Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant–Microbe Interactions
Abstract
1. Introduction
2. Results
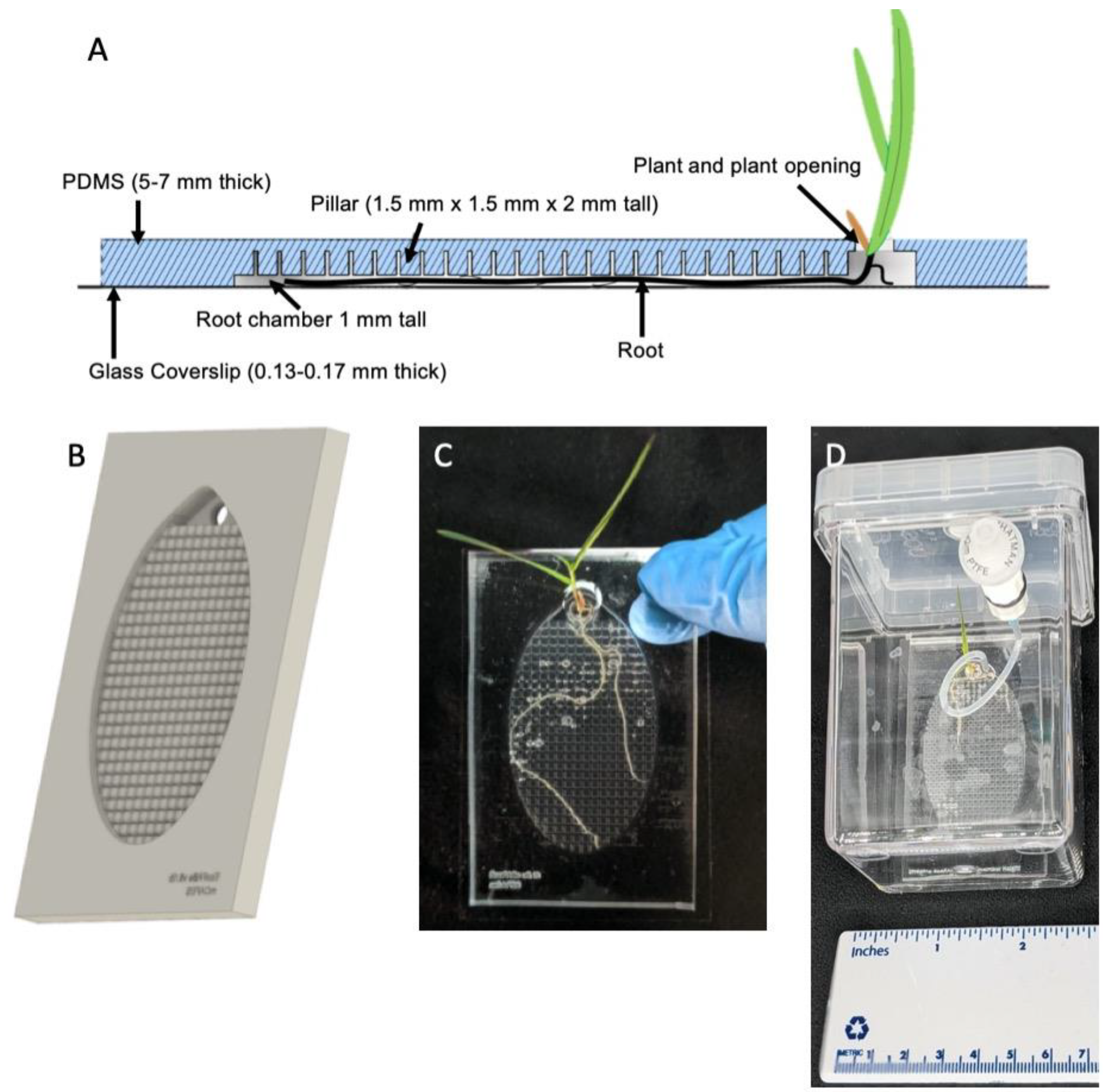
2.1. Imaging EcoFAB Design and the Modified Magenta Box for Better Sterility Control
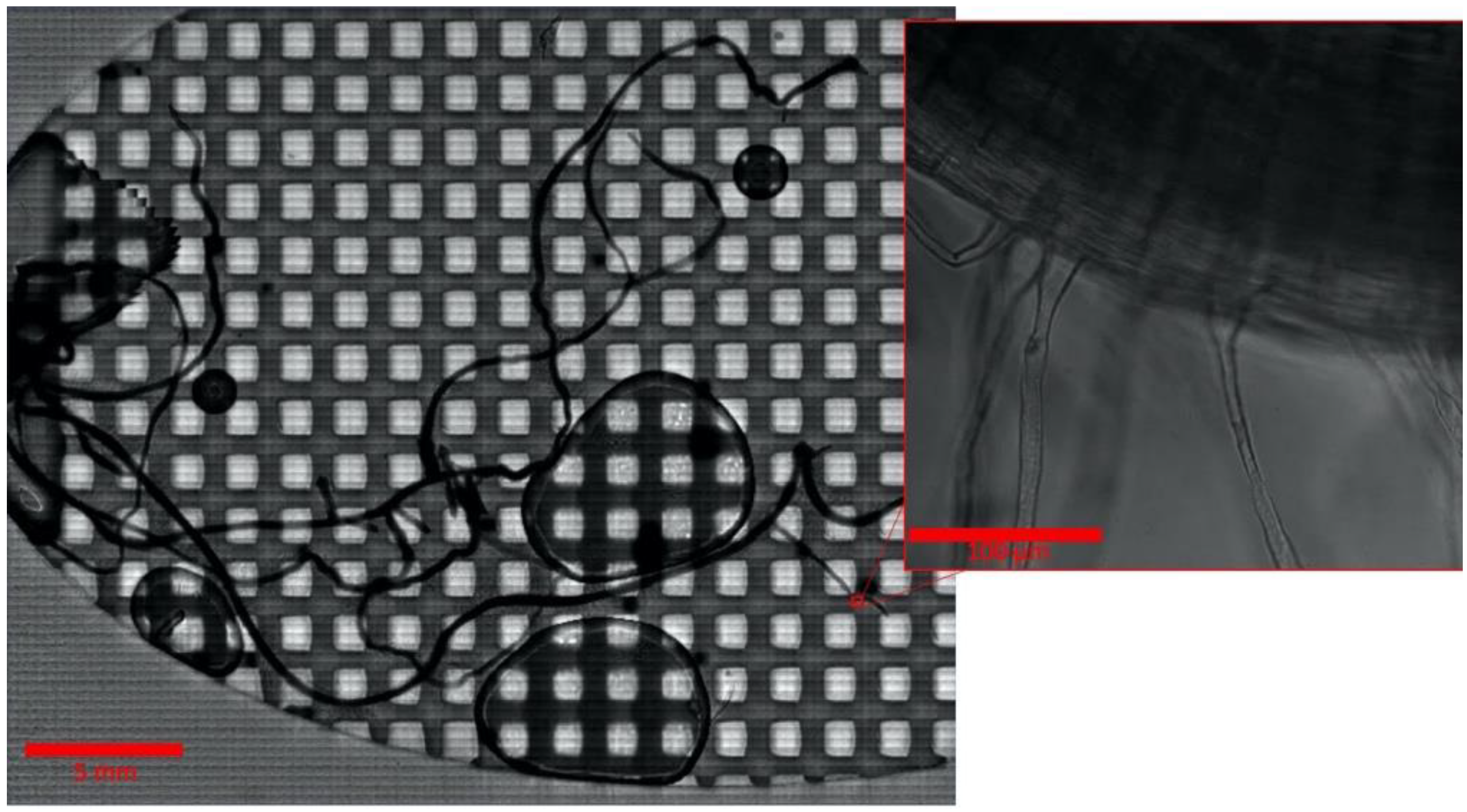
2.2. Imaging of the Entire B. distachyon Root at High Magnification
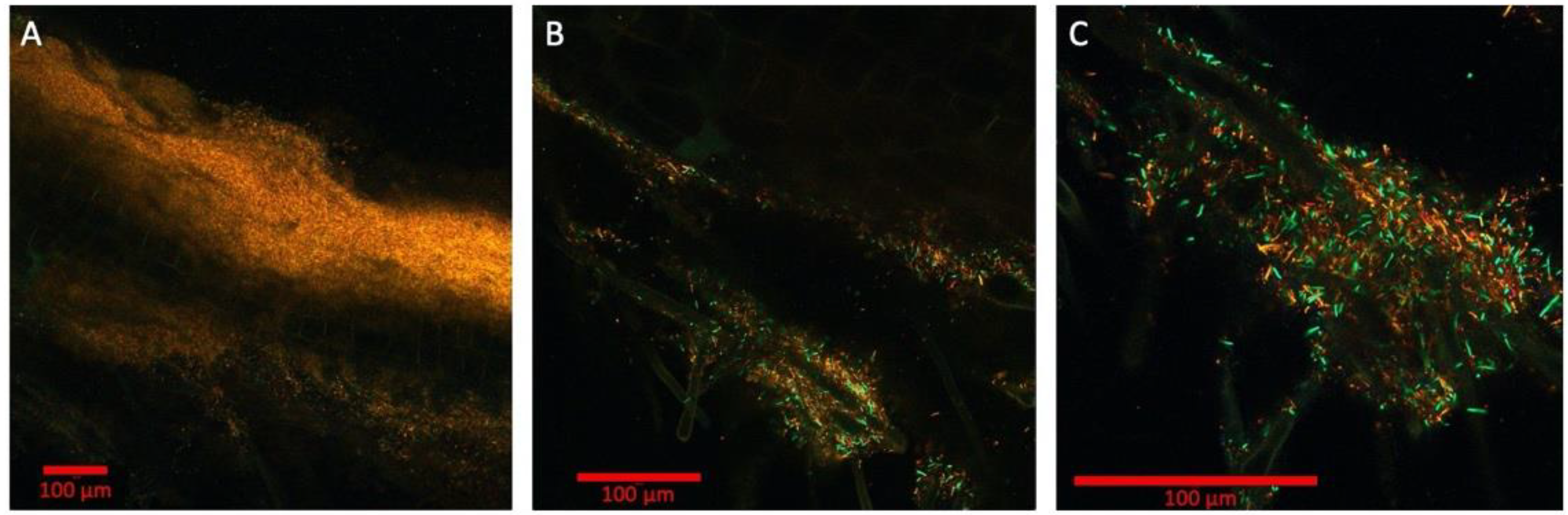
2.3. Multispectral Imaging of Fluorescent Protein Expressing P. simiae on B. distachyon Root
2.4. Bacterial Succession Study with Two Fluorescent P. simiae Strains
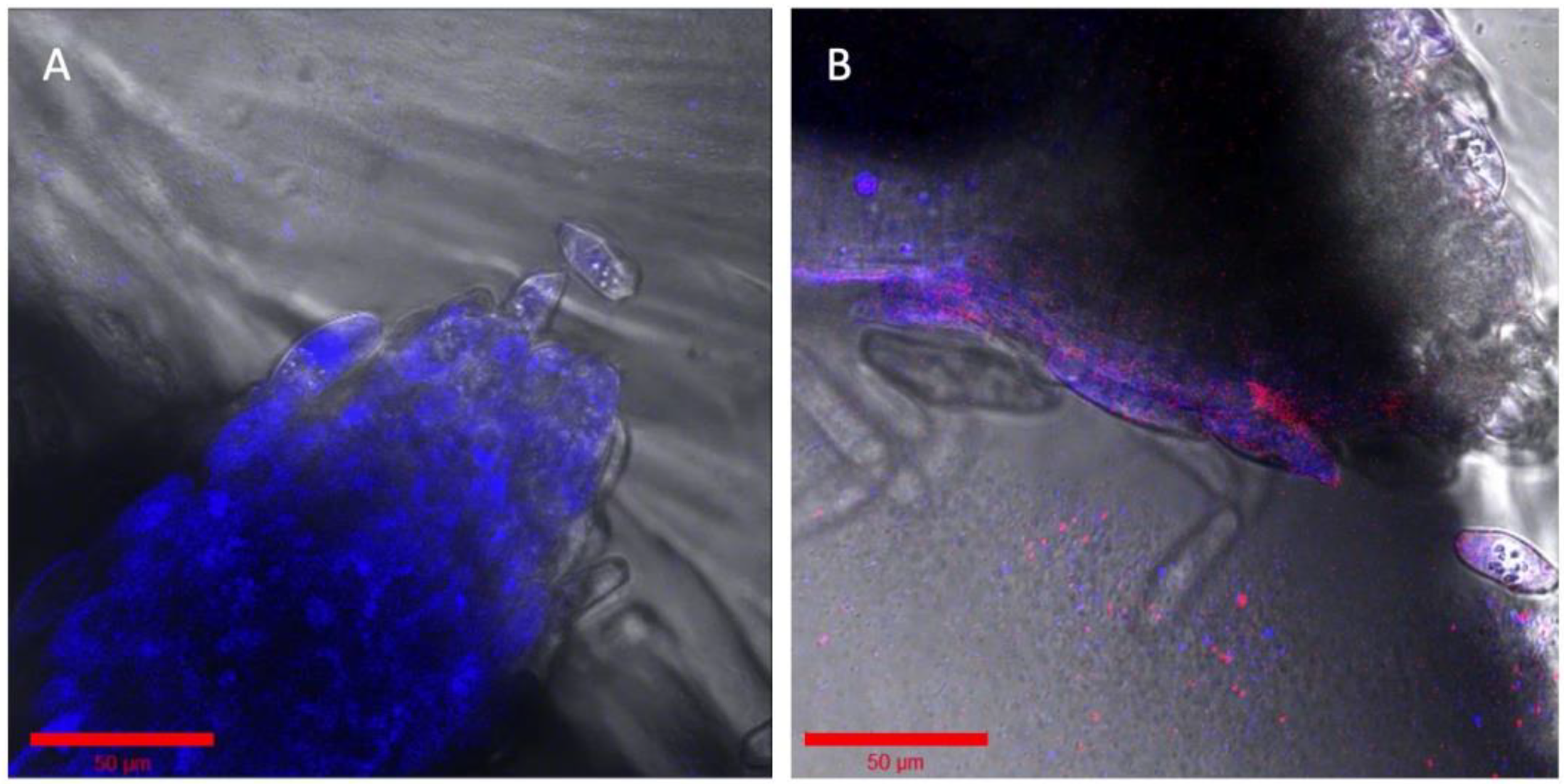
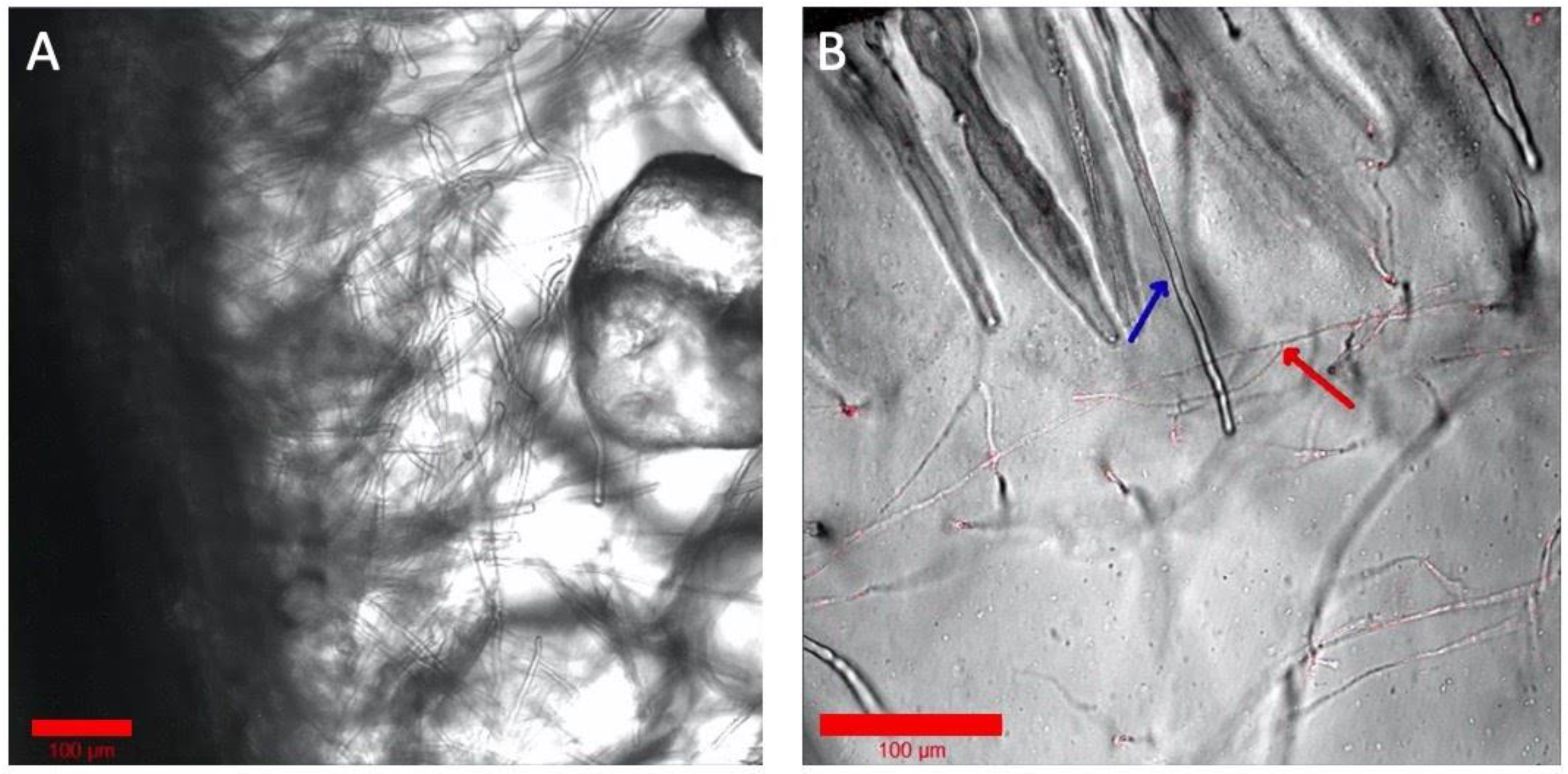
2.5. Imaging with Various Solid Substrates in the Root Chamber and Fungal Hyphae
3. Discussion
4. Materials and Methods
4.1. Fabrication of the Imaging EcoFABs
4.2. Magenta Box Housing Modification
4.3. Brachypodium Distachyon Seed Sterilization and Germination
4.4. Pseudomonas Simiae Inoculation
4.5. Neurospora Crassa Inoculation
4.6. Imaging and Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nguyen, C. Rhizodeposition of Organic C by Plants: Mechanisms and Controls. Agronomie 2003, 23, 375–396. