Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori
Abstract
1. Introduction
2. Results
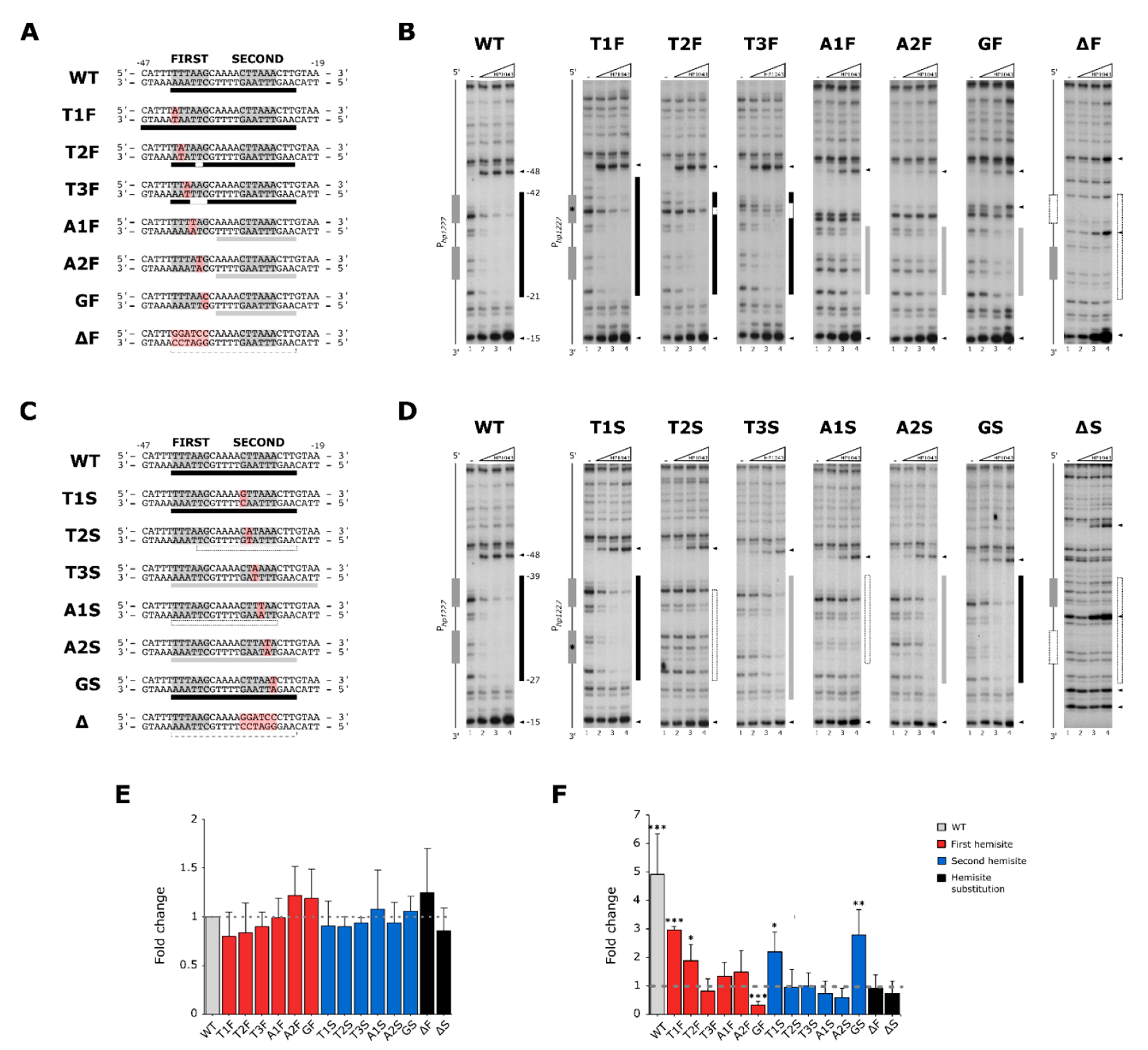
2.1. Molecular Characterization of HP1043 Consensus Binding Sequence
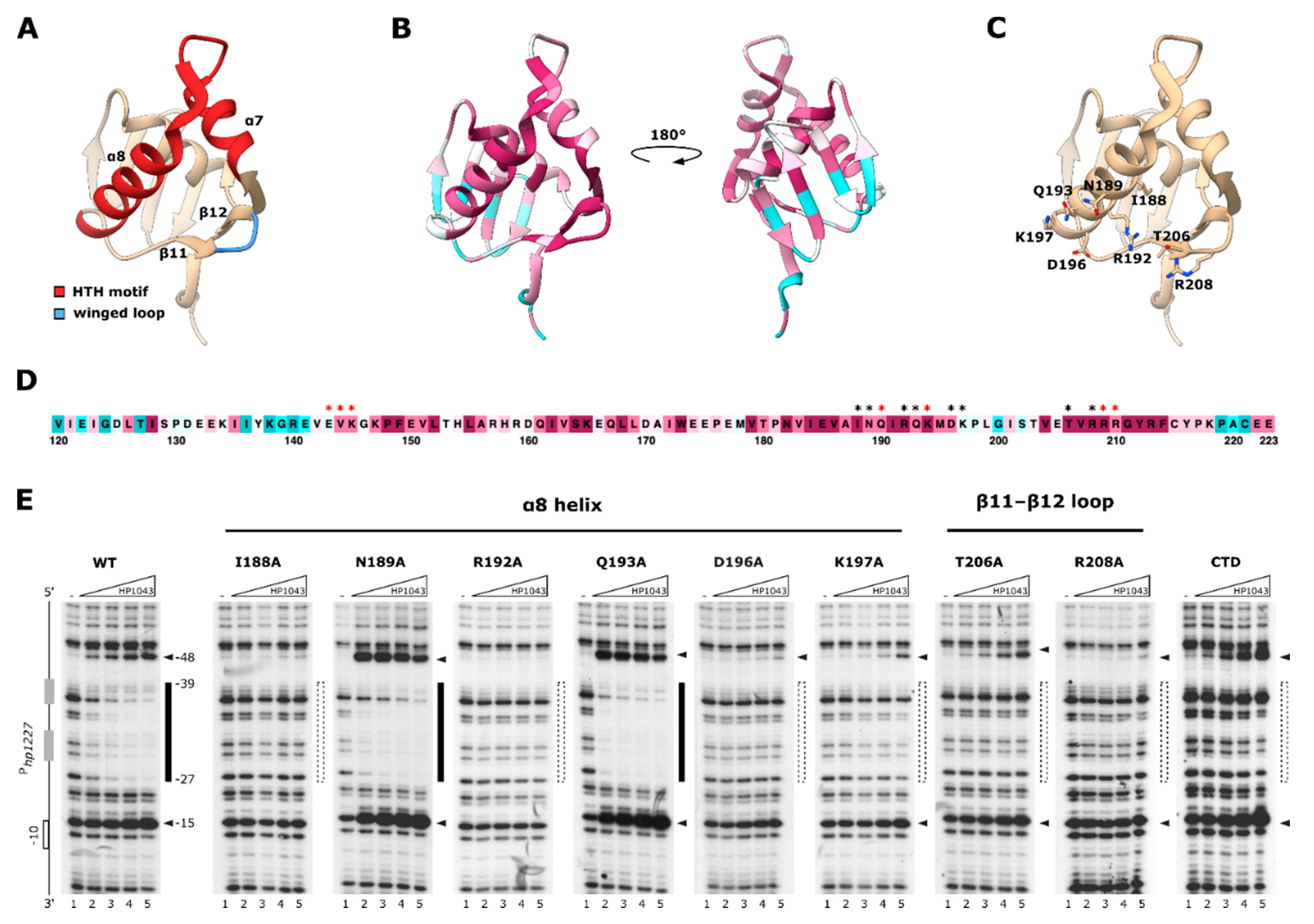
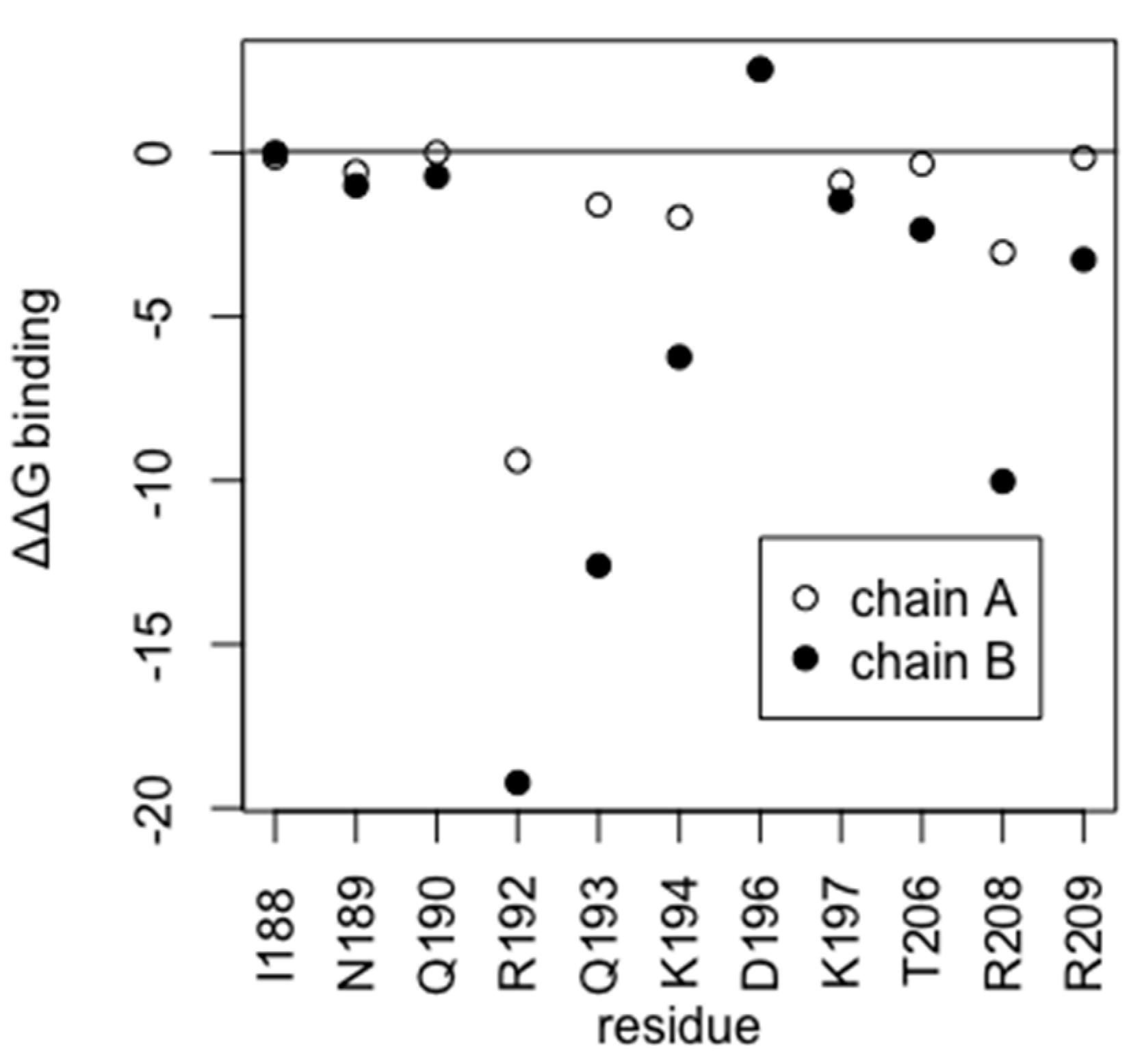
2.2. Identification of Amino Acid Residues Fundamental for Target DNA Recognition by HP1043
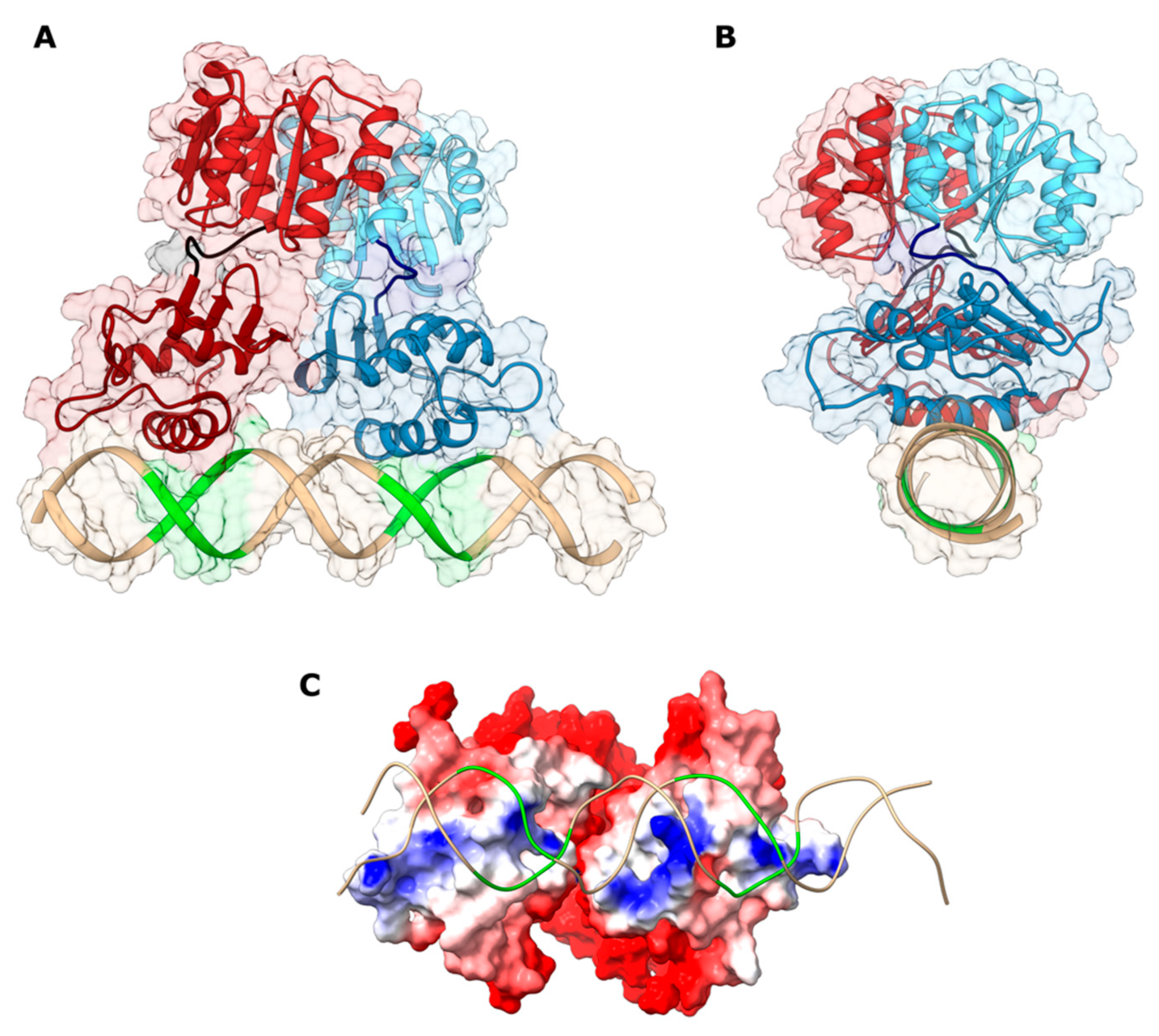
2.3. Structural Modeling of the HP1043_DBD-DNA Interaction
2.4. Validation of the HP1043-Php1227 Docking Model
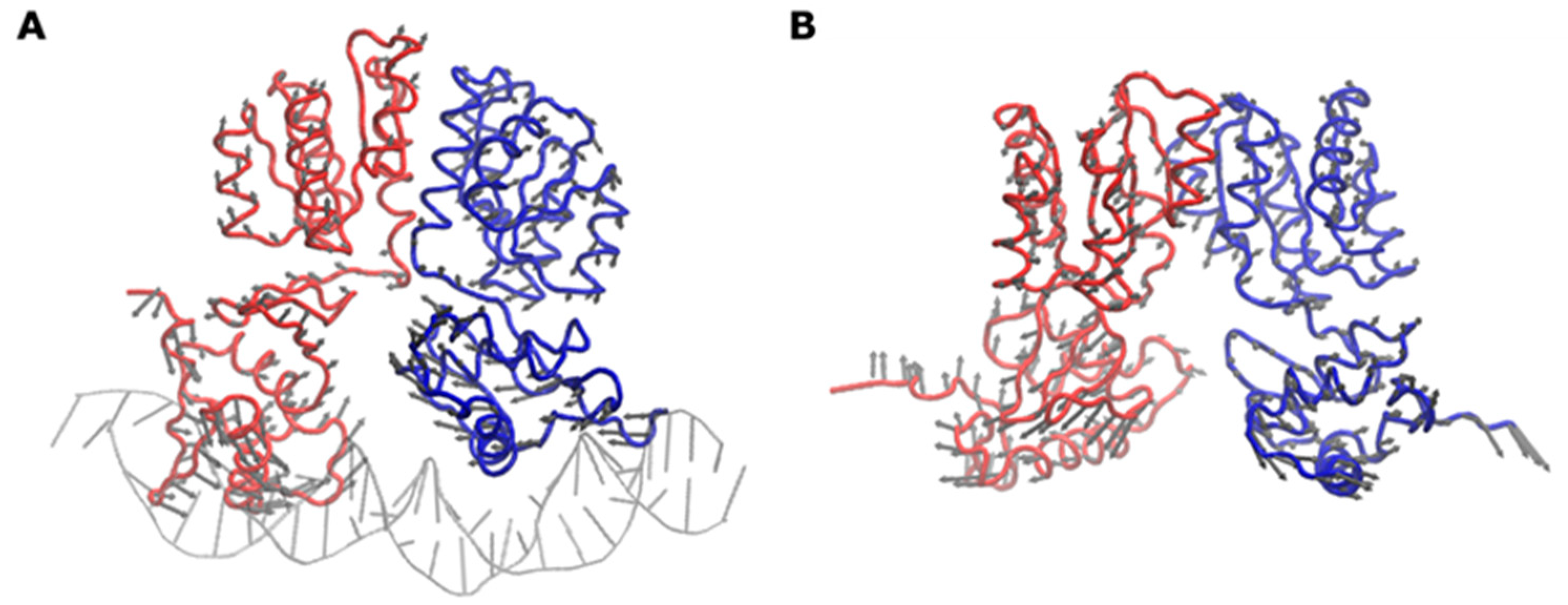
2.5. Conformational and Interaction Analysis of HP1043-DNA Full Model
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. DNA Manipulations
4.3. Overexpression and Purification of Recombinant His6-HP1043
4.4. DNase I Footprinting
4.5. Generation of Probes for DNase I Footprinting and Templates for In Vitro Transcription Experiments
4.6. In Vitro Transcription and cDNA Synthesis
4.7. qRT-PCR Analysis
4.8. Protein-DNA Docking and HP1043 Structure Reconstruction
4.9. Electrophoretic Mobility Shift Assays
4.10. Molecular Dynamics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Salama, N.R.; Hartung, M.L.; Muller, A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Micro. 2013, 11, 385–399. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Calvet, X. Review article: Common misconceptions in the management of Helicobacter pylori-associated gastric MALT-lymphoma. Aliment. Pharmacol. Ther. 2011, 34, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Scarlato, V.; Delany, I.; Spohn, G.; Beier, D. Regulation of transcription in Helicobacter pylori: Simple systems or complex circuits? Int. J. Med. Microbiol. 