Mouse Models of Achromatopsia in Addressing Temporal “Point of No Return” in Gene-Therapy
Abstract
1. Introduction
2. Results
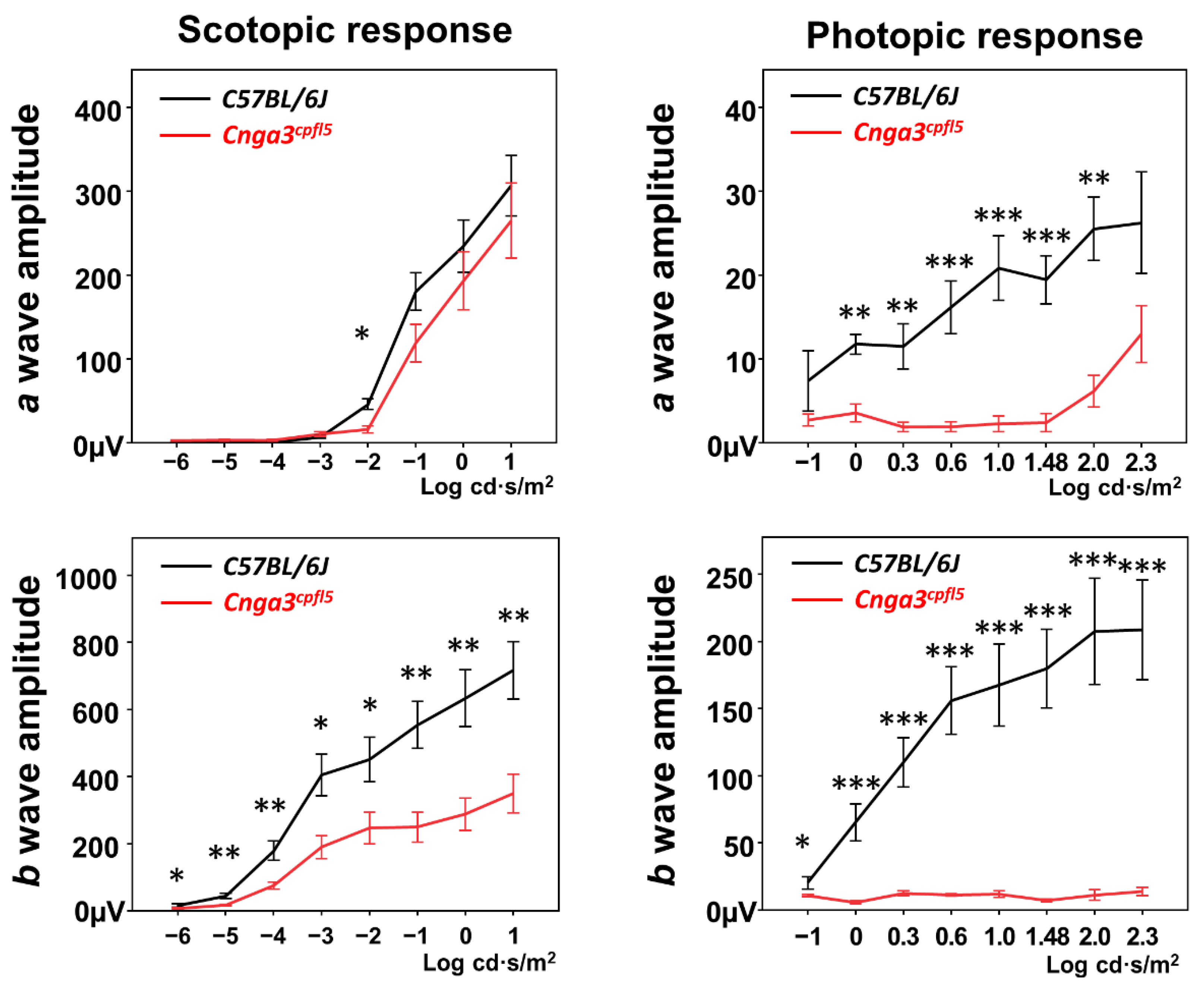
2.1. Functional Electroretinography of Cnga3cpfl5 Mice
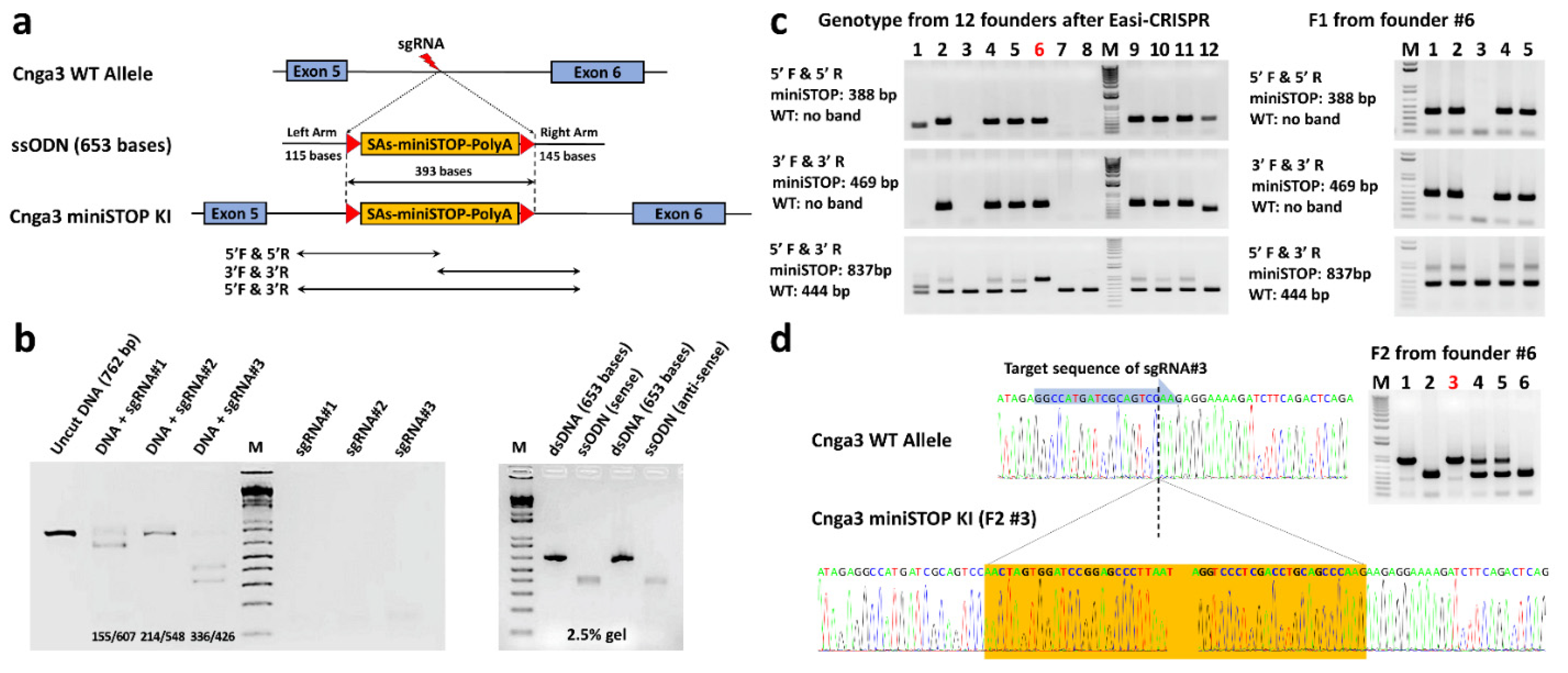
2.2. Generation of Cnga3floxed-miniSTOP Mice Using Easi-CRISPR
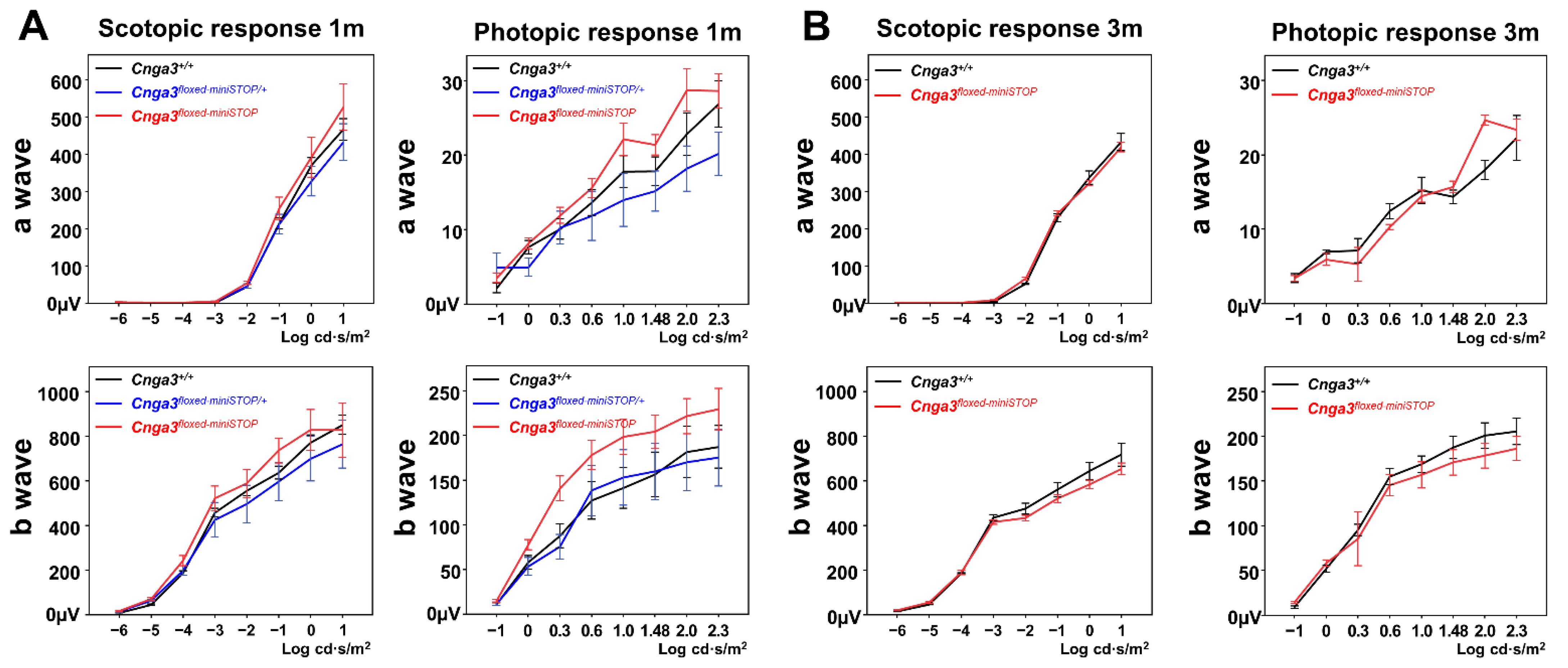
2.3. Functional Electroretinography and Histology of Cnga3floxed-miniSTOP Mice
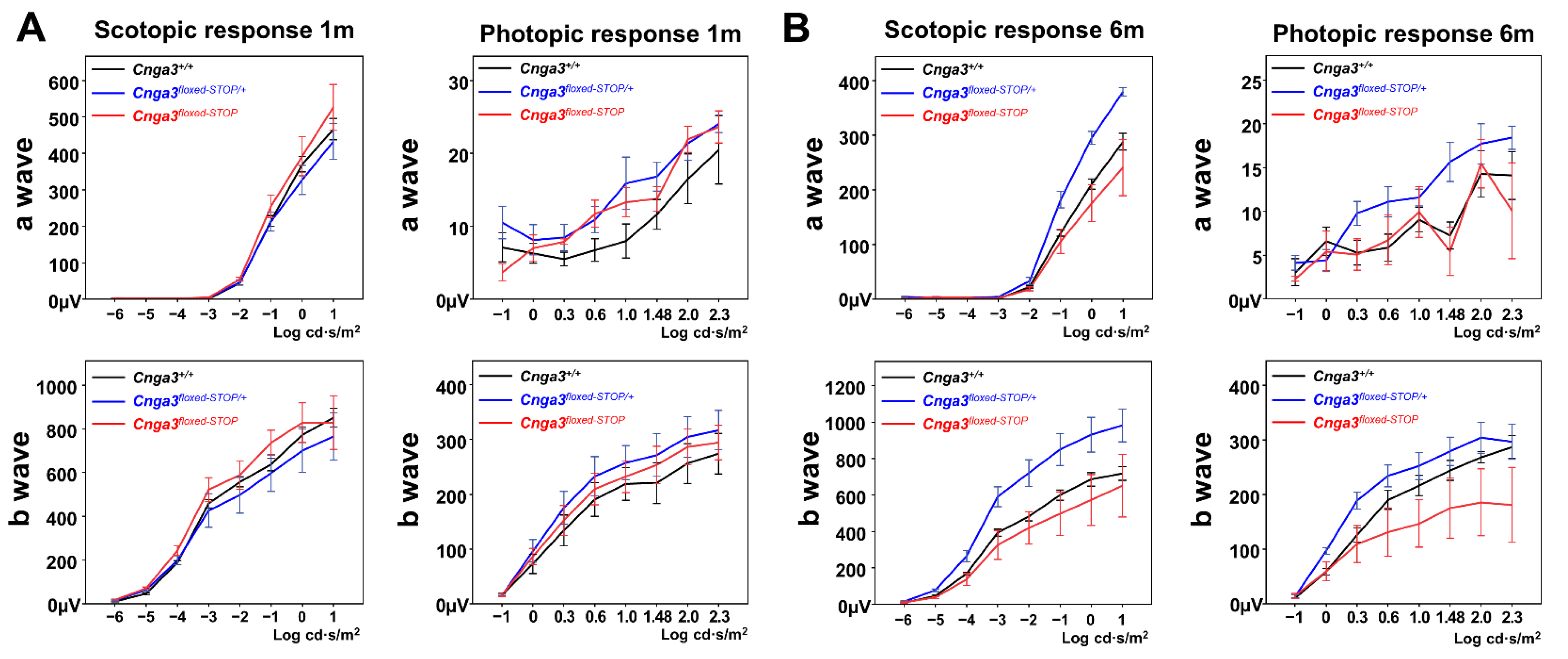
2.4. Functional Electroretinography of Cnga3floxed-STOP Mice
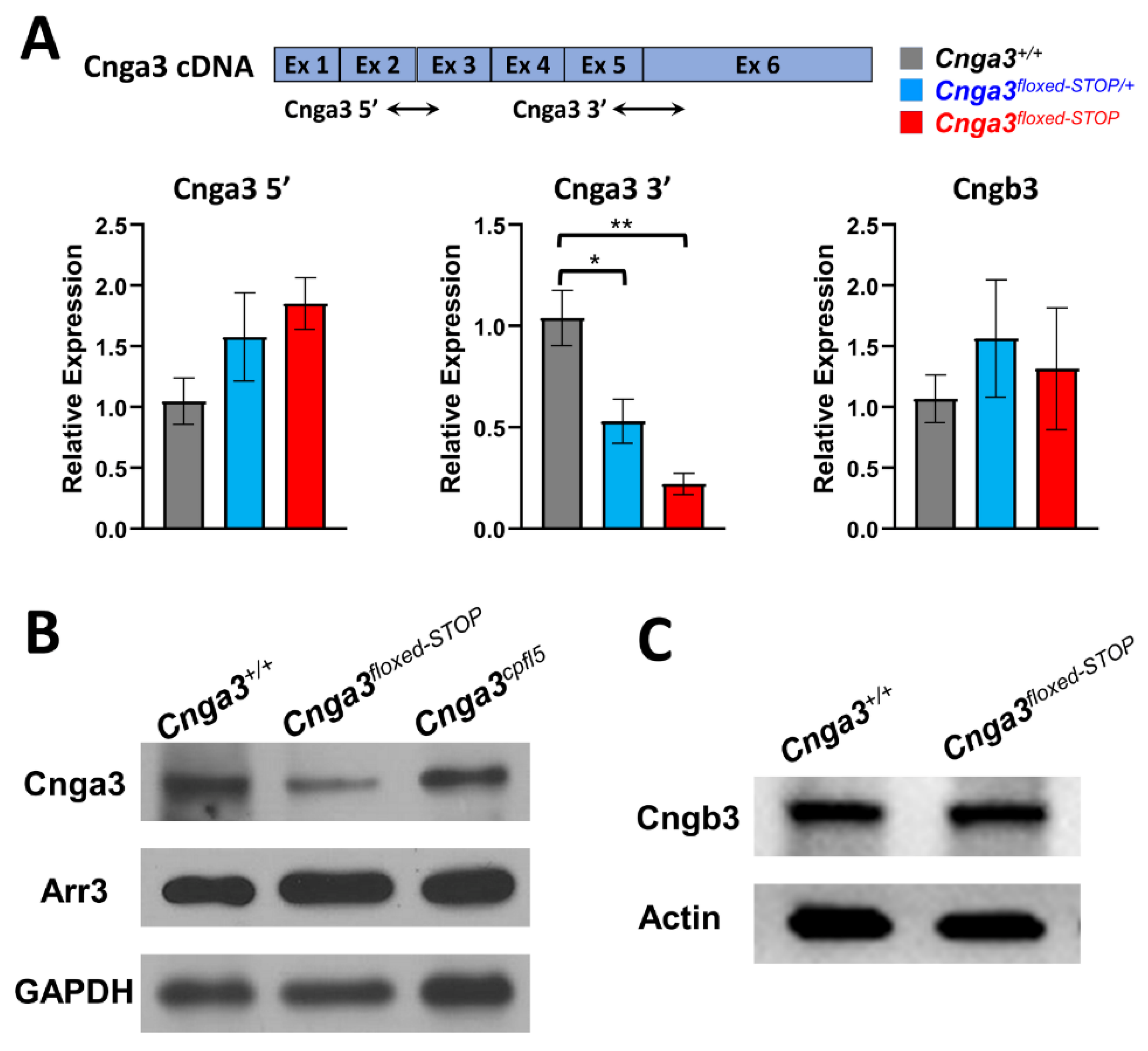
2.5. Quantitative Real-Time RT-PCR of Cnga3floxed-STOP Mice
2.6. Immunoblotting of CNGA3 in Cnga3floxed-STOP Mice
2.7. Functional Electroretinography of Compound Heterozygous Cnga3cpfl5/floxed-STOP Mice
3. Discussion
3.1. Easi-CRISPR
3.2. ERG Setting for Testing Mouse Models
3.3. Splicing Acceptors and Score Prediction
3.4. Ion-Conducting Subunit of the Channel
3.5. ERG Cone Response Corresponds to CNG Channel Function?
4. Materials and Methods
4.1. Mouse Care and Housing
4.2. Generation of Cnga3floxed-miniSTOP Mice Using Easi-CRISPR
4.2.1. sgRNAs
4.2.2. In Vitro Digestion (IVD) Protocols
4.2.3. Single-Stranded Oligo Donor (ssODN) Generation:
4.3. Generation of Cnga3floxed-STOP Mice Using ES Cell Gene Targeting
4.4. Genotype
4.5. Electroretinography
4.6. Histology
4.7. Immunoblotting
4.8. Quantitative Real-Time RT-PCR (qRT-PCR)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Easi-CRISPR | Efficient additions with ssDNA inserts-CRISPR |