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon Flow in the Rhizosphere: Carbon Trading at the Soil–Root Interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic Root Exudate Chemistry and Microbial Substrate Preferences Drive Patterns in Rhizosphere Microbial Community Assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Zhalnina, K.; Zengler, K.; Newman, D.; Northen, T.R. Need for Laboratory Ecosystems To Unravel the Structures and Functions of Soil Microbial Communities Mediated by Chemistry. mBio 2018, 9, e01175-18. [Google Scholar] [CrossRef] [PubMed]
- Yee, M.O.; Kim, P.; Li, Y.; Singh, A.K.; Northen, T.R.; Chakraborty, R. Specialized Plant Growth Chamber Designs to Study Complex Rhizosphere Interactions. Front. Microbiol. 2021, 12, 507. [Google Scholar] [CrossRef] [PubMed]
- Massalha, H.; Korenblum, E.; Malitsky, S.; Shapiro, O.H.; Aharoni, A. Live Imaging of Root–Bacteria Interactions in a Microfluidics Setup. Proc. Natl. Acad. Sci. USA 2017, 114, 4549–4554. [Google Scholar] [CrossRef] [PubMed]
- Huck, M.G.; Taylor, H.M. The rhizotron as a tool for root research. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1982; Volume 35, pp. 1–35. ISBN 0065-2113. [Google Scholar]
