A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters
Abstract
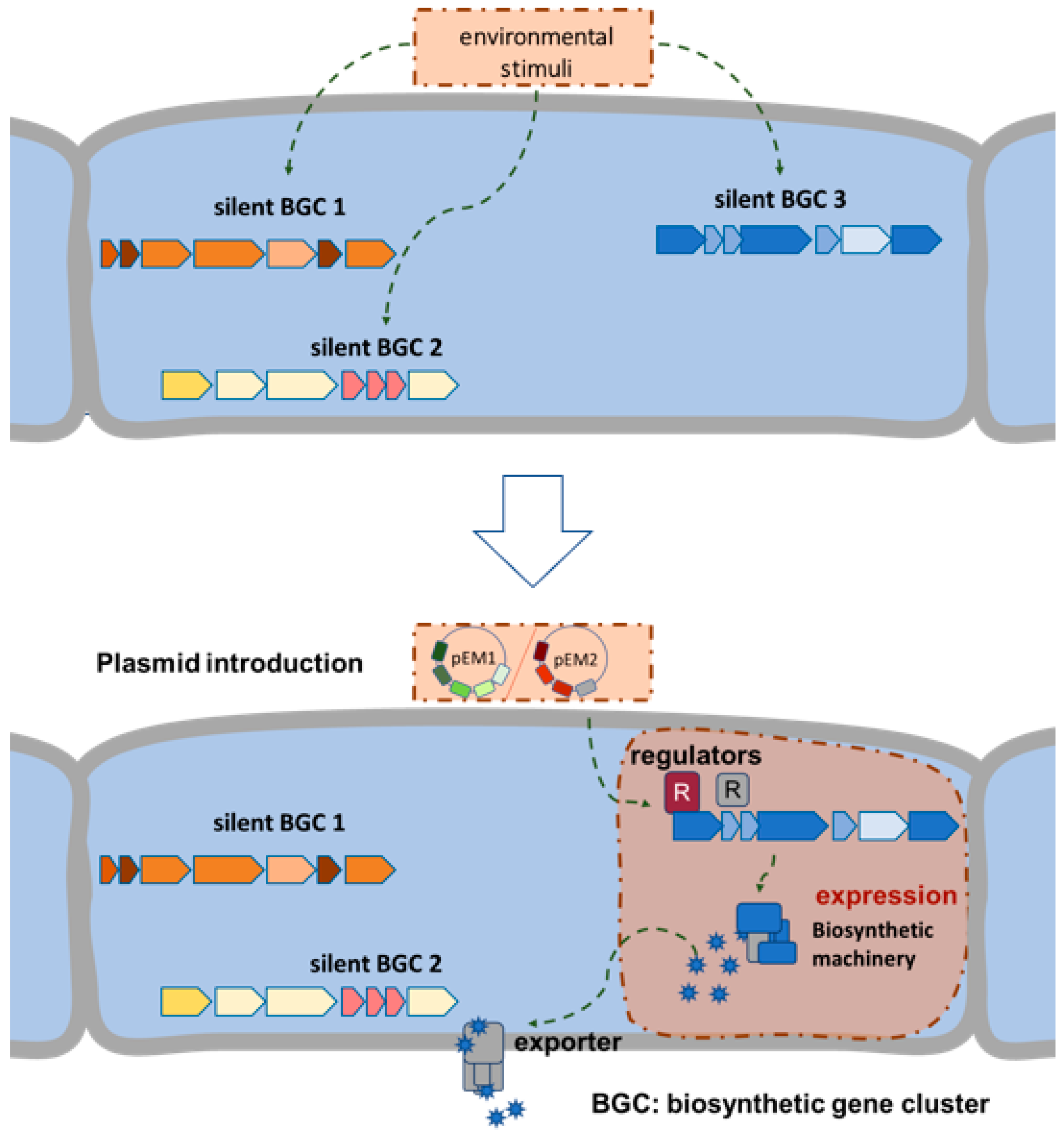
1. Introduction
2. Results and Discussion
2.1. Selection and Cloning of Regulatory Genes for Activation of Silent Clusters
2.2. Activation of Silent BGCs in Strains of the Tübingen Strain Collection
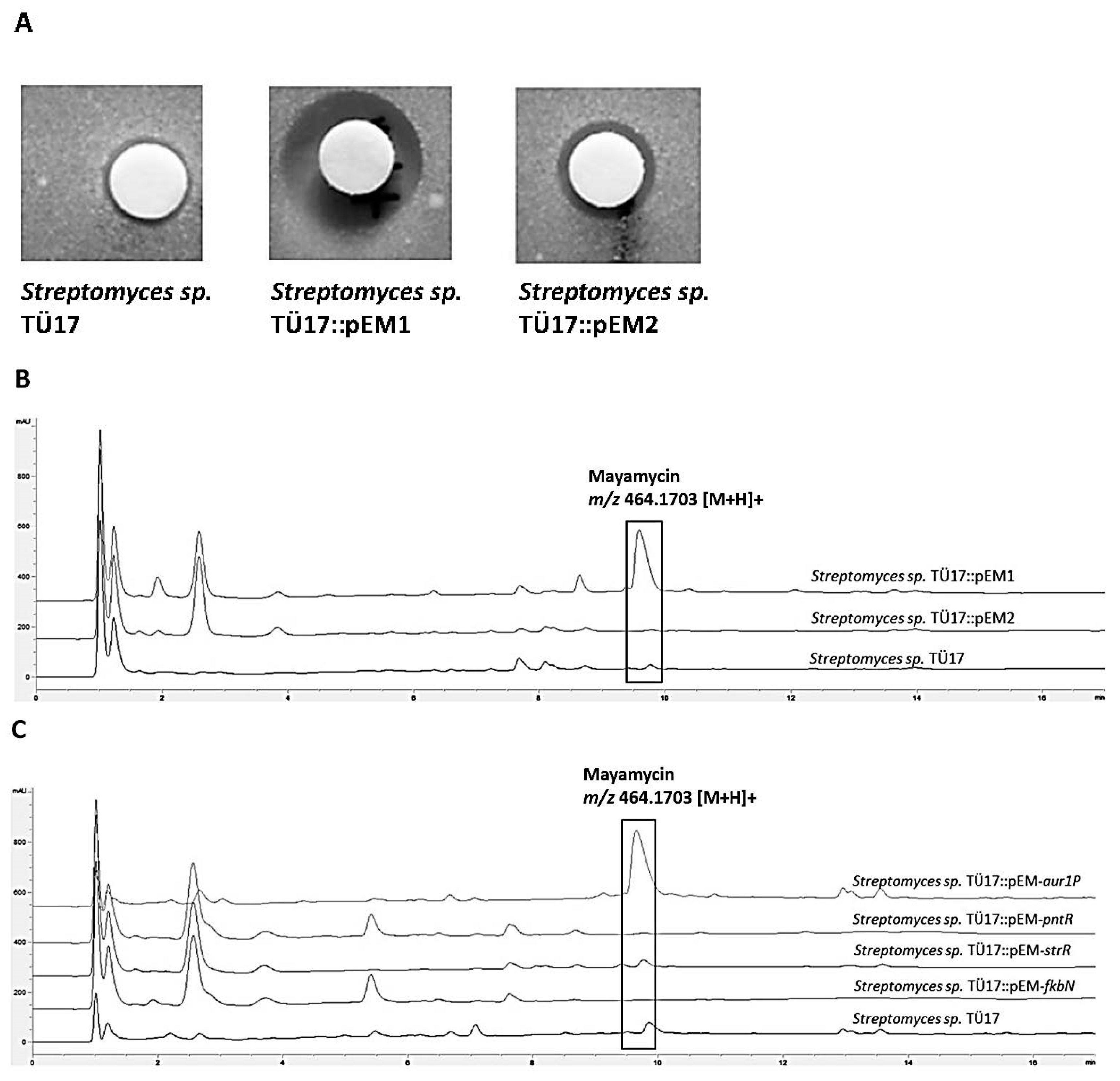
2.2.1. Metabolites Produced by Recombinant Streptomyces sp. TÜ17 Strains
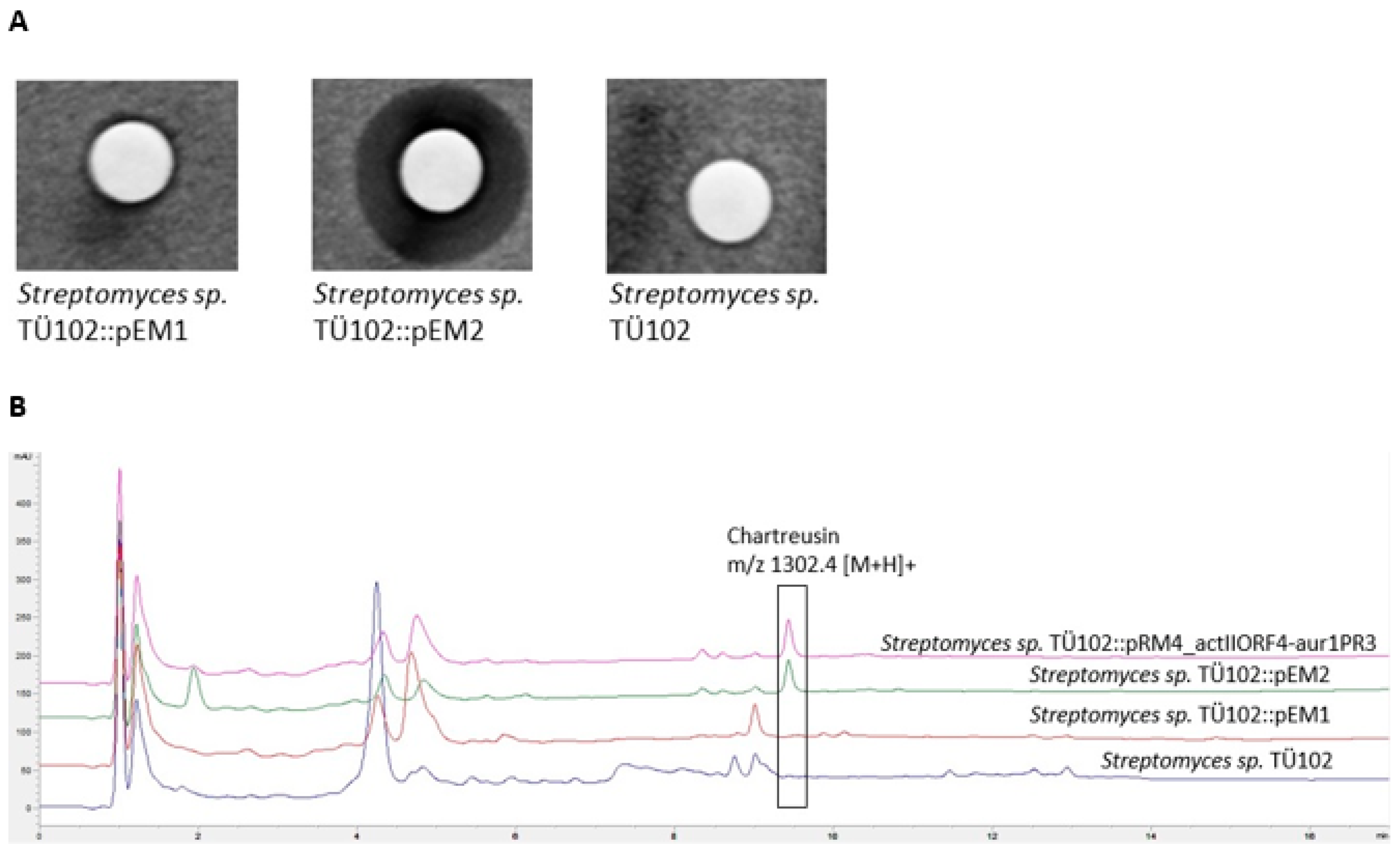
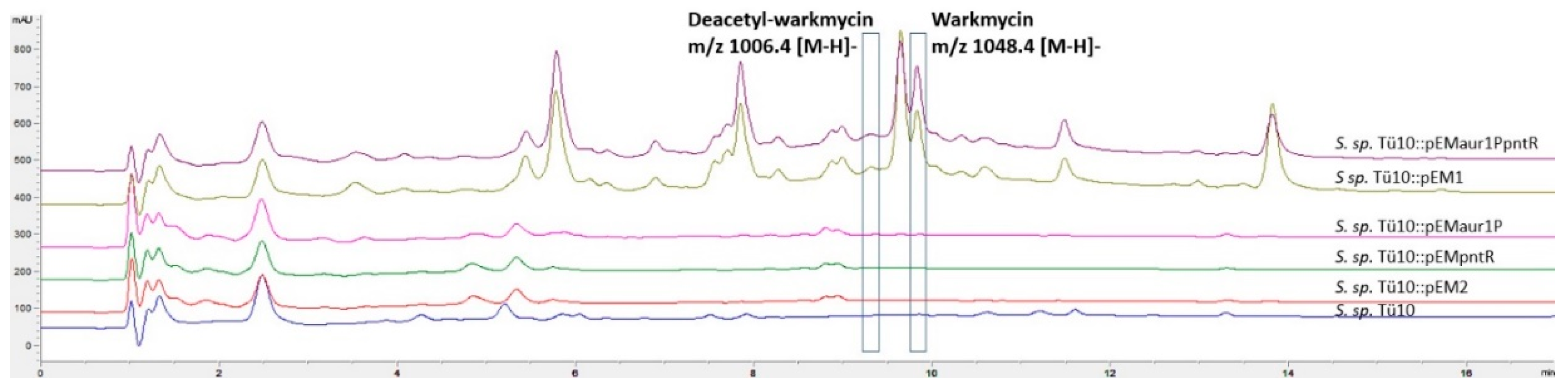
2.2.2. Metabolites Produced by Recombinant Strains Streptomyces sp. TÜ102 and Streptomyces sp. TÜ10
3. Conclusions
4. Materials and Methods
4.1. Bacterial Strains and Cultivation Conditions
4.2. DNA Manipulations, Plasmids
4.3. Constructions of Plasmid pEM1 and pEM2
4.4. Phylogenetic Analysis
4.5. Multiple Sequence Alignment
4.6. Conjugation
4.7. Sample Collection and Extraction
4.8. Bioassay
4.9. HPLC-MS/MS Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Musiol-Kroll, E.M.; Tocchetti, A.; Sosio, M.; Stegmann, E. Challenges and advances in genetic manipulation of filamentous actinomycetes—the remarkable producers of specialised metabolites. Nat. Prod. Rep. 2019, 36, 1351–1369. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Kazempour, D.; Wohlleben, W.; Weber, T. Improved Lanthipeptide Detection and Prediction for antiSMASH. PLoS ONE 2014, 9, e89420. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The Natural Product Domain Seeker NaPDoS: A Phylogeny Based Bioinformatic Tool to Classify Secondary Metabolite Gene Diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Luzhetskyy, A. Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 2019, 36, 1281–1294. [Google Scholar] [CrossRef]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces Species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Yamazaki, H.; Kato, J.