M.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, A.; Van Den Bogaard, P.; Rifkin, S.A.; Van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [Green Version]
- Rhee, H.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef] [Green Version]
- Lam, S.S.; Martell, J.D.; Kamer, K.J.; Deerinck, T.J.; Ellisman, M.H.; Mootha, V.K.; Ting, A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–54. [Google Scholar] [CrossRef]
- Wu, J.; Zaccara, S.; Khuperkar, D.; Kim, H.; Tanenbaum, M.E.; Jaffrey, S.R. Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat. Methods 2019, 16, 862–865. [Google Scholar] [CrossRef]
- Fazal, F.M.; Han, S.; Parker, K.R.; Kaewsapsak, P.; Xu, J.; Boettiger, A.N.; Chang, H.Y.; Ting, A.Y. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 2019, 178, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Mowel, W.K.; Kotzin, J.J.; McCright, S.J.; Neal, V.D.; Henao-Mejia, J. Control of immune cell homeostasis and function by lncRNAs. Trends Immunol. 2018, 39, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601. [Google Scholar] [CrossRef] [Green Version]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Phillips, J.E.; Corces, V.G. CTCF: Master weaver of the genome. Cell 2009, 137, 1194–1211. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.; Yin, Q.; Chen, T.; Zhang, Y.; Zhang, X.; Wu, Z.; Zhang, S.; Wang, H.; Ge, J.; Lu, X. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparmann, A.; van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 2006, 6, 846–856. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.; Hung, T.; Argani, P.; Rinn, J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Krecic, A.M.; Swanson, M.S. hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 1999, 11, 363–371. [Google Scholar] [CrossRef]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Xu, P.; Meng, C.; Song, C.; Blackwell, T.S.; Li, R.; Li, H.; Zhang, J.; Lv, C. lncITPF promotes pulmonary fibrosis by targeting hnRNP-L depending on its host gene ITGBL. Mol. Ther. 2019, 27, 380–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Feng, Y.; Li, F.; Jia, Y.; Peng, Y.; Zhao, W.; Hu, J.; He, A. lncRNA ST3GAL6-AS1 promotes invasion by inhibiting hnRNPA2B1-mediated ST3GAL6 expression in multiple myeloma. Int. J. Oncol. 2021, 58, 5. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Liu, M.; Duan, R.; Tao, Y.; Zhou, J.; Fang, W.; Zhu, J.; Niu, L.; Sun, J. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol. Sin. 2020, 41, 22–33. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Yan, X.; Cheng, X.; He, X.; Zheng, W. LncRNA MALAT1 regulates sepsis-induced cardiac inflammation and dysfunction via interaction with miR-125b and p38 MAPK/NFκB. Int. Immunopharmacol. 2018, 55, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Zhang, G.; Cheng, Z.; Wang, X.; Jia, L.; Jing, X.; Wang, H.; Zhang, R.; Liu, M.; Jiang, T. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up-regulating miR-146a in LPS-induced acute lung injury. Connect. Tissue Res. 2018, 59, 581–592. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, L.; Wang, D.; Yu, J.; Zhu, Y.; Xu, Q.; Zheng, X.; Zhan, R. Long noncoding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via the modulation of a miR-199b/IKKβ/NF-κB signaling pathway. Am. J. Physiol.-Cell Physol. 2018, 315, C52–C61. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; He, L.; Zhang, Y. LincRNA-Cox2 promotes pulmonary arterial hypertension by regulating the let-7a-mediated STAT3 signaling pathway. Mol. Cell. Biochem. 2020, 475, 239–247. [Google Scholar] [CrossRef]
