Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase
Abstract
1. Introduction
2. Mitochondrial Protein Translocases
3. Discovery of Tim50
4. Tim50 Primary Structure and Membrane Topology
4.1. Tim50 tertiary Structure
4.2. Role of Tim50 in Mitochondrial Protein Import
5. The Role of Tim50 in Other Cellular Processes
5.1. The Role of TIMM50 in Steroidogenesis
5.2. Tim50 Mutations in Genetic Disorders
5.3. TIMM50 in Cardiac Function
5.4. TIMM50 in Cancer
5.5. The Role of TbTim50 in T. brucei Biology
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medini, H.; Cohen, T.; Mishmar, D. Mitochondria are fundamental for the emergence of metazoans: On metabolism, genomic regulation, and the birth of complex organisms. Annu. Rev. Genet. 2020, 54, 151–166. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Gincel, D. The voltage-dependent anion channel: Characterization, modulation, and role in cell life and death. Cell. Biochem. Biophys. 2003, 39, 279–292. [Google Scholar] [CrossRef]
- Urbani, A.; Prosdocimi, E.; Carrer, A.; Checchetto, V.; Szabò, I. Mitochondrial ion channels of the inner membrane and their regulation in cell death signaling. Front. Cell Dev. Biol. 2021, 8, 620081. [Google Scholar] [CrossRef]
- Wollweber, F.; von der Malsburg, K.; van der Laan, M. Mitochondrial contact site and cristae organizing system: A central player in membrane shaping and crosstalk. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1481–1489. [Google Scholar] [CrossRef]
- Javadov, S.; Jang, S.; Chapa-Dubocq, X.R.; Khuchua, Z.; Camara, A.K. Mitochondrial respiratory supercomplexes in mammalian cells: Structural versus functional role. J. Mol. Med. 2021, 99, 57–73. [Google Scholar] [CrossRef]
- Palmfeldt, J.; Bross, P. Proteomics of human mitochondria. Mitochondrion 2017, 33, 2–14. [Google Scholar] [CrossRef]
- Rao, R.S.; Salvato, F.; Thal, B.; Eubel, H.; Thelen, J.J.; Møller, I.M. The proteome of higher plant mitochondria. Mitochondrion 2017, 33, 22–37. [Google Scholar] [CrossRef]
- Schmidt, O.; Pfanner, N.; Meisinger, C. Mitochondrial protein import: From proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 655–667. [Google Scholar] [CrossRef]
- Neupert, W.; Herrmann, J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007, 76, 723–749. [Google Scholar] [CrossRef]
- Baker, M.J.; Frazier, A.E.; Gulbis, J.M.; Ryan, M.T. Mitochondrial protein import machinery: Correlating structure with function. Trends Cell Biol. 2007, 17, 456–464. [Google Scholar] [CrossRef]
- Diederichs, K.A.; Buchanan, S.K.; Botos, I. Building better barrels-beta-barrel biogenesis and insertion in bacteria and mitochondria. J. Mol. Biol. 2021, 24, 166894. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Eaglesfield, R.; Tokatlidis, K. The mitochondrial intermembrane space: The most constricted mitochondrial sub-compartment with the largest variety of protein import pathways. Open Biol. 2021, 11, 210002. [Google Scholar] [CrossRef] [PubMed]
- Chacinska, A.; Rehling, P.; Guiard, B.; Frazier, A.E.; Schulze-Specking, A.; Pfanner, N.; Voos, W.; Meisinger, C. Mitochondrial translocation contact sites: Separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 2003, 22, 5370–5381. [Google Scholar] [CrossRef] [PubMed]
- Roise, D.J. Recognition and binding of mitochondrial presequence during the import of proteins into mitochondria. Bioenerg. Biomembr. 1997, 29, 19–27. [Google Scholar] [CrossRef]
- Bauer, M.F.; Gempel, K.; Reichert, A.S.; Rappold, G.A.; Lichtner, P.; Gerbitz, K.D.; Neupert, W.; Brunner, M.; Hofmann, S. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 1999, 289, 69–82. [Google Scholar] [CrossRef]