-Y.; Tomono, A.; Horinouchi, S. AdpA, a central transcriptional regulator in the A-Factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 2005, 69, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Uguru, G.C.; Stephens, K.E.; Stead, J.A.; Towle, J.E.; Baumberg, S.; McDowall, K.J. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 2005, 58, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Higo, A.; Horinouchi, S.; Ohnishi, Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 2011, 81, 1607–1622. [Google Scholar] [CrossRef]
- Iqbal, M.; Mast, Y.; Amin, R.; Hodgson, D.A.; Wohlleben, W.; Burroughs, N.J. Stream Consortium Extracting regulator activity profiles by integration of de novo motifs and expression data: Characterizing key regulators of nutrient depletion responses in Streptomyces coelicolor. Nucleic Acids Res. 2012, 40, 5227–5239. [Google Scholar] [CrossRef]
- Hou, B.; Zhu, X.; Kang, Y.; Wang, R.; Wu, H.; Ye, J.; Zhang, H. LmbU, a Cluster-Situated Regulator for Lincomycin, Consists of a DNA-Binding Domain, an Auto-Inhibitory Domain, and Forms Homodimer. Front. Microbiol. 2019, 10, 989. [Google Scholar] [CrossRef]
- Zou, Z.; Du, D.; Zhang, Y.; Zhang, J.; Niu, G.; Tan, H. A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol. Microbiol. 2014, 94, 490–505. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, L.; He, X.; Tian, Y.; Liu, G.; Tan, H. SabR enhances nikkomycin production via regulating the transcriptional level of sanG, a pathway-specific regulatory gene in Streptomyces ansochromogenes. BMC Microbiol. 2011, 11, 164. [Google Scholar] [CrossRef]
- Li, R.; Xie, Z.; Tian, Y.; Yang, H.; Chen, W.; You, D.; Liu, G.; Deng, Z.; Tan, H. polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology 2009, 155, 1819–1831. [Google Scholar] [CrossRef][Green Version]