| ES | embryonic stem |
| CNG | cyclic nucleotide-gated |
| AAV | adeno-associated virus |
| ERG | electroretinogram |
| Cas9-RNPs | Cas9 protein/sgRNA ribonucleoprotein complexes |
| ssODN | single-stranded oligodeoxynucleotides |
| IVD | In vitro digestion |
| PCR | polymerase chain reaction |
| WT | wild-type |
| ISCEV | International society for clinical electrophysiology of vision |
References
- Tsang, S.H.; Sharma, T. Rod Monochromatism (Achromatopsia). Adv. Exp. Med. Biol. 2018, 1085, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Hunt, D.M.; Moore, A.T. The cone dysfunction syndromes. Br. J. Ophthalmol. 2004, 88, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Hamel, C. Clinical utility gene card for: Achromatopsia-update 2013. Eur. J. Hum. Genet. 2013, 21, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Coppieters, F.; Meire, F.; Schaich, S.; Roosing, S.; Brennenstuhl, C.; Bolz, S.; van Genderen, M.M.; Riemslag, F.C.; European Retinal Disease, C.; et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am. J. Hum. Genet. 2012, 91, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Thiadens, A.A.; den Hollander, A.I.; Roosing, S.; Nabuurs, S.B.; Zekveld-Vroon, R.C.; Collin, R.W.; De Baere, E.; Koenekoop, R.K.; van Schooneveld, M.J.; Strom, T.M.; et al. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am. J. Hum. Genet. 2009, 85, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.; Michaelides, M.; Aligianis, I.A.; Ainsworth, J.R.; Mollon, J.D.; Maher, E.R.; Moore, A.T.; Hunt, D.M. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J. Med. Genet. 2004, 41, e20. [Google Scholar] [CrossRef]