- Lister, R.; Hulett, J.M.; Lithgow, T.; Whelan, J. Protein import into mitochondria: Origins and functions today. Mol. Membr. Biol. 2005, 22, 87–100. [Google Scholar] [CrossRef]
- Harsman, A.; Schneider, A. Mitochondrial protein import in trypanosomes: Expect the unexpected. Traffic 2017, 18, 96–109. [Google Scholar] [CrossRef]
- Chaudhuri, M.; Darden, C.; Gonzalez, F.S.; Singha, U.K.; Quinones, L.; Tripathi, A. Tim17 updates: A comprehensive review of an ancient mitochondrial protein translocator. Biomolecules 2020, 10, 1643. [Google Scholar] [CrossRef]
- Rada, P.; Doležal, P.; Jedelský, P.L.; Bursac, D.; Perry, A.J.; Šedinová, M.; Smíšková, K.; Novotný, M.; Beltrán, N.C.; Hrdý, I.; et al. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS ONE 2011, 6, e24428. [Google Scholar] [CrossRef]
- Dolezal, P.; Dagley, M.J.; Kono, M.; Wolynec, P.; Likić, V.A.; Foo, J.H.; Sedinová, M.; Tachezy, J.; Bachmann, A.; Bruchhaus, I.; et al. The essentials of protein import in the degggerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 2010, 6, e1000812. [Google Scholar] [CrossRef]
- Dekker, P.J.T.; Ryan, M.T.; Brix, J.; Müller, H.; Hönlinger, A.; Pfanner, N. Preprotein translocase of the outer mitochondrial membrane: Molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 1998, 18, 6515–6524. [Google Scholar] [CrossRef]
- Kozjak, V.; Wiedemann, N.; Milenkovic, D.; Lohaus, C.; Meyer, H.E.; Guiard, B.; Meisinger, C.; Pfanner, N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 2003, 278, 48520–48523. [Google Scholar] [CrossRef]
- Becker, T.; Pfannschmidt, S.; Guiard, B.; Stojanovski, D.; Milenkovic, D.; Kutik, S.; Pfanner, N.; Meisinger, C.; Wiedemann, N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 2008, 283, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Bomer, U.; Rassow, J.; Zufall, N.; Pfanner, N.; Meijer, M.; Maarse, A.C. The preprotein translocase of the inner mitochondrial membrane: Evolutionary conservation of targeting and assembly of Tim17. J. Mol. Biol. 1996, 262, 389–395. [Google Scholar] [CrossRef]
- Sirrenberg, C.; Bauer, M.F.; Guiard, B.; Neupert, W.; Brunner, M. Import of carrier proteins into the mitochondria inner membrane mediated by Tim22. Nature 1996, 384, 582–585. [Google Scholar] [CrossRef]
- Schilke, B.A.; Hayashi, M.; Craig, E.A. Genetic analysis. Of complex interactions among. Components of the mitochondrial import motor and translocon in Saccharomyces cerevisiae. Genetics 2012, 190, 1341–1353. [Google Scholar] [CrossRef][Green Version]
- McDowell, M.A.; Heimes, M.; Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by Oxa1 superfamily. Nat. Struct. Mol. Biol. 2021, 28, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Künkele, K.P.; Heins, S.; Dembowski, M.; Nargang, F.E.; Benz, R.; Thieffry, M.; Walz, J.; Lill, R.; Nussberger, S.; Neupert, W. The preprotein translocation channel of the outer membrane of mitochondria. Cell 1998, 93, 1009–1019. [Google Scholar] [CrossRef]
- Suzuki, H.; Okazawa, Y.; Komiya, T.; Saeki, K.; Mekada, E.; Kitada, S.; Ito, A.; Mihara, K. Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J. Biol. Chem. 2000, 275, 37930–37936. [Google Scholar] [CrossRef]
- Court, D.A.; Nargang, F.E.; Steiner, H.; Hodges, R.S.; Neupert, W.; Lill, R. Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 1996, 16, 4035–4042. [Google Scholar] [CrossRef][Green Version]
- Moczko, M.; Bömer, U.; Kübrich, M.; Zufall, N.; Hönlinger, A.; Pfanner, N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 1997, 17, 6574–6584. [Google Scholar] [CrossRef] [PubMed]
- Truscott, K.N.; Kovermann, P.; Geissler, A.; Merlin, A.; Meijer, M.; Driessen, A.J.M.; Rassow, J.; Pfanner, N.; Wagner, R. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 2001, 8, 1074–1082. [Google Scholar] [CrossRef]