- Hu, J.; Yang, Z.; Wu, H.; Wang, D. Rhein attenuates renal inflammatory injury of uric acid nephropathy via lincRNA-Cox2/miR-150-5p/STAT1 axis. Int. Immunopharmacol. 2020, 85, 106620. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Pu, J.; Hao, B.; Huang, L.; Chen, J.; Hong, W.; Zhou, Y.; Li, B.; Ran, P. LncRNA RP11-86H7. 1 promotes airway inflammation induced by TRAPM2. 5 by acting as a ceRNA of miRNA-9-5p to regulate NFKB1 in HBECS. Sci. Rep. 2020, 10, 11587. [Google Scholar] [CrossRef]
- Shen, Q.; Zheng, J.; Wang, X.; Hu, W.; Jiang, Y.; Jiang, Y. LncRNA SNHG5 regulates cell apoptosis and inflammation by miR-132/PTEN axis in COPD. Biomed. Pharmacother. 2020, 126, 110016. [Google Scholar] [CrossRef] [PubMed]
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference—A crash course. Trends Genet. 2005, 21, 339–345. [Google Scholar] [CrossRef] [Green Version]
- Ebisuya, M.; Yamamoto, T.; Nakajima, M.; Nishida, E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008, 10, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ruan, X.; Auerbach, R.K.; Sandhu, K.S.; Zheng, M.; Wang, P.; Poh, H.M.; Goh, Y.; Lim, J.; Zhang, J. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012, 148, 84–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paralkar, V.R.; Taborda, C.C.; Huang, P.; Yao, Y.; Kossenkov, A.V.; Prasad, R.; Luan, J.; Davies, J.O.; Hughes, J.R.; Hardison, R.C. Unlinking an lncRNA from its associated cis element. Mol. Cell 2016, 62, 104–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, L.; Wang, Y.; Luo, W.; Li, F.; Xiao, J.; Qin, S.; Wang, Z.; Song, X.; Wang, Y. Single-cell RNA-sequencing analysis identifies host long noncoding RNA MAMDC2-AS1 as a co-factor for HSV-1 nuclear transport. Int. J. Biol. Sci. 2020, 16, 1586–1603. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef] [Green Version]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long non-coding RNAs and microRNAs interplay in osteogenic differentiation of mesenchymal stem cells. Front. Cell Dev. Biol. 2021, 9, 742. [Google Scholar] [CrossRef]
- Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 2010, 238, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Beutler, B. Innate immunity: An overview. Mol. Immunol. 2004, 40, 845–859. [Google Scholar] [CrossRef]
- Locati, M.; Mantovani, A.; Sica, A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv. Immunol. 2013, 120, 163–184. [Google Scholar]
- Chen, M.; Lin, H.; Shen, C.; Ma, Y.; Wang, F.; Zhao, H.; Yu, J.; Zhang, J. PU. 1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p. Mol. Cell. Biol. 2015, 35, 3212–3224. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Li, J.; Yen, J.; Lai, I.; Ho, Y.; Chen, Y.; Lan, J.; Chang, J. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int. J. Mol. Sci. 2018, 19, 2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Beirão, B.C.; Raposo, T.; Pang, L.Y.; Argyle, D.J. Canine mammary cancer cells direct macrophages toward an intermediate activation state between M1/M. BMC Vet. Res. 2015, 11, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Su, Z.; Mookhtiar, A.K. Long non-coding RNA: A versatile regulator of the nuclear factor-κB signalling circuit. Immunology 2017, 150, 379–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, A.; Shen, J.; Zuo, Z. Expression and regulation of long noncoding RNAs in TLR4 signaling in mouse macrophages. BMC Genom. 2015, 16, 45. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Han, X.; Huang, S.; Xue, P.; Fu, J.; Liu, L.; Zhang, C.; Yang, L.; Xia, L.; Sun, L.; Huang, S. LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci. Adv. 2019, 5, x9230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Xu, Y.; Lai, Y.U.; He, W.; Li, Y.; Wang, R.; Luo, X.; Chen, R.; Chen, T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J. Cell. Biochem. 