- Kohl, S.; Baumann, B.; Rosenberg, T.; Kellner, U.; Lorenz, B.; Vadala, M.; Jacobson, S.G.; Wissinger, B. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am. J. Hum. Genet. 2002, 71, 422–425. [Google Scholar] [CrossRef]
- Kohl, S.; Zobor, D.; Chiang, W.C.; Weisschuh, N.; Staller, J.; Gonzalez Menendez, I.; Chang, S.; Beck, S.C.; Garcia Garrido, M.; Sothilingam, V.; et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat. Genet. 2015, 47, 757–765. [Google Scholar] [CrossRef]
- Aboshiha, J.; Dubis, A.M.; Carroll, J.; Hardcastle, A.J.; Michaelides, M. The cone dysfunction syndromes. Br. J. Ophthalmol. 2016, 100, 115–121. [Google Scholar] [CrossRef]
- Poloschek, C.M.; Kohl, S. [Achromatopsia]. Ophthalmologe 2010, 107, 571–582. [Google Scholar] [CrossRef]
- Kohl, S.; Varsanyi, B.; Antunes, G.A.; Baumann, B.; Hoyng, C.B.; Jagle, H.; Rosenberg, T.; Kellner, U.; Lorenz, B.; Salati, R.; et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur. J. Hum. Genet. 2005, 13, 302–308. [Google Scholar] [CrossRef]
- Hirano, A.A.; Hack, I.; Wassle, H.; Duvoisin, R.M. Cloning and immunocytochemical localization of a cyclic nucleotide-gated channel alpha-subunit to all cone photoreceptors in the mouse retina. J. Comp. Neurol. 2000, 421, 80–94. [Google Scholar] [CrossRef]
- Rabson, A.R.; Whiting, D.A.; Anderson, R.; Glover, A.; Koornhof, H.J. Depressed neutrophil motility in patients with recurrent herpes simplex virus infections: In vitro restoration with levamisole. J. Infect. Dis. 1977, 135, 113–116. [Google Scholar] [CrossRef]