- Rehling, P.; Model, K.; Brandner, K.; Kovermann, P.; Sickmann, A.; Meyer, H.E.; Kühlbrandt, W.; Wagner, R.; Truscott, K.N.; Pfanner, N. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 2003, 299, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Caballero, S.; Grigoriev, S.M.; Herrmann, J.M.; Campo, M.L.; Kinnally, K.W. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 2007, 282, 3584–3593. [Google Scholar] [CrossRef] [PubMed]
- Mokranjac, D.; Sichting, M.; Popov-Celeketić, D.; Mapa, K.; Gevorkyan-Airapetov, L.; Zohary, K.; Hell, K.; Azem, A.; Neupert, W. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell 2009, 20, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Tamura, Y.; Harada, Y.; Shiota, T.; Yamano, K.; Watanabe, K.; Yokota, M.; Yamamoto, H.; Sesaki, H.; Endo, T. Tim23–Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 2009, 184, 129–141. [Google Scholar] [CrossRef]
- Ting, S.Y.; Schilke, B.A.; Hayashi, M.; Craig, E.A. Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor. J. Biol. Chem. 2014, 289, 28689–28696. [Google Scholar] [CrossRef]
- Chacinska, A.; Rehling, P. Moving proteins from the cytosol into mitochondria. Biochem. Soc. Trans. 2004, 32, 774–776. [Google Scholar] [CrossRef]
- Van der Laan, M.; Hutu, D.P.; Rehling, P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 732–739. [Google Scholar] [CrossRef]
- Koehler, C.M.; Jarosch, E.; Tokatlidis, K.; Schmid, K.; Schweyen, R.J.; Schatz, G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 1998, 279, 369–373. [Google Scholar] [CrossRef]
- Kerscher, O.; Holder, J.; Srinivasan, M.; Leung, R.S.; Jensen, R.E. The Tim54p-Tim22 complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 1997, 139, 1663–1675. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N.; Chacinska, A. Chaperoning through the mitochondrial intermembrane space. Mol. Cell 2006, 21, 145–148. [Google Scholar] [CrossRef]
- Weinhäupl, K.; Lindau, C.; Hessel, A.; Wang, Y.; Schütze, C.; Jores, T.; Melchionda, L.; Schönfisch, B.; Kalbacher, H.; Bersch, B.; et al. Structural basis of membrane protein chaperoning through the mitochondrial intermembrane space. Cell 2018, 175, 1365–1379. [Google Scholar] [CrossRef]
- Sternberg, J.M.; MacLean, L. A spectrum of disease in Human African trypanosomiasis: The host and parasite genetics of virulence. Parasitology 2010, 137, 2007–2015. [Google Scholar] [CrossRef]
- De Koning, H.P. The drugs of sleeping sickness: Their mechanisms of action and resistance, and a brief history. Trop. Med. Infect. Dis. 2015, 5, 14. [Google Scholar] [CrossRef]
- Peña-Diaz, P.; Lukeš, J. Fe-S cluster assembly in the supergroup Excavata. J. Biol. Chem. 2018, 23, 521–524. [Google Scholar] [CrossRef]
- Haanstra, J.R.; Gerding, A.; Dolga, A.M.; Sorgdrager, F.J.H.; Buist-Homan, M.; du Toit, F.; Faber, K.N.; Holzhütter, H.G.; Szöör, B.; Matthews, K.R.; et al. Targeting pathogen metabolism without collateral damage to the host. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shlomai, J. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 2005, 4, 623–647. [Google Scholar] [CrossRef]
- Schneider, A. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 2001, 1403–1415. [Google Scholar] [CrossRef]
- Jensen, R.E.; Englund, P.T. Network news: The replication of kinetoplast DNA. Annu. Rev. Microbiol. 2012, 66, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Acestor, N.; Panigrahi, A.K.; Ogata, Y.; Anupama, A.; Stuart, K.D. Protein composition of Trypanosoma brucei mitochondrial membranes. Proteomics 2009, 9, 5497–5508. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, J.; Nilsson, D.; Gunasekera, K.; Chanfon, A.; Song, X.; Wang, H.; Xu, Y.; Ochsenreiter, T. The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 2010, 38, 7378–7387. [Google Scholar] [CrossRef]