2001, 291, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Foynes, S.; Dorrell, N.; Ward, S.J.; Stabler, R.A.; McColm, A.A.; Rycroft, A.N.; Wren, B.W. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 2000, 68, 2016–2023. [Google Scholar] [CrossRef]
- Waidner, B.; Melchers, K.; Stähler, F.N.; Kist, M.; Bereswill, S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J. Bacteriol. 2005, 187, 4683–4688. [Google Scholar] [CrossRef]
- Niehus, E.; Gressmann, H.; Ye, F.; Schlapbach, R.; Dehio, M.; Dehio, C.; Stack, A.; Meyer, T.F.; Suerbaum, S.; Josenhans, C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 2004, 52, 947–961. [Google Scholar] [CrossRef]
- Pflock, M.; Finsterer, N.; Joseph, B.; Meyer, T.F.; Beier, D.; Mollenkopf, H. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 2006, 188, 3449–3462. [Google Scholar] [CrossRef]
- McDaniel, T.K.; DeWalt, K.C.; Salama, N.R.; Falkow, S. New Approaches for Validation of Lethal Phenotypes and Genetic Reversion in Helicobacter pylori. Helicobacter. 2001, 6, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Schär, J.; Sickmann, A.; Beier, D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 2005, 187, 3100–3109. [Google Scholar] [CrossRef]
- Delany, I.; Spohn, G.; Rappuoli, R.; Scarlato, V. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 2002, 184, 4800–4810. [Google Scholar] [CrossRef]