2018, 119, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Ding, B.; Zhang, L.; Zhang, J.; Wang, J.; Zhang, W. lncRNA GAS5 promotes M1 macrophage polarization via miR-455-5p/SOCS3 pathway in childhood pneumonia. J. Cell. Physiol. 2019, 234, 13242–13251. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, Y.; Cai, J. LncRNA MIR155HG regulates M1/M2 macrophage polarization in chronic obstructive pulmonary disease. Biomed. Pharmacother. 2019, 117, 109015. [Google Scholar] [CrossRef]
- Du, M.; Yuan, L.; Tan, X.; Huang, D.; Wang, X.; Zheng, Z.; Mao, X.; Li, X.; Yang, L.; Huang, K. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 2017, 8, 2049. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Dong, R.; Jiang, L.; Gong, Y.; Yuan, M.; You, J.; Meng, W.; Chen, Z.; Zhang, N.; Weng, Q. LncRNA-MM2P identified as a modulator of macrophage M2 polarization. Cancer Immunol. Res. 2019, 7, 292–305. [Google Scholar] [CrossRef]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Zheng, Y.; Jin, X.; Liu, M.; Li, S.; Zhao, Q.; Liu, X.; Wang, Y.; Shi, M. The long noncoding RNA MALAT1 induces tolerogenic dendritic cells and regulatory T cells via miR155/dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin/IL10 axis. Front. Immunol. 2018, 9, 1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113, 2526–2534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Ni, F.; Fu, B.; Wu, Y.; Sun, R.; Tian, Z.; Wei, H. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget 2016, 7, 72546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, P.; Xiang, L.; Chen, W.; Li, S.; Huang, S.; Li, J.; Zhuge, L.; Jin, L.; Feng, W.; Chen, Y. LncRNA GAS5 enhanced the killing effect of NK cell on liver cancer through regulating miR-544/RUNX. Innate Immun. 2019, 25, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhu, A.; Ren, K.; Li, S.; Chen, L. IFNβ-induced exosomal linc-EPHA6-1 promotes cytotoxicity of NK cells by acting as a ceRNA for hsa-miR-4485-5p to up-regulate NKp46 expression. Life Sci. 2020, 257, 118064. [Google Scholar] [CrossRef]
- Steinman, R.M.; Hemmi, H. Dendritic Cells: Translating Innate to Adaptive Immunity; Springer: Berlin/Heidelberg, Germany, 2006; pp. 17–58. [Google Scholar]
- Schmidt, S.V.; Nino-Castro, A.C.; Schultze, J.L. Regulatory dendritic cells: There is more than just immune activation. Front. Immunol. 2012, 3, 274. [Google Scholar] [CrossRef] [Green Version]
- Akam, M. Hox genes and the evolution of diverse body plans. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1995, 349, 313–319. [Google Scholar]
- Bei, L.; Lu, Y.; Bellis, S.L.; Zhou, W.; Horvath, E.; Eklund, E.A. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding β3 integrin during myeloid differentiation. J. Biol. Chem. 2007, 282, 16846–16859. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.L.; Licht, J.D. HOX deregulation in acute myeloid leukemia. J. Clin. Investig. 2007, 117, 865–868. [Google Scholar] [CrossRef] [Green Version]
- Caligiuri, M.A. Human natural killer cells. Blood 2008, 112, 461–469. [Google Scholar] [CrossRef]
- Van Acker, H.H.; Capsomidis, A.; Smits, E.L.; Van Tendeloo, V.F. CD56 in the immune system: More than a marker for cytotoxicity? Front. Immunol. 2017, 8, 892. [Google Scholar] [CrossRef]
- Bianchi, M.E. DAMPs, PAMPs and alarmins: All we need to know about danger. J. Leukoc. Biol. 2007, 81, 1–5. [Google Scholar] [CrossRef]