- Laureti, L.; Song, L.; Huang, S.; Corre, C.; Leblond, P.; Challis, G.L.; Aigle, B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 2011, 108, 6258–6263. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Han, L.; Vining, L.C. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: Involvement of a repressor gene, jadR2. J. Bacteriol. 1995, 177, 6111–6117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Spohn, M.; Kirchner, N.; Kulik, A.; Jochim, A.; Wolf, F.; Muenzer, P.; Borst, O.; Gross, H.; Wohlleben, W.; Stegmann, E. Overproduction of ristomycin a by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 2014, 58, 6185–6196. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.A. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 2007, 107, 3467–3497. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Robledo-Casados, I.; Sánchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1849, 1017–1039. [Google Scholar] [CrossRef]
- Shadel, G.S.; Baldwin, T.O. The Vibrio fischeri LuxR protein is capable of bidirectional stimulation of transcription and both positive and negative regulation of the luxR gene. J. Bacteriol. 1991, 173, 568–574. [Google Scholar] [CrossRef]
- Takano, E.; Chakraburtty, R.; Nihira, T.; Yamada, Y.; Bibb, M.J. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 2008, 41, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Antón, N.; Santos-Aberturas, J.; Mendes, M.V.; Guerra, S.M.; Martín, J.F.; Aparicio, J.F. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 2007, 153, 3174–3183. [Google Scholar] [CrossRef]
- Goranovič, D.; Blažič, M.; Magdevska, V.; Horvat, J.; Kuščer, E.; Polak, T.; Santos-Aberturas, J.; Martínez-Castro, M.; Barreiro, C.; Mrak, P.; et al. FK506 biosynthesis is regulated by two positive regulatory elements in Streptomyces tsukubaensis. BMC Microbiol. 2012, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez-Robles, M.; Rodríguez-García, A.; Martín, J.F. Target genes of the Streptomyces tsukubaensis FkbN regulator include most of the tacrolimus biosynthesis genes, a phosphopantetheinyl transferase and other PKS genes. Appl. Microbiol. Biotechnol. 2016, 100, 8091–8103. [Google Scholar] [CrossRef]