- Hassall, M.M.; Barnard, A.R.; Orlans, H.O.; McClements, M.E.; Charbel Issa, P.; Aslam, S.A.; MacLaren, R.E. A Novel Achromatopsia Mouse Model Resulting From a Naturally Occurring Missense Change in Cngb3. Investig. Ophthalmol. Vis. Sci. 2018, 59, 6102–6110. [Google Scholar] [CrossRef]
- Kaupp, U.B.; Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002, 82, 769–824. [Google Scholar] [CrossRef]
- Michalakis, S.; Becirovic, E.; Biel, M. Retinal Cyclic Nucleotide-Gated Channels: From Pathophysiology to Therapy. Int. J. Mol. Sci. 2018, 19, 749. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.J.; Alexander, J.; Lei, B.; Deng, W.; Zhang, K.; Li, Q.; Chang, B.; Hauswirth, W.W. Achromatopsia as a potential candidate for gene therapy. Adv. Exp. Med. Biol. 2010, 664, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Michalakis, S.; Muhlfriedel, R.; Tanimoto, N.; Krishnamoorthy, V.; Koch, S.; Fischer, M.D.; Becirovic, E.; Bai, L.; Huber, G.; Beck, S.C.; et al. Restoration of cone vision in the CNGA3-/- mouse model of congenital complete lack of cone photoreceptor function. Mol. Ther. 2010, 18, 2057–2063. [Google Scholar] [CrossRef]
- Reichel, F.F.; Michalakis, S.; Wilhelm, B.; Zobor, D.; Muehlfriedel, R.; Kohl, S.; Weisschuh, N.; Sothilingam, V.; Kuehlewein, L.; Kahle, N.; et al. Three-year results of phase I retinal gene therapy trial for CNGA3-mutated achromatopsia: Results of a non randomised controlled