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA sensing by the cGAS–STING pathway in health and disease. Nat. Rev. Genet. 2019, 20, 657–674. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.X.; Kirschning, C.J.; Mancinelli, R.; Xu, X.; Jin, Y.; Faure, E.; Mantovani, A.; Rothe, M.; Muzio, M.; Arditi, M. Bacterial lipopolysaccharide activates nuclear factor-κB through interleukin-1 signaling mediators in cultured human dermal endothelial cells and mononuclear phagocytes. J. Biol. Chem. 1999, 274, 7611–7614. [Google Scholar] [CrossRef] [Green Version]
- Nathan, C. Points of control in inflammation. Nature 2002, 420, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhong, Z.; Darnell, J.E., Jr. Maximal activation of transcription by Statl and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Yu, G.; Temkin, V.; Ogata, H.; Kuntzen, C.; Sakurai, T.; Sieghart, W.; Peck-Radosavljevic, M.; Leffert, H.L.; Karin, M. Hepatocyte IKKβ/NF-κB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell 2010, 17, 286–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, R.; Rajabi, H.; Kosugi, M.; Joshi, M.D.; Alam, M.; Vasir, B.; Kawano, T.; Kharbanda, S.; Kufe, D. MUC1-C oncoprotein promotes STAT3 activation in an autoinductive regulatory loop. Sci. Signal. 2011, 4, a9. [Google Scholar] [CrossRef] [Green Version]
- Freire, M.O.; Van Dyke, T.E. Natural resolution of inflammation. Periodontology 2013, 63, 149–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.; Wang, Y.; Ma, S.; Chen, X.; Chen, J.; Su, C.; Shibata, A.; Strauss-Soukup, J.K.; Drescher, K.M. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J. Immunol. 2016, 196, 2799–2808. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Chao, T.; Chang, K.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.J.; Zeng, X.Y.; Jiang, S.L.; Tan, H.Y.; Yan, M.Y.; Yang, H.Z. Long non-coding RNA MALAT1 sponges miR-149 to promote inflammatory responses of LPS-induced acute lung injury by targeting MyD88. Cell Biol. Int. 2020, 44, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Jiang, Y.; Xu, X.; Su, X.; Liu, Y.; Ma, Y.; Zhao, Y.; Shen, Z.; Huang, B.; Cao, X. Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019, 20, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Maarouf, M.; Chen, B.; Chen, Y.; Wang, X.; Rai, K.R.; Zhao, Z.; Liu, S.; Li, Y.; Xiao, M.; Chen, J.L. Identification of lncRNA-155 encoded by MIR155HG as a novel regulator of innate immunity against influenza A virus infection. Cell. Microbiol. 2019, 21, e13036. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, J.; Liu, S.; Chen, B.; Xiao, M.; Li, Y.; Liao, Y.; Rai, K.R.; Zhao, Z.; Ouyang, J. RDUR, a lncRNA, Promotes Innate Antiviral Responses and Provides Feedback Control of NF-κB Activation. Front. Immunol. 2021, 12, 1849. [Google Scholar]
- Zhang, P.; Cao, L.; Zhou, R.; Yang, X.; Wu, M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 2019, 10, 1495. [Google Scholar] [CrossRef] [Green Version]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Jiang, M.; Liu, L.; Yang, Z.; Ma, Z.; Liu, S.; Ma, Y.; Zhang, L.; Cao, X. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 2019, 20, 812–823. [Google Scholar] [CrossRef]