- Pusnik, M.; Schmidt, O.; Perry, A.J.; Oeljeklaus, S.; Niemann, M.; Warscheid, B.; Lithgow, T.; Meisinger, C.; Schneider, A. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 2011, 21, 1738–1743. [Google Scholar] [CrossRef]
- Harsman, A.; Niemann, M.; Pusnik, M.; Schmidt, O.; Burmann, B.M.; Hiller, S.; Meisinger, C.; Schneider, A.; Wagner, R. Bacterial origin of a mitochondrial outer membrane protein translocase: New perspectives from comparative single channel electrophysiology. J. Biol. Chem. 2012, 287, 31437–31445. [Google Scholar] [CrossRef]
- Singha, U.K.; Hamilton, V.N.; Duncan, M.R.; Weems, E.; Tripathi, M.K.; Chaudhuri, M. Protein translocase of mitochondrial inner membrane in Trypanosoma brucei. J. Biol. Chem. 2012, 287, 14480–14493. [Google Scholar] [CrossRef] [PubMed]
- Harsman, A.; Oeljeklaus, S.; Wenger, C.; Huot, J.L.; Warscheid, B.; Schneider, A. The non-canonical mitochondrial inner membrane presequence translocase of trypanosomatids contains two essential rhomboid-like proteins. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Weems, E.; Singha, U.K.; Hamilton, V.N.; Smith, J.T.; Waegemann, K.; Mokranjac, D.; Chaudhuri, M. Functional complementation analyses reveal that the single PRAT family protein of trypanosoma brucei is a divergent homolog of Tim17 in Saccharomyces cerevisiae. Eukaryot. Cell 2015, 14, 286–296. [Google Scholar] [CrossRef]
- Duncan, M.R.; Fullerton, M.; Chaudhuri, M. Tim50 in Trypanosoma brucei possesses a dual specificity phosphatase activity and is critical for mitochondrial protein import. J. Biol. Chem. 2013, 288, 3184–3197. [Google Scholar] [CrossRef] [PubMed]
- Singha, U.K.; Hamilton, V.; Chaudhuri, M. Tim62, a novel mitochondrial protein in Trypanosoma brucei, is essential for assembly and stability of the TbTim17 protein complex. J. Biol. Chem. 2015, 290, 23226–23239. [Google Scholar] [CrossRef] [PubMed]
- Singha, U.K.; Tripathi, A.; Smith, J.T.; Quinones, L.; Saha, A.; Singha, T.; Chaudhuri, M. Novel IM-associated protein Tim54 plays a role in the mitochondrial import of internal signal-containing proteins in Trypanosoma brucei. Biol. Cell 2021, 113, 39–57. [Google Scholar] [CrossRef]
- Wenger, C.; Oeljeklaus, S.; Warscheid, B.; Schneider, A.; Harsman, A. A trypanosomal orthologue of an intermembrane space chaperone has a non-canonical function in biogenesis of the single mitochondrial inner membrane protein translocase. PLoS Pathog. 2017, 13, e1006550. [Google Scholar] [CrossRef]
- Smith, J.T.; Singha, U.K.; Misra, S.; Chaudhuri, M. Divergent small Tim homologues are associated with TbTim17 and critical for the biogenesis of TbTim17 protein complexes in Trypanosoma brucei. mSphere 2018, 3, e00204-18. [Google Scholar] [CrossRef]
- Gebert, N.; Chacinska, A.; Wagner, K.; Guiard, B.; Koehler, C.M.; Rehling, P.; Pfanner, N.; Wiedemann, N. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep. 2008, 9, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Webb, C.T.; Stroud, D.A.; Palmer, C.S.; Frazier, A.E.; Guiard, B.; Chacinska, A.; Gulbis, J.M.; Ryan, M.T. Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol. Biol. Cell 2009, 20, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Geissler, A.; Chacinska, A.; Truscott, K.N.; Wiedemann, N.; Brandner, K.; Sickmann, A.; Meyer, H.E.; Meisinger, C.; Pfanner, N.; Rehling, P. The mitochondrial presequence translocase: An essential role of Tim50 in directing preproteins to the import channel. Cell 2002, 111, 507–518. [Google Scholar] [CrossRef]