- Müller, S.; Pflock, M.; Schär, J.; Kennard, S.; Beier, D. Regulation of expression of atypical orphan response regulators of Helicobacter pylori. Microbiol. Res. 2007, 162, 1–14. [Google Scholar] [CrossRef]
- Croxen, M.A.; Ernst, P.B.; Hoffman, P.S. Antisense RNA Modulation of Alkyl Hydroperoxide Reductase Levels in Helicobacter pylori Correlates with Organic Peroxide Toxicity but Not Infectivity. J. Bacteriol. 2007, 189, 3359–3368. [Google Scholar] [CrossRef][Green Version]
- Pelliciari, S.; Pinatel, E.; Vannini, A.; Peano, C.; Puccio, S.; De Bellis, G.; Danielli, A.; Scarlato, V.; Roncarati, D. Insight into the essential role of the Helicobacter pylori HP1043 orphan response regulator: Genome-wide identification and characterization of the DNA-binding sites. Sci. Rep. 2017, 7, 41063. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J.W. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016, 14, 638–650. [Google Scholar] [CrossRef]
- Hong, E.; Jung, J.-W.; Shin, J.; Kim, J.H.; Jeon, Y.-H.; Yamazaki, T.; Lee, W. Letter to the Editor: Backbone 1 H, 13 C and 15 N resonance assignments of the response regulator HP1043 from Helicobactor pylori. J. Biomol. NMR 2004, 28, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Hyang, M.L.; Ko, H.; Kim, D.U.; Jeon, B.Y.; Jung, J.; Shin, J.; Lee, S.A.; Kim, Y.; Young, H.J.; et al. Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J. Biol. Chem. 2007, 282, 20667–20675. [Google Scholar] [CrossRef]