trial. Br. J. Ophthalmol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.D.; Michalakis, S.; Wilhelm, B.; Zobor, D.; Muehlfriedel, R.; Kohl, S.; Weisschuh, N.; Ochakovski, G.A.; Klein, R.; Schoen, C.; et al. Safety and Vision Outcomes of Subretinal Gene Therapy Targeting Cone Photoreceptors in Achromatopsia: A Nonrandomized Controlled Trial. JAMA Ophthalmol. 2020, 138, 643–651. [Google Scholar] [CrossRef]
- Kahle, N.A.; Peters, T.; Zobor, D.; Kuehlewein, L.; Kohl, S.; Zhour, A.; Werner, A.; Seitz, I.P.; Sothilingam, V.; Michalakis, S.; et al. Development of Methodology and Study Protocol: Safety and Efficacy of a Single Subretinal Injection of rAAV.hCNGA3 in Patients with CNGA3-Linked Achromatopsia Investigated in an Exploratory Dose-Escalation Trial. Hum. Gene Ther. Clin. Dev. 2018, 29, 121–131. [Google Scholar] [CrossRef]
- Blair, K.; Cibis, G.; Gulani, A.C. Amblyopia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK430890/ (accessed on 26 June 2021).
- Holmes, J.M.; Lazar, E.L.; Melia, B.M.; Astle, W.F.; Dagi, L.R.; Donahue, S.P.; Frazier, M.G.; Hertle, R.W.; Repka, M.X.; Quinn, G.E.; et al. Effect of age on response to amblyopia treatment in children. Arch. Ophthalmol. 2011, 129, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Hawes, N.L.; Wang, X.; Hurd, R.E.; Wang, J.; Davisson, M.T.; Nusinowitz, S.; Heckenlively, J.R.; Chang, B. A Point Mutation in the Cnga3 Gene Causes Cone Photoreceptor Function Loss (cpfl5) in Mice. ARVO Annual Meeting Abstract. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4579. [Google Scholar]