- Yamamoto, H.; Esaki, M.; Kanamori, T.; Tamura, Y.; Nishikawa, S.I.; Endo, T. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 2002, 111, 519–528. [Google Scholar] [CrossRef]
- Mokranjac, D.; Paschen, S.A.; Kozany, C.; Prokisch, H.; Hoppins, S.C.; Nargang, F.E.; Neupert, W.; Hell, K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003, 22, 816–825. [Google Scholar] [CrossRef]
- Sugiyama, S.; Moritoh, S.; Furukawa, Y.; Mizuno, T.; Lim, Y.M.; Tsuda, L.; Nishida, Y. Involvement of the mitochondrial protein translocator component Tim50 in growth, cell proliferation and the modulation of respiration in drosophila. Genetics 2007, 176, 927–936. [Google Scholar] [CrossRef]
- Guo, Y.; Cheong, N.E.; Zhang, Z.J.; de Rose, R.; Deng, Y.; Farber, S.A.; Fernandes-Alnemri, T.; Alnemri, E.S. Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J. Biol. Chem. 2004, 279, 24813–24825. [Google Scholar] [CrossRef]
- Xu, H.; Somers, Z.B.; Robinson, M.L.; Hebert, M.D. Tim50a, a nuclear isoform of the mitochondrial Tim50, interacts with proteins involved in snRNP biogenesis. BMC Cell Biol. 2005, 6, 1–14. [Google Scholar] [CrossRef]
- Staněk, D. Cajal bodies and snRNPs—Friends with benefits. RNA Biol. 2017, 14, 671–679. [Google Scholar] [CrossRef]
- Kamenski, T.; Heilmeier, S.; Meinhart, A.; Cramer, P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 2004, 15, 399–407. [Google Scholar] [CrossRef]
- Ghosh, A.; Shuman, S.; Lima, C.D. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell 2008, 32, 478–490. [Google Scholar] [CrossRef]
- Tripathi, A.; Singha, U.K.; Cooley, A.; Gillyard, T.; Krystofiak, E.; Pratap, S.; Davis, J.; Chaudhuri, M. Trypanosoma brucei Tim50 plays a critical role in cell cycle regulation and parasite infectivity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nejad, L.D.; Serricchio, M.; Jelk, J.; Hemphill, A.; Bütikofer, P. TbLpn, a key enzyme in lipid droplet formation and phospholipid metabolism, is essential for mitochondrial integrity and growth of Trypanosoma brucei. Mol. Microbiol. 2018, 109, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Han, G.S.; Wu, W.I.; Carman, G.M. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006, 281, 9210–9218. [Google Scholar] [CrossRef]
- Seifried, A.; Schultz, J.; Gohla, A. Human HAD phosphatases: Structure, mechanism, and roles in health and disease. FEBS J. 2013, 280, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Rallabandi, H.R.; Ganesan, P.; Kim, Y.J. Targeting the C-terminal domain small phosphatase 1. Life 2020, 10, 57. [Google Scholar] [CrossRef]
- Sun, S.; Liu, S.; Zhang, Z.; Zeng, W.; Sun, C.; Tao, T.; Lin, X.; Feng, X.H. Phosphatase UBLCP1 controls proteasome assembly. Open Biol. 2017, 7, 170042. [Google Scholar] [CrossRef]
- Hayata, T.; Chiga, M.; Ezura, Y.; Asashima, M.; Katabuchi, H.; Nishinakamura, R.; Noda, M. Dullard deficiency causes hemorrhage in the adult ovarian follicles. Genes Cells 2018, 23, 345–356. [Google Scholar] [CrossRef]
- Qian, X.; Gebert, M.; Höpker, J.; Yan, M.; Li, J.; Wiedemann, N.; van der Laan, M.; Pfanner, N.; Sha, B. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 2011, 411, 513–519. [Google Scholar] [CrossRef]
- Li, J.; Sha, B. The structure of Tim50(164–361) suggests the mechanism by which Tim50 receives mitochondrial presequences. Acta Crystallogr. Sect. Struct. Biol. Commun. 2015, 71, 1146–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.; Zhao, Q.; Ji, Z.; Li, Q.; Li, S.J. Expression and structural characterization of human translocase of inner membrane of mitochondria Tim50. Protein Expr. Purif. 2011, 80, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Lytovchenko, O.; Melin, J.; Schulz, C.; Kilisch, M.; Hutu, D.P.; Rehling, P. Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 2013, 32, 886–898. [Google Scholar] [CrossRef]