- Jeong, K.W.; Ko, H.; Lee, S.A.; Hong, E.; Ko, S.; Cho, H.S.; Lee, W.; Kim, Y. Backbone dynamics of an atypical orphan response regulator protein, Helicobacter pylori 1043. Mol. Cells 2013, 35, 158–165. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Van Dijk, M.; Van Dijk, A.D.J.; Hsu, V.; Rolf, B.; Bonvin, A.M.J.J. Information-driven protein-DNA docking using HADDOCK: It is a matter of flexibility. Nucleic Acids Res. 2006, 34, 3317–3325. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.G.; Sola, M.; Gomis-Rüth, F.X.; Coll, M. Tandem DNA Recognition by PhoB, a Two-Component Signal Transduction Transcriptional Activator. Structure 2002, 10, 701–713. [Google Scholar] [CrossRef]
- Li, Y.-C.; Chang, C.; Chang, C.-F.; Cheng, Y.-H.; Fang, P.-J.; Yu, T.; Chen, S.-C.; Li, Y.-C.; Hsiao, C.-D.; Huang, T. Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element. Nucleic Acids Res. 2014, 42, 8777–8788. [Google Scholar] [CrossRef]
- Lou, Y.-C.; Weng, T.-H.; Li, Y.-C.; Kao, Y.-F.; Lin, W.-F.; Peng, H.-L.; Chou, S.-H.; Hsiao, C.-D.; Chen, C. Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA. Nat. Commun. 2015, 6, 8838. [Google Scholar] [CrossRef]
- Martínez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors 1 1Edited by M. Gottesman. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Tanaka, I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 1997, 24, 665–667. [Google Scholar] [CrossRef]
- Rhee, J.E.; Sheng, W.; Morgan, L.K.; Nolet, R.; Liao, X.; Kenney, L.J. Amino Acids Important for DNA Recognition by the Response Regulator OmpR. J. Biol. Chem. 2008, 283, 8664–8677. [Google Scholar] [CrossRef]