- Wang, Y.; Huo, Z.; Lin, Q.; Lin, Y.; Chen, C.; Huang, Y.; Huang, C.; Zhang, J.; He, J.; Liu, C. Positive Feedback Loop of Long Noncoding RNA OASL-IT1 and Innate Immune Response Restricts the Replication of Zika Virus in Epithelial A549 Cells. J. Innate Immun. 2021, 13, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Fernandez-Jimenez, N.; Kratchmarov, R.; Luo, X.; Bhagat, G.; Green, P.H.; Schneider, R.; Kiledjian, M.; Bilbao, J.R.; Ghosh, S. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016, 352, 91–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2013, 2, e762. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Su, Z.; Song, D.; Mao, Y.; Mao, X. The long noncoding RNA MALAT 1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016, 590, 2884–2895. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Cheng, M.; Chi, X.; Liu, X.; Yang, W. A Human Long Non-coding RNA LncATV Promotes Virus Replication Through Restricting RIG-I–Mediated Innate Immunity. Front. Immunol. 2019, 10, 1711. [Google Scholar] [CrossRef] [Green Version]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 2018, 173, 906–919. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Wei, Y. Modulators of MicroRNA function in the immune system. Int. J. Mol. Sci. 2020, 21, 2357. [Google Scholar] [CrossRef]
- Zgheib, C.; Hodges, M.M.; Hu, J.; Liechty, K.W.; Xu, J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS ONE 2017, 12, e177453. [Google Scholar]
- Rehwinkel, J.; E Sousa, C.R. RIGorous detection: Exposing virus through RNA sensing. Science 2010, 327, 284–286. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohto, U.; Ishida, H.; Shibata, T.; Sato, R.; Miyake, K.; Shimizu, T. Toll-like receptor 9 contains two DNA binding sites that function cooperatively to promote receptor dimerization and activation. Immunity 2018, 48, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Chow, K.T.; Gale Jr, M.; Loo, Y. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018, 36, 667–694. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Röhl, I.; Hopfner, K.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef] [Green Version]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, L.; Yang, Z.; Liu, S.; Ma, Z.; Liu, Y.; Ma, Y.; Zhang, L.; Zhang, X. LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Proc. Natl. Acad. Sci. USA 2020, 117, 23695–23706. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, L.; Fabbri, E. Inflammageing: Chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018, 15, 505–522. [Google Scholar] [CrossRef]
- Kumar, V.; Westra, H.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Võsa, U. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013, 9, e1003201. [Google Scholar] [CrossRef] [Green Version]
- Ricaño-Ponce, I.; Wijmenga, C. Mapping of immune-mediated disease genes. Annu. Rev. Genom. Hum. Genet. 2013, 14, 325–353. [Google Scholar] [CrossRef] [PubMed]
- Manz, M.G.; Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 2014, 14, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Jung, S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 392–404. [Google Scholar] [CrossRef]
- Kotzin, J.J.; Spencer, S.P.; McCright, S.J.; Kumar, D.B.U.; Collet, M.A.; Mowel, W.K.; Elliott, E.N.; Uyar, A.; Makiya, M.A.; Dunagin, M.C. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 2016, 537, 239–243. [Google Scholar] [CrossRef]
- Zeng, C.; Xu, Y.; Xu, L.; Yu, X.; Cheng, J.; Yang, L.; Chen, S.; Li, Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014, 14, 693. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Chen, W.; Lin, J. LncRNA: An all-rounder in rheumatoid arthritis. J. Transl. Intern. Med. 2019, 7, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef]
- Liu, C.; Guo, X.; Bai, S.; Zeng, G.; Wang, H. lncRNA CASC2 downregulation participates in rheumatoid arthritis, and CASC2 overexpression promotes the apoptosis of fibroblast-like synoviocytes by downregulating IL-17. Mol. Med. Rep. 2020, 21, 2131–2137. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.; Bot, I.; Kovanen, P.T. Mast cells in human and experimental cardiometabolic diseases. Nat. Rev. Cardiol. 2015, 12, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.J.; Xu, N.; Yutzey, K.E. Macrophage lineages in heart valve development and disease. Cardiovasc. Res. 2021, 117, 663–673. [Google Scholar] [CrossRef]