- Grove, A. MarR family transcription factors. Curr. Biol. 2013, 23, R142–R143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Wang, Y.; Zhang, M.; Ikeda, H.; Deng, Z.; Cane, D.E. Product-mediated regulation of pentalenolactone biosynthesis in Streptomyces species by the MarR/SlyA family activators PenR and PntR. J. Bacteriol. 2013, 195, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Beppu, T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism inStreptomyces griseus. Mol. Microbiol. 1994, 12, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Tsigkinopoulou, A.; Takano, E.; Breitling, R. Unravelling the γ-butyrolactone network in Streptomyces coelicolor by computational ensemble modelling. PLoS Comput. Biol. 2020, 16, e1008039. [Google Scholar] [CrossRef]
- Retzlaff, L.; Distler, J. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 1995, 18, 151–162. [Google Scholar] [CrossRef]
- Krause, J.; Handayani, I.; Blin, K.; Kulik, A.; Mast, Y. Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in Streptomycetes. Front. Microbiol. 2020, 11, 225. [Google Scholar] [CrossRef]
- Wietzorrek, A.; Bibb, M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 1997, 25, 1181–1184. [Google Scholar] [CrossRef]
- Arias, P.; Moreno, M.A.F.; Malpartida, F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-Binding Protein. J. Bacteriol. 1999, 181, 6958–6968. [Google Scholar] [CrossRef]
- Wang, R.; Mast, Y.; Wang, J.; Zhang, W.; Zhao, G.; Wohlleben, W.; Lu, Y.; Jiang, W. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol. 2012, 87, 30–48. [Google Scholar] [CrossRef]
- Wang, L.; Tian, X.; Wang, J.; Yang, H.; Fan, K.; Xu, G.; Yang, K.; Tan, H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. USA 2009, 106, 8617–8622. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Homerova, D.; Feckova, L.; Kormanec, J.; Naylor, S.W.; Roe, A.J.; Nart, P.; Spears, K.; Smith, D.G.E.; Low, J.C.; et al. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 2005, 151, 2693–2706. [Google Scholar] [CrossRef] [PubMed]