- Waegemann, K.; Popov-Čeleketić, D.; Neupert, W.; Azem, A.; Mokranjac, D. Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria. J. Mol. Biol. 2015, 427, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Gevorkyan-Airapetov, L.; Zohary, K.; Popov-Celeketic, D.; Mapa, K.; Hell, K.; Neupert, W.; Azem, A.; Mokranjac, D. Intraction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 2009, 284, 4865–4872. [Google Scholar] [CrossRef]
- Günsel, U.; Paz, E.; Gupta, R.; Mathes, I.; Azem, A.; Mokranjac, D. In vivo dissection of the intrinsically disordered receptor domain of Tim23. J. Mol. Biol. 2020, 432, 3326–3337. [Google Scholar] [CrossRef]
- Dayan, D.; Bandel, M.; Günsel, U.; Nussbaum, I.; Prag, G.; Mokranjac, D.; Neupert, W.; Azem, A. A mutagenesis analysis of Tim50, the major receptor of the TIM23 complex, identifies regions that affect its interaction with Tim23. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Alder, N.N.; Jensen, R.E.; Johnson, A.E. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 2008, 134, 439–450. [Google Scholar] [CrossRef]
- Malhotra, K.; Modak, A.; Nangia, S.; Daman, T.H.; Gunsel, U.; Robinson, V.L.; Mokranjac, D.; May, E.R.; Alder, N.N. Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci. Adv. 2017, 3, e1700532. [Google Scholar] [CrossRef]
- Caumont-Sarcos, A.; Moulin, C.; Poinot, L.; Guiard, B.; van der Laan, M.; Ieva, R. Transmembrane coordination of preprotein recognition and motor coupling by the mitochondrial presequence receptor Tim50. Cell Rep. 2020, 30, 3092–3104. [Google Scholar] [CrossRef]
- Matta, S.K.; Kumar, A.; D’Silva, P. Mgr2 regulates mitochondrial preprotein import by associating with channel-forming Tim23 subunit. Mol. Biol. Cell 2020, 31, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Fullerton, M.; Singha, U.K.; Duncan, M.; Chaudhuri, M. Down regulation of Tim50 in Trypanosoma brucei increases tolerance to oxidative stress. Mol. Biochem. Parasitol. 2015, 199, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Singha, U.K.; Paromov, V.; Hill, S.; Pratap, S.; Rose, K.; Chaudhuri, M. The cross talk between TbTim50 and PIP39, two aspartate-based protein phosphatases, maintains cellular homeostasis in Trypanosoma brucei. mSphere 2019, 4, e00353-19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, S.; Zhou, H.; Cai, L.; Li, J.; Liu, N.; Liu, Y.; Wang, L.; Fan, C.; Li, A.; et al. TIMM50 promotes tumor progression via ERK signaling and predicts poor prognosis of non-small cell lung cancer patients. Mol. Carcinog. 2019, 58, 767–776. [Google Scholar] [CrossRef]
- Kumar, S.; Yoshizumi, T.; Hongo, H.; Yoneda, A.; Hara, H.; Hamasaki, H.; Takahashi, N.; Nagata, N.; Shimada, H.; Matsui, M. Arabidopsis mitochondrial protein TIM50 affects hypocotyl cell elongation through intracellular ATP level. Plant Sci. 2012, 183, 212–217. [Google Scholar] [CrossRef]
- Meinecke, M.; Wagner, R.; Kovermann, P.; Guiard, B.; Mick, D.U.; Hutu, D.P.; Voos, W.; Truscott, K.N.; Chacinska, A.; Pfanner, N.; et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 2006, 312, 1523–1526. [Google Scholar] [CrossRef] [PubMed]
- Bose, H.S.; Gebrail, F.; Marshall, B.; Perry, E.W.; Whittal, R.M. Inner mitochondrial translocase Tim50 is central in adrenal and testicular steroid synthesis. Mol. Cell. Biol. 2018, 39, e00484-18. [Google Scholar] [CrossRef]