- Mohamed, A.; Monnier, Y.; Mao, Z.; Lobet, G.; Maeght, J.-L.; Ramel, M.; Stokes, A. An Evaluation of Inexpensive Methods for Root Image Acquisition When Using Rhizotrons. Plant Methods 2017, 13, 11. [Google Scholar] [CrossRef]
- Gao, J.; Sasse, J.; Lewald, K.M.; Zhalnina, K.; Cornmesser, L.T.; Duncombe, T.A.; Yoshikuni, Y.; Vogel, J.P.; Firestone, M.K.; Northen, T.R. Ecosystem Fabrication (EcoFAB) Protocols for The Construction of Laboratory Ecosystems Designed to Study Plant-Microbe Interactions. J. Vis. Exp. 2018, e57170. [Google Scholar] [CrossRef]
- Sasse, J.; Kant, J.; Cole, B.J.; Klein, A.P.; Arsova, B.; Schlaepfer, P.; Gao, J.; Lewald, K.; Zhalnina, K.; Kosina, S. Multilab EcoFAB Study Shows Highly Reproducible Physiology and Depletion of Soil Metabolites by a Model Grass. New Phytol. 2019, 222, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Heller, J.; Jabusch, L.K.; Kim, P.W.; Northen, T.R.; Glass, N.L. Devices for Studying Plant-Fungus Interactions and for Imaging Plant Roots. U.S. Patent Application 16/886,849, 10 December 2020. [Google Scholar]
- Wang, B.; Zhao, Z.; Jabusch, L.K.; Chiniquy, D.M.; Ono, K.; Conway, J.M.; Zhang, Z.; Wang, G.; Robinson, D.; Cheng, J.-F.; et al. CRAGE-Duet Facilitates Modular Assembly of Biological Systems for Studying Plant–Microbe Interactions. ACS Synth. Biol. 2020, 9, 2610–2615. [Google Scholar] [CrossRef]