- Sun, C.; Fu, Y.; Gu, X.; Xi, X.; Peng, X.; Wang, C.; Sun, Q.; Wang, X.; Qian, F.; Qin, Z. Macrophage-enriched lncRNA RAPIA: A novel therapeutic target for atherosclerosis. Arterioscl. Throm. Vas. 2020, 40, 1464–1478. [Google Scholar] [CrossRef]
- Simion, V.; Zhou, H.; Haemmig, S.; Pierce, J.B.; Mendes, S.; Tesmenitsky, Y.; Pérez-Cremades, D.; Lee, J.F.; Chen, A.F.; Ronda, N. A macrophage-specific lncRNA regulates apoptosis and atherosclerosis by tethering HuR in the nucleus. Nat. Commun. 2020, 11, 1–16. [Google Scholar]
- Ye, Z.; Yang, S.; Xia, Y.; Hu, R.; Chen, S.; Li, B.; Chen, S.; Luo, X.; Mao, L.; Li, Y.; et al. LncRNA MIAT sponges miR-149-5p to inhibit efferocytosis in advanced atherosclerosis through CD47 upregulation. Cell Death Dis. 2019, 10, 138. [Google Scholar] [CrossRef] [Green Version]
- Hung, J.; Scanlon, J.P.; Mahmoud, A.D.; Rodor, J.; Ballantyne, M.; Fontaine, M.A.C.; Temmerman, L.; Kaczynski, J.; Connor, K.L.; Bhushan, R.; et al. Novel Plaque Enriched Long Noncoding RNA in Atherosclerotic Macrophage Regulation (PELATON). Arterioscl. Throm. Vas. 2020, 40, 697–713. [Google Scholar] [CrossRef]
- Yang, S.; Sun, J. LncRNA SRA deregulation contributes to the development of atherosclerosis by causing dysfunction of endothelial cells through repressing the expression of adipose triglyceride lipase. Mol. Med. Rep. 2018, 18, 5207–5214. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Wu, S.; Chen, F.; Wu, Q.; Wu, C.; Kang, C.; He, X.; Zhang, R.; Lu, Z.; Li, X. Atorvastatin inhibits pyroptosis through the lncRNA NEXN-AS1/NEXN pathway in human vascular endothelial cells. Atherosclerosis 2020, 293, 26–34. [Google Scholar] [CrossRef]
- Khyzha, N.; Khor, M.; DiStefano, P.V.; Wang, L.; Matic, L.; Hedin, U.; Wilson, M.D.; Maegdefessel, L.; Fish, J.E. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 16410–16419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Chao, T.C.; Patil, V.S.; Qin, Y.; Tiwari, S.K.; Chiou, J.; Dobin, A.; Tsai, C.M.; Li, Z.; Dang, J. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019, 38, e100041. [Google Scholar] [CrossRef]
- Quan, Y.; Song, K.; Zhang, Y.; Zhu, C.; Shen, Z.; Wu, S.; Luo, W.; Tan, B.; Yang, Z.; Wang, X. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem. Biophys. Res. Commun. 2018, 501, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Yang, L.; Wan, J.; Liu, X.; Pang, C.; You, W.; Zhao, G. Long noncoding RNA ANRIL contributes to the development of ulcerative colitis by miR-323b-5p/TLR4/MyD88/NF-κB pathway. Biochem. Biophys. Res. Commun. 2019, 508, 217–224. [Google Scholar] [CrossRef]
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193–1202. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Jimenez, N.; Castellanos-Rubio, A.; Plaza-Izurieta, L.; Irastorza, I.; Elcoroaristizabal, X.; Jauregi-Miguel, A.; Lopez-Euba, T.; Tutau, C.; de Pancorbo, M.M.; Vitoria, J.C. Coregulation and modulation of NFκB-related genes in celiac disease: Uncovered aspects of gut mucosal inflammation. Hum. Mol. Genet. 2014, 23, 1298–1310. [Google Scholar] [CrossRef] [PubMed]
- Gnodi, E.; Mancuso, C.; Elli, L.; Ballarini, E.; Meneveri, R.; Beaulieu, J.F.; Barisani, D. Gliadin, through the Activation of Innate Immunity, Triggers lncRNA NEAT1 Expression in Celiac Disease Duodenal Mucosa. Int. J. Mol. Sci. 2021, 22, 1289. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Wen, F.; Jiang, M.; Liu, Q.; Nie, Y. LncRNA uc. 48+ is involved in the diabetic immune and inflammatory responses mediated by P2X7 receptor in RAW264. 7 macrophages. Int. J. Mol. Med. 2018, 42, 1152–1160. [Google Scholar]