- Quadros, R.M.; Miura, H.; Harms, D.W.; Akatsuka, H.; Sato, T.; Aida, T.; Redder, R.; Richardson, G.P.; Inagaki, Y.; Sakai, D.; et al. Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017, 18, 92. [Google Scholar] [CrossRef] [PubMed]
- Benchling [Biology Software]. 2018. Available online: https://benchling.com (accessed on 26 June 2021).
- Ding, X.Q.; Harry, C.S.; Umino, Y.; Matveev, A.V.; Fliesler, S.J.; Barlow, R.B. Impaired cone function and cone degeneration resulting from CNGB3 deficiency: Down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum. Mol. Genet. 2009, 18, 4770–4780. [Google Scholar] [CrossRef]
- Michalakis, S.; Geiger, H.; Haverkamp, S.; Hofmann, F.; Gerstner, A.; Biel, M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Morris, L.; Fliesler, S.J.; Sherry, D.M.; Ding, X.Q. Early-onset, slow progression of cone photoreceptor dysfunction and degeneration in CNG channel subunit CNGB3 deficiency. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3557–3566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- McCulloch, D.L.; Marmor, M.F.; Brigell, M.G.; Hamilton, R.; Holder, G.E.; Tzekov, R.; Bach, M. ISCEV Standard for full-field clinical electroretinography (2015 update). Doc. Ophthalmol. 2015, 130, 1–12. [Google Scholar] [CrossRef]
- Perlman, I. The Electroretinogram: ERG. In Webvision: The Organization of the Retina and Visual System; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2007; Available online: http://webvision.med.utah.edu/book/electrophysiology/the-electroretinogram-erg/. (accessed on 26 June 2021).