- Matulova, M.; Feckova, L.; Novakova, R.; Mingyar, E.; Csolleiova, D.; Zduriencikova, M.; Sedlak, J.; Patoprsty, V.; Sasinkova, V.; Uhliarikova, I.; et al. A structural analysis of the angucycline-like antibiotic auricin from Streptomyces lavendulae subsp. lavendulae CCM 3239 revealed its high similarity to griseusins. Antibiotics 2019, 8, 102. [Google Scholar] [CrossRef]
- Hiard, S.; Marée, R.; Colson, S.; Hoskisson, P.A.; Titgemeyer, F.; van Wezel, G.P.; Joris, B.; Wehenkel, L.; Rigali, S. PREDetector: A new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 2007, 357, 861–864. [Google Scholar] [CrossRef]
- Daniel-Ivad, M.; Hameed, N.; Tan, S.; Dhanjal, R.; Socko, D.; Pak, P.; Gverzdys, T.; Elliot, M.A.; Nodwell, J.R. An engineered allele of afsQ1 facilitates the discovery and investigation of cryptic natural products. ACS Chem. Biol. 2017, 12, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hosaka, T.; Ochi, K. Rare earth elements activate the secondary metabolite–biosynthetic gene clusters in Streptomyces coelicolor A3(2). J. Antibiot. 2010, 63, 477–481. [Google Scholar] [CrossRef]
- Wang, B.; Guo, F.; Dong, S.-H.; Zhao, H. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 2018, 15, 111–114. [Google Scholar] [CrossRef]
- Rutledge, P.; Challis, G. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Genet. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Sun, Y.-Q.; Busche, T.; Rückert, C.; Paulus, C.; Rebets, Y.; Novakova, R.; Kalinowski, J.; Luzhetskyy, A.; Kormanec, J.; Sekurova, O.N.; et al. Development of a biosensor concept to detect the production of cluster-specific secondary metabolites. ACS Synth. Biol. 2017, 6, 1026–1033. [Google Scholar] [CrossRef]
- Menges, R.; Muth, G.; Wohlleben, W.; Stegmann, E. The ABC transporter Tba of Amycolatopsis balhimycina is required for efficient export of the glycopeptide antibiotic balhimycin. Appl. Microbiol. Biotechnol. 2007, 77, 125–134. [Google Scholar] [CrossRef]