- Pawlak, K.J.; Prasad, M.; Thomas, J.L.; Whittal, R.M.; Bose, H.S. Inner mitochondrial translocase tim50 interacts with 3β-hydroxysteroid dehydrogenase type 2 to regulate adrenal and gonadal steroidogenesis. J. Biol. Chem. 2011, 286, 39130–39140. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.L.; Bose, H.S. Regulation of human 3-beta-hydroxysteroid dehydrogenase type-2 (3βHSD2) by molecular chaperones and the mitochondrial environment affects steroidogenesis. J. Steroid Biochem. Mol. Biol. 2015, 151, 74–84. [Google Scholar] [CrossRef]
- Tang, K.; Zhao, Y.; Li, H.; Zhu, M.; Li, W.; Liu, W.; Zhu, G.; Xu, D.; Peng, W.; Xu, Y.W. Translocase of inner membrane 50 functions as a novel protective regulator of pathological cardiac hypertrophy. J. Am. Heart Assoc. 2017, 6, e004346. [Google Scholar] [CrossRef]
- Sankala, H.; Vaughan, C.; Wang, J.; Deb, S.; Graves, P.R. Upregulation of the mitochondrial transport protein, Tim50, by mutant p53 contributes to cell growth and chemoresistance. Arch. Biochem. Biophys. 2011, 512, 52–60. [Google Scholar] [CrossRef]
- Gao, S.P.; Sun, H.F.; Jiang, H.L.; Li, L.D.; Hu, X.; Xu, X.E.; Jin, W. Loss of TIM50 suppresses proliferation and induces apoptosis in breast cancer. Tumor Biol. 2016, 37, 1279–1287. [Google Scholar] [CrossRef]
- Reyes, A.; Melchionda, L.; Burlina, A.; Robinson, A.J.; Ghezzi, D.; Zeviani, M. Mutations in TIMM50 compromise cell survival in OxPhos-dependent metabolic conditions. EMBO Mol. Med. 2018, 10, e8698. [Google Scholar] [CrossRef]
- Shahrour, M.A.; Staretz-Chacham, O.; Dayan, D.; Stephen, J.; Weech, A.; Damseh, N.; Pri Chen, H.; Edvardson, S.; Mazaheri, S.; Saada, A.; et al. Mitochondrial epileptic encephalopathy, 3-methylglutaconic aciduria and variable complex V deficiency associated with TIMM50 mutations. Clin. Genet. 2017, 91, 690–696. [Google Scholar] [CrossRef]
- Mir, A.; Hadab, S.; Sammak, M.; Alhazmi, R.; Housawi, Y.; Bashir, S. Complete resolution of epileptic spasms with vigabatrin in a patient with 3-methylglutaconic aciduria caused by TIMM50 gene mutation. Clin. Genet. 2020, 98, 102–103. [Google Scholar] [CrossRef] [PubMed]
- Tort, F.; Ugarteburu, O.; Texidó, L.; Gea-Sorlí, S.; García-Villoria, J.; Ferrer-Cortès, X.; Arias, Á.; Matalonga, L.; Gort, L.; Ferrer, I.; et al. Mutations in TIMM50 cause severe mitochondrial dysfunction by targeting key aspects of mitochondrial physiology. Hum. Mutat. 2019, 40, 1700–1712. [Google Scholar] [CrossRef] [PubMed]
- Niemi, N.M.; Wilson, G.M.; Overmyer, K.A.; Vögtle, F.N.; Myketin, L.; Lohman, D.C.; Schueler, K.L.; Attie, A.D.; Meisinger, C.; Coon, J.J.; et al. Pptc7 is an essential phosphatase for promoting mammalian mitochondrial metabolism and biogenesis. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L.; Bose, H.S. Early steps in steroidogenesis: Intracellular cholesterol trafficking. J. Lipid Res. 2011, 52, 2111–2135. [Google Scholar] [CrossRef]
- Miller, W.L.; Auchus, R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011, 32, 81–151. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef]
- Chang, T.Y.; Reid, P.C.; Sugii, S.; Ohgami, N.; Cruz, J.C.; Chang, C.C.Y. Niemann-Pick type C disease and intracellular cholesterol trafficking. J. Biol. Chem. 2005, 280, 20917–20920. [Google Scholar] [CrossRef] [PubMed]
- Clark, B.J.; Stocco, D.M. Expression of the steroidogenic acute regulatory (stAR) protein: A novel LH-induced mitochondrial protein required for the acute regulation of steroidogenesis in mouse leydig tumor cells. Endocr. Res. 1995, 21, 243–257. [Google Scholar] [CrossRef]
- Vamecq, J.; Papegay, B.; Nuyens, V.; Boogaerts, J.; Leo, O.; Kruys, V. Mitochondrial dysfunction, AMPK activation and peroxisomal metabolism: A coherent scenario for non-canonical 3-methylglutaconic acidurias. Biochimie 2020, 168, 53–82. [Google Scholar] [CrossRef] [PubMed]