- Benchorin, G.; Calton, M.A.; Beaulieu, M.O.; Vollrath, D. Assessment of Murine Retinal Function by Electroretinography. Bio Protoc. 2017, 7, e2218. [Google Scholar] [CrossRef]
- Fu, Y. Phototransduction in Rods and Cones. In Webvision: The Organization of the Retina and Visual System; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 2007; Available online: https://webvision.med.utah.edu/book/part-v-phototransduction-in-rods-and-cones/phototransduction-in-rods-and-cones/ (accessed on 26 June 2021).
- Matulef, K.; Zagotta, W.N. Cyclic nucleotide-gated ion channels. Annu. Rev. Cell Dev. Biol. 2003, 19, 23–44. [Google Scholar] [CrossRef]
- Zhong, H.; Molday, L.L.; Molday, R.S.; Yau, K.W. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 2002, 420, 193–198. [Google Scholar] [CrossRef]
- Zheng, J.; Trudeau, M.C.; Zagotta, W.N. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 2002, 36, 891–896. [Google Scholar] [CrossRef]
- Weitz, D.; Ficek, N.; Kremmer, E.; Bauer, P.J.; Kaupp, U.B. Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron 2002, 36, 881–889. [Google Scholar] [CrossRef]
- Peng, C.; Rich, E.D.; Varnum, M.D. Subunit configuration of heteromeric cone cyclic nucleotide-gated channels. Neuron 2004, 42, 401–410. [Google Scholar] [CrossRef]
- Gerstner, A.; Zong, X.; Hofmann, F.; Biel, M. Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J. Neurosci. 2000, 20, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Faillace, M.P.; Bernabeu, R.O.; Korenbrot, J.I. Cellular processing of cone photoreceptor cyclic GMP-gated ion channels: A role for the S4 structural motif. J. Biol. Chem. 2004, 279, 22643–22653. [Google Scholar] [CrossRef] [PubMed]
- Mattapallil, M.J.; Wawrousek, E.F.; Chan, C.C.; Zhao, H.; Roychoudhury, J.; Ferguson, T.A.; Caspi, R.R. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- Errijgers, V.; Van Dam, D.; Gantois, I.; Van Ginneken, C.J.; Grossman, A.W.; D’Hooge, R.; De Deyn, P.P.; Kooy, R.F. FVB.129P2-Pde6b(+) Tyr(c-ch)/Ant, a sighted variant of the FVB/N mouse strain suitable for behavioral analysis. Genes Brain Behav. 2007, 6, 552–557. [Google Scholar] [CrossRef]
- Wu, W.H.; Tsai, Y.T.; Justus, S.; Lee, T.T.; Zhang, L.; Lin, C.S.; Bassuk, A.G.; Mahajan, V.B.; Tsang, S.H. CRISPR Repair Reveals Causative Mutation in a Preclinical Model of Retinitis Pigmentosa. Mol. Ther. 2016, 24, 1388–1394. [Google Scholar] [CrossRef]
- Wang, N.K.; Tosi, J.; Kasanuki, J.M.; Chou, C.L.; Kong, J.; Parmalee, N.; Wert, K.J.; Allikmets, R.; Lai, C.C.; Chien, C.L.; et al. Transplantation of reprogrammed embryonic stem cells improves visual function in a mouse model for retinitis pigmentosa. Transplantation 2010, 89, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Burns, M.E.; Calvert, P.D.; Gouras, P.; Baylor, D.A.; Goff, S.P.; Arshavsky, V.Y. Role of the Target Enzyme in Deactivation of Photoreceptor G Protein in Vivo. Science 1998, 282, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Gouras, P.; Yamashita, C.K.; Kjeldbye, H.; Fisher, J.; Farber, D.B.; Goff, S.P. Retinal degeneration in mice lacking the γ subunit of rod cGMP phosphodiesterase. Science 1996, 272, 1026–1029. [Google Scholar] [CrossRef] [PubMed]


| sgRNA | Strand | Sequence (5′-3′) | PAM | Off-Target Score | On-Target Score |