- Reddy, M.A.; Chen, Z.; Park, J.T.; Wang, M.; Lanting, L.; Zhang, Q.; Bhatt, K.; Leung, A.; Wu, X.; Putta, S. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 2014, 63, 4249–4261. [Google Scholar] [CrossRef] [Green Version]
- Sathishkumar, C.; Prabu, P.; Mohan, V.; Balasubramanyam, M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genom. 2018, 12, 41. [Google Scholar] [CrossRef]
| Model | LncRNA | Functional Consequences | Mechanism | Reference |
|---|---|---|---|---|
| Positive pattern | LincRNA-Cox2 | Transactivates inflammatory genes | Incorporates NF-κB into the SWI/SNF complex | [125] |
| THRIL | Promotes TLR2-mediated cytokines and chemokines expression | Forms an RNA-protein complex with hnRNP | [126] | |
| MALAT1 | Promotes IL-1β, IL-6 and TNF-α expression | Sponges miR-149 | [127] | |
| LncRNA Sros1 | Promotes IFN-γ-STAT1-mediated innate immunity | Frees STAT1 mRNA from the RBP CAPRIN1 | [128] | |
| LncRNA-155 | Promotes IFN-β and ISGs production | Inhibits PTP1B production | [129] | |
| RDUR | Upregulates IFNs and ISGs expression, alleviates inflammation | Inactivates NF-κB | [130] | |
| NEAT1 | Promotes inflammasomes assembly, initiates antiviral gene IL-8 transcription | Transfers the SFPQ from IL-8 promoter, maintains caspase-1 maturation | [131,132] | |
| Lnczc3h7a | Activates TRIM25-mediated RIG-I antiviral response | Forms trimeric complex with RIG-I and TRIM25 | [133] | |
| OASL-IT1 | Triggers IFN-β and ISGs expression, inhibits ZIKV infection | Activates p38 MAPK, IRF3, and NF-κB | [134] | |
| Negative pattern | LincRNA-Cox2 | Represses inflammatory response | Interaction with hnRNP-A/B and hnRNP-A2/B1 | [11] |
| LincRNA-EPS | Represses inflammatory genes expression | Interaction with chromatin, hnRNPD or histone | [10] | |
| Lnc13 | Decreases inflammatory regulators expression | Binds to hnRNPD p42 and Hdac1 on chromatin | [135] | |
| Lethe | Prevents proinflammatory cytokines production | Prevents RelA-mediated transcription | [136] | |
| MALAT1 | Prevents proinflammatory cytokines and IFN-I production | Binding to NF-κB, prevents IRF3 degradation | [137] | |
| Mirt2 | Inhibits cytokine (e.g., IL-6, CXCL9) production | Inactivates MAPK/NF-κB pathways | [99] | |
| LncATV | Inhibits IFNs and ISGs production | Induces a mono-allelic mutation in the CARD of RIG-I | [138] | |
| Lnc-Lsm3b | Terminates type I IFNs production | Limits RIG-I ubiquitination and phosphorylation | [139] | |
| NRAV | Inhibits transcription of ISGs | Regulation on histone modification | [140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Wei, Y. A Novel Regulatory Player in the Innate Immune System: Long Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 9535. https://doi.org/10.3390/ijms22179535
Xie Y, Wei Y. A Novel Regulatory Player in the Innate Immune System: Long Non-Coding RNAs. International Journal of Molecular Sciences. 2021; 22(17):9535. https://doi.org/10.3390/ijms22179535
Chicago/Turabian StyleXie, Yuhuai, and Yuanyuan Wei. 2021. "A Novel Regulatory Player in the Innate Immune System: Long Non-Coding RNAs" International Journal of Molecular Sciences 22, no. 17: 9535. https://doi.org/10.3390/ijms22179535
APA StyleXie, Y., & Wei, Y. (2021). A Novel Regulatory Player in the Innate Immune System: Long Non-Coding RNAs. International Journal of Molecular Sciences, 22(17), 9535. https://doi.org/10.3390/ijms22179535