- Walthers, D.; Tran, V.K.; Kenney, L.J. Interdomain Linkers of Homologous Response Regulators Determine Their Mechanism of Action. J. Bacteriol. 2003, 185, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.J.; Moughon, S.; Wang, C.; Schueler-Furman, O.; Kuhlman, B.; Rohl, C.A.; Baker, D. Protein–Protein Docking with Simultaneous Optimization of Rigid-body Displacement and Side-chain Conformations. J. Mol. Biol. 2003, 331, 281–299. [Google Scholar] [CrossRef]
- Marze, N.A.; Roy Burman, S.S.; Sheffler, W.; Gray, J.J. Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics 2018, 34, 3461–3469. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Tsui, V.; Case, D.A. Molecular Dynamics Simulations of Nucleic Acids with a Generalized Born Solvation Model. J. Am. Chem. Soc. 2000, 122, 2489–2498. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Gonzalez, A.; Fillat, M.F.; Lanas, Á. Transcriptional regulators: Valuable targets for novel antibacterial strategies. Future Med. Chem. 2018, 10, 541–560. [Google Scholar] [CrossRef]
- González, A.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Fillat, M.F.; Sancho, J.; Lanas, Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- González, A.; Casado, J.; Chueca, E.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Bénejat, L.; Guignard, J.; Giese, A.; Sancho, J.; et al. Repurposing Dihydropyridines for Treatment of Helicobacter pylori Infection. Pharmaceutics 2019, 11, 681. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, NY, USA, 1989; pp. 1–1546. [Google Scholar]