- Reynolds, D.M.; Schatz, A.; Waksman, S.A. Grisein, a new antibiotic produced by a strain of Streptomyces griseus. Exp. Biol. Med. 1947, 64, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Corbaz, R.; Ettlinger, L.; Gäumann, E.; Keller-Schierlein, W.; Kradolfer, F.; Neipp, L.; Prelog, V.; Zahner, H. Stoffwechselprodukte von Actinomyceten. 3. Mitteilung. Nonactin. Helvetica Chim. Acta 1955, 38, 1445–1448. [Google Scholar] [CrossRef]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a cytotoxic polyketide from a Streptomyces strain isolated from the Marine SpongeHalichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Bo, S.T.; Xu, Z.F.; Yang, L.; Cheng, P.; Tan, R.X.; Jiao, R.H.; Ge, H.M. Structure and biosynthesis of mayamycin B, a new polyketide with antibacterial activity from Streptomyces sp. 120454. J. Antibiot. 2018, 71, 601–605. [Google Scholar] [CrossRef]
- Xu, Z.; Jakobi, K.; Welzel, K.; Hertweck, C. Biosynthesis of the Antitumor Agent Chartreusin Involves the Oxidative Rearrangement of an Anthracyclic Polyketide. Chem. Biol. 2005, 12, 579–588. [Google Scholar] [CrossRef]
- Denny, W.A. Anti Cancer: DNA Topoisomerase Inhibitors. In Comprehensive Medicinal Chemistry II; Elsevier: London, UK, 2007; Volume 7, pp. 111–128. [Google Scholar]
- Helaly, S.E.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Süssmuth, R.D.; Fiedler, H.-P. Warkmycin, a novel angucycline antibiotic produced by Streptomyces sp. Acta 2930. J. Antibiot. 2013, 66, 669–674. [Google Scholar] [CrossRef]
- Malmierca, M.G.; González-Montes, L.; Pérez-Victoria, I.; Sialer, C.A.; Braña, A.F.; Salcedo, R.G.; Martín, J.; Reyes, F.; Méndez, C.; Olano, C.; et al. Searching for glycosylated natural products in actinomycetes and identification of novel macrolactams and angucyclines. Front. Microbiol. 2018, 9, 39. [Google Scholar] [CrossRef]
- Kormanec, J.; Novakova, R.; Mingyar, E.; Feckova, L. Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl. Microbiol. Biotechnol. 2014, 98, 45–60. [Google Scholar] [CrossRef]