- Freitag, M.; Selker, E. Expression and Visualization of Red Fluorescent Protein (RFP) in Neurospora Crassa. Fungal Genet. Newsl. 2005, 52. [Google Scholar] [CrossRef][Green Version]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Day, R.N.; Davidson, M.W. The Fluorescent Protein Palette: Tools for Cellular Imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.J.; Lagarias, J.C. Harnessing Phytochrome’s Glowing Potential. Proc. Natl. Acad. Sci. USA 2004, 101, 17334–17339. [Google Scholar] [CrossRef] [PubMed]
- Hinsinger, P.; Gobran, G.R.; Gregory, P.J.; Wenzel, W.W. Rhizosphere Geometry and Heterogeneity Arising from Root-Mediated Physical and Chemical Processes. New Phytol. 2005, 168, 293–303. [Google Scholar] [CrossRef]
- Donn, S.; Kirkegaard, J.A.; Perera, G.; Richardson, A.E.; Watt, M. Evolution of Bacterial Communities in the Wheat Crop Rhizosphere. Environ. Microbiol. 2015, 17, 610–621. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere Microbiome Mediates Systemic Root Metabolite Exudation by Root-to-Root Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and Exudates of the Root and Rhizosphere of Brachypodium Distachyon, a Model for Wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Razavi, B.S. Rhizosphere Size and Shape: Temporal Dynamics and Spatial Stationarity. Soil Biol. Biochem. 2019, 135, 343–360. [Google Scholar] [CrossRef]
- WATT, M.; SILK, W.K.; PASSIOURA, J.B. Rates of Root and Organism Growth, Soil Conditions, and Temporal and Spatial Development of the Rhizosphere. Ann. Bot. 2006, 97, 839–855. [Google Scholar] [CrossRef]
- York, L.M.; Carminati, A.; Mooney, S.J.; Ritz, K.; Bennett, M.J. The Holistic Rhizosphere: Integrating Zones, Processes, and Semantics in the Soil Influenced by Roots. J. Exp. Bot. 2016, 67, 3629–3643. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.J. Roots, Rhizosphere and Soil: The Route to a Better Understanding of Soil Science? Eur. J. Soil Sci. 2006, 57, 2–12. [Google Scholar] [CrossRef]
- de la Porte, A.; Schmidt, R.; Yergeau, É.; Constant, P. A Gaseous Milieu: Extending the Boundaries of the Rhizosphere. Trends Microbiol. 2020, 28, 536–542. [Google Scholar] [CrossRef]