|---|---|---|---|---|---|
| sgRNA#1 | Forward | gcagtcacactagtatccac | agg | 42.8263215 | 66.09532194 |
| sgRNA#2 | Forward | cttgtctcaggcgagcctca | ggg | 42.2064369 | 55.07114307 |
| sgRNA#3 | Forward | ggccatgatcgcagtccaag | agg | 45.198453 | 63.29196792 |
| Gene | Direction/Exon | Nucleotide Sequence (5′-3′) |
|---|---|---|
| Cnga3 | Forward/5 | ATGAGCTACAATCAGAGCACC |
| Reverse/6 | TTTCCACAGCCTCTTGGTATC | |
| Cnga3 | Forward/2 | CCCCGACCCAACTTTCAATA |
| Reverse/3 | TTCATCGTGTAAGTGCCTGG | |
| Cngb3 | Forward/2 | GCAGGACACCAATCACATTTG |
| Reverse/3 | CCTAGTTTTCTCCATCTCTGCC | |
| Rhodopsin | Forward/2 | GGGAGAATCACGCTATCATGG |
| Reverse/3 | GTCAATCCCGCATGAACATTG | |
| Beta-Actin | Forward/3 | ACCTTCTACAATGAGCTGCG |
| Reverse/4 | CTGGATGGCTACGTACATGG |
| Cnga3 | |||
| Exon | Position | Score 1 * | Score 2 # |
| 1 | 1–278 | 0 | 0 |
| 2 | 12949–13085 | 0.95 | 0 |
| 3 | 25573–25752 | 0.65 | 0.97 |
| 4 | 34697–34804 | 0.96 | 0.92 |
| 5 | 38733–38839 | 0.95 | 0.86 |
| 6 | 41437–44124 | 0.97 | 0.99 |
| Cnga3 floxed-STOP | |||
| Exon | Position | Score 1 * | Score 2 # |
| 1 | 1–278 | 0 | 0 |
| 2 | 12949–13085 | 0.95 | 0 |
| 3 | 25573–25752 | 0.65 | 0.97 |
| 4 | 34697–34804 | 0.96 | 0.92 |
| 5 | 38733–38839 | 0.95 | 0.86 |
| floxed-STOP cassette | 41121–44006 | 0.97 | 1 |
| 6 | 44323–47010 | 0.97 | 0.99 |
| Gene | Direction | Nucleotide Sequence (5′-3′) | Size |
|---|---|---|---|
| Floxed-miniSTOP | 5′ Forward | GTATGAATGGCCTCTGCAGCAGTCA | 388 |
| 5′ Reverse | ACTGGAAAGACCGCGAAGAGTTTGT | ||
| Floxed-miniSTOP | 3′ Forward | CTCTTCGCGGTCTTTCCAGTTAACT | 469 |
| 3′ Reverse | GAAGCCGTATTGTGTTGTGTAGGGA | ||
| Floxed-STOP | 5′ Forward | CACTGTTTCCCTGAACACTACT | 285 |
| 5′ Reverse | AGACAGGGTCTTGTATAGGCTACATTC | ||
| Floxed-STOP | 3′ Forward | GGGAGGATTGGGAAGACAATAG | 355 |
| 3′ Reverse | TTTCTCTTCTGGGAATGGAACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.-K.; Liu, P.-K.; Kong, Y.; Levi, S.R.; Huang, W.-C.; Hsu, C.-W.; Wang, H.-H.; Chen, N.; Tseng, Y.-J.; Quinn, P.M.J.; et al. Mouse Models of Achromatopsia in Addressing Temporal “Point of No Return” in Gene-Therapy. Int. J. Mol. Sci. 2021, 22, 8069. https://doi.org/10.3390/ijms22158069
Wang N-K, Liu P-K, Kong Y, Levi SR, Huang W-C, Hsu C-W, Wang H-H, Chen N, Tseng Y-J, Quinn PMJ, et al. Mouse Models of Achromatopsia in Addressing Temporal “Point of No Return” in Gene-Therapy. International Journal of Molecular Sciences. 2021; 22(15):8069. https://doi.org/10.3390/ijms22158069
Chicago/Turabian StyleWang, Nan-Kai, Pei-Kang Liu, Yang Kong, Sarah R. Levi, Wan-Chun Huang, Chun-Wei Hsu, Hung-Hsi Wang, Nelson Chen, Yun-Ju Tseng, Peter M. J. Quinn, and et al. 2021. "Mouse Models of Achromatopsia in Addressing Temporal “Point of No Return” in Gene-Therapy" International Journal of Molecular Sciences 22, no. 15: 8069. https://doi.org/10.3390/ijms22158069
APA StyleWang, N.-K., Liu, P.-K., Kong, Y., Levi, S. R., Huang, W.-C., Hsu, C.-W., Wang, H.-H., Chen, N., Tseng, Y.-J., Quinn, P. M. J., Tai, M.-H., Lin, C.-S., & Tsang, S. H. (2021). Mouse Models of Achromatopsia in Addressing Temporal “Point of No Return” in Gene-Therapy. International Journal of Molecular Sciences, 22(15), 8069. https://doi.org/10.3390/ijms22158069