- Endo, G.; Silver, S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1995, 177, 4437–4441. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. (Eds.) Current Protocols in Molecular Biology; Wiley: New York, NY, USA, 1995. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]

| Strain | Regulatory Genes |
|---|---|
| Streptomyces griseus DSM 40236 | griR, strR |
| Streptomyces coelicolor M145 | actIIORF4, redD |
| Streptomyces tsukubaensis | fkbN |
| Streptomyces arenae TÜ469 | pntR |
| Streptomyces pristinaespiralis | papR2 |
| Cosmid pCos51 (Novakova et al., 2005) | aur1P and aur1PR3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mingyar, E.; Mühling, L.; Kulik, A.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Blin, K.; Weber, T.; Wohlleben, W.; Stegmann, E. A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters. Int. J. Mol. Sci. 2021, 22, 7567. https://doi.org/10.3390/ijms22147567
Mingyar E, Mühling L, Kulik A, Winkler A, Wibberg D, Kalinowski J, Blin K, Weber T, Wohlleben W, Stegmann E. A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters. International Journal of Molecular Sciences. 2021; 22(14):7567. https://doi.org/10.3390/ijms22147567
Chicago/Turabian StyleMingyar, Erik, Lucas Mühling, Andreas Kulik, Anika Winkler, Daniel Wibberg, Jörn Kalinowski, Kai Blin, Tilmann Weber, Wolfgang Wohlleben, and Evi Stegmann. 2021. "A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters" International Journal of Molecular Sciences 22, no. 14: 7567. https://doi.org/10.3390/ijms22147567
APA StyleMingyar, E., Mühling, L., Kulik, A., Winkler, A., Wibberg, D., Kalinowski, J., Blin, K., Weber, T., Wohlleben, W., & Stegmann, E. (2021). A Regulator Based “Semi-Targeted” Approach to Activate Silent Biosynthetic Gene Clusters. International Journal of Molecular Sciences, 22(14), 7567. https://doi.org/10.3390/ijms22147567