- Gonzalez Alzate, A.; Sevillano, X.; Betegón Putze, I.; Blasco, D.; Ferrer, M.; Caño-Delgado, A. MyROOT 2.0: An Automatic Tool for High Throughput and Accurate Primary Root Length Measurement. Comput. Electron. Agric. 2020, 168, 105125. [Google Scholar] [CrossRef]
- Yasrab, R.; Atkinson, J.A.; Wells, D.M.; French, A.P.; Pridmore, T.P.; Pound, M.P. RootNav 2.0: Deep Learning for Automatic Navigation of Complex Plant Root Architectures. GigaScience 2019, 8, giz123. [Google Scholar] [CrossRef]
- Van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. Scikit-Image: Image Processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef] [PubMed]
- Dukovski, I.; Bajić, D.; Chacón, J.M.; Quintin, M.; Vila, J.C.; Sulheim, S.; Pacheco, A.R.; Bernstein, D.B.; Rieh, W.J.; Korolev, K.S. Computation Of Microbial Ecosystems in Time and Space (COMETS): An Open Source Collaborative Platform for Modeling Ecosystems Metabolism. arXiv 2020, arXiv:200901734. [Google Scholar]
- Vogel, J.P.; Garvin, D.F.; Leong, O.M.; Hayden, D.M. Agrobacterium-Mediated Transformation and Inbred Line Development in the Model Grass Brachypodium Distachyon. Plant Cell Tissue Organ Cult. 2006, 84, 199–211. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabusch, L.K.; Kim, P.W.; Chiniquy, D.; Zhao, Z.; Wang, B.; Bowen, B.; Kang, A.J.; Yoshikuni, Y.; Deutschbauer, A.M.; Singh, A.K.; et al. Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant–Microbe Interactions. Int. J. Mol. Sci. 2021, 22, 7880. https://doi.org/10.3390/ijms22157880
Jabusch LK, Kim PW, Chiniquy D, Zhao Z, Wang B, Bowen B, Kang AJ, Yoshikuni Y, Deutschbauer AM, Singh AK, et al. Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant–Microbe Interactions. International Journal of Molecular Sciences. 2021; 22(15):7880. https://doi.org/10.3390/ijms22157880
Chicago/Turabian StyleJabusch, Lauren K., Peter W. Kim, Dawn Chiniquy, Zhiying Zhao, Bing Wang, Benjamin Bowen, Ashley J. Kang, Yasuo Yoshikuni, Adam M. Deutschbauer, Anup K. Singh, and et al. 2021. "Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant–Microbe Interactions" International Journal of Molecular Sciences 22, no. 15: 7880. https://doi.org/10.3390/ijms22157880
APA StyleJabusch, L. K., Kim, P. W., Chiniquy, D., Zhao, Z., Wang, B., Bowen, B., Kang, A. J., Yoshikuni, Y., Deutschbauer, A. M., Singh, A. K., & Northen, T. R. (2021). Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant–Microbe Interactions. International Journal of Molecular Sciences, 22(15), 7880. https://doi.org/10.3390/ijms22157880