- Liu, S.-T.; Hong, G.-F. Three-Minute G+A Specific Reaction for DNA Sequencing. Anal. Biochem. 1998, 255, 158–159. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K.; Swift, S.; Hill, P.J.; Sims, C.M.; Griesmayr, G.; Bycroft, B.W.; Williams, P.; Stewart, G.S.A. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn 5 constructs. FEMS Microbiol. Lett. 1998, 163, 193–202. [Google Scholar] [CrossRef]
- de Vries, S.J.; van Dijk, A.D.J.; Krzeminski, M.; van Dijk, M.; Thureau, A.; Hsu, V.; Wassenaar, T.; Bonvin, A.M.J.J. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins Struct. Funct. Bioinforma 2007, 69, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Agriesti, F.; Roncarati, D.; Musiani, F.; Del Campo, C.; Iurlaro, M.; Sparla, F.; Ciurli, S.; Danielli, A.; Scarlato, V. FeON-FeOFF: The Helicobacter pylori Fur regulator commutates iron-responsive transcription by discriminative readout of opposed DNA grooves. Nucleic Acids Res. 2014, 42, 3138–3151. [Google Scholar] [CrossRef]
- Mazzei, L.; Dobrovolska, O.; Musiani, F.; Zambelli, B.; Ciurli, S. On the interaction of Helicobacter pylori NikR, a Ni(II)-responsive transcription factor, with the urease operator: In solution and in silico studies. J. Biol. Inorg. Chem. 2015, 20, 1021–1037. [Google Scholar] [CrossRef]
- van Dijk, M.; Bonvin, A.M.J.J. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009, 37, W235–W239. [Google Scholar] [CrossRef]
- Musiani, F.; Ciurli, S.; Dikiy, A. Interaction of Selenoprotein W with 14-3-3 Proteins: A Computational Approach. J. Proteome Res. 2011, 10, 968–976. [Google Scholar] [CrossRef]