- Zegallai, H.M.; Hatch, G.M. Barth syndrome: Cardiolipin, cellular pathophysiology, management, and novel therapeutic targets. Mol. Cell. Biochem. 2021, 476, 1605–1629. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Hartmann, M.; Rehling, P. The role of mitochondrial cardiolipin in heart function and its implication in cardiac disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, R.; Gonzalez, B.; Johnson, S.C. Mitochondrial pathways in human health and aging. Mitochondrion 2020, 54, 72–84. [Google Scholar] [CrossRef]
- Sedlák, E.; Kožár, T.; Musatov, A. The interplay among subunit composition, cardiolipin content, and aggregation state of bovine heart cytochrome c oxidase. Cells 2020, 9, 2588. [Google Scholar] [CrossRef]
- Gonzalvez, F.; D’Aurelio, M.; Boutant, M.; Moustapha, A.; Puech, J.P.; Landes, T.; Arnauné-Pelloquin, L.; Vial, G.; Taleux, N.; Slomianny, C.; et al. Barth syndrome: Cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1194–1206. [Google Scholar] [CrossRef]
- Spencer, C.T.; Bryant, R.M.; Day, J.; Gonzalez, I.L.; Colan, S.D.; Thompson, W.R.; Berthy, J.; Redfearn, S.P.; Byrne, B.J. Cardiac and clinical phenotype in Barth syndrome. Pediatrics 2006, 118, e337–e346. [Google Scholar] [CrossRef]
- Jordens, E.Z.; Palmieri, L.; Huizing, M.; van den Heuvel, L.P.; Sengers, R.C.A.; Dörner, A.; Ruitenbeek, W.; Trijbels, F.J.; Valsson, J.; Sigfusson, G.; et al. Adenine nucleotide translocator 1 deficiency associated with Sengers syndrome. Ann. Neurol. 2002, 52, 95–99. [Google Scholar] [CrossRef]
- Mayr, J.A.; Haack, T.B.; Graf, E.; Zimmermann, F.A.; Wieland, T.; Haberberger, B.; Superti-Furga, A.; Kirschner, J.; Steinmann, B.; Baumgartner, M.R.; et al. Lack of the mitochondrial protein acylglycerol kinase causes sengers syndrome. Am. J. Hum. Genet. 2012, 90, 314–320. [Google Scholar] [CrossRef]
- Claypool, S.M. Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim. Biophys. Acta Biomembr. 2009, 1788, 2059–2068. [Google Scholar] [CrossRef]
- Vaughan, C.; Pearsall, I.; Yeudall, A.; Deb, S.P.; Deb, S. p53: Its mutations and their impact on transcription. Subcell. Biochem. 2014, 85, 71–90. [Google Scholar] [CrossRef]
- Yang, L.; Kong, D.; He, M.; Gong, J.; Nie, Y.; Tai, S.; Teng, C.-B. miR-7 induces regulated cell death in rhabdomyosarcoma though its targets in mitochondria. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Wei, A.W.; Li, L.F. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed. Pharmacother. 2017, 96, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Quintana, J.F.; Zoltner, M.; Field, M.C. Evolving differentiation in African trypanosomes. Trends Parasitol. 2021, 37, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Szöőr, B.; Silvester, E.; Matthews, K.R. A leap into the unknown—Early events in african trypanosome transmission. Trends Parasitol. 2020, 36, 266–278. [Google Scholar] [CrossRef]
- Chaudhuri, M.; Ott, R.D.; Hill, G.C. Trypanosome alternative oxidase: From molecule to function. Trends Parasitol. 2006, 22, 484–491. [Google Scholar] [CrossRef]
- Verner, Z.; Basu, S.; Benz, C.; Dixit, S.; Dobáková, E.; Faktorová, D.; Hashimi, H.; Horáková, E.; Huang, Z.; Paris, Z.; et al. Malleable mitochondrion of Trypanosoma brucei. Int. Rev. Cell Mol. Biol. 2015, 315, 73–151. [Google Scholar] [PubMed]
- Schnaufer, A.; Clark-Walker, G.D.; Steinberg, A.G.; Stuart, K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005, 24, 4029–4040. [Google Scholar] [CrossRef]
- Brown, S.V.; Hosking, P.; Li, J.; Williams, N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot. Cell 2006, 5, 45–53. [Google Scholar] [CrossRef][Green Version]
- Szöőr