- Shen, M.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Tomb, J.-F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Dougherty, B.A.; et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Cheatham, T.E.; Case, D.A. Twenty-five years of nucleic acid simulations. Biopolymers 2013. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins Struct. Funct. Bioinforma 2004, 55, 383–394. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zannoni, A.; Pelliciari, S.; Musiani, F.; Chiappori, F.; Roncarati, D.; Scarlato, V. Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori. Int. J. Mol. Sci. 2021, 22, 7848. https://doi.org/10.3390/ijms22157848
Zannoni A, Pelliciari S, Musiani F, Chiappori F, Roncarati D, Scarlato V. Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori. International Journal of Molecular Sciences. 2021; 22(15):7848. https://doi.org/10.3390/ijms22157848
Chicago/Turabian StyleZannoni, Annamaria, Simone Pelliciari, Francesco Musiani, Federica Chiappori, Davide Roncarati, and Vincenzo Scarlato. 2021. "Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori" International Journal of Molecular Sciences 22, no. 15: 7848. https://doi.org/10.3390/ijms22157848
APA StyleZannoni, A., Pelliciari, S., Musiani, F., Chiappori, F., Roncarati, D., & Scarlato, V. (2021). Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori. International Journal of Molecular Sciences, 22(15), 7848. https://doi.org/10.3390/ijms22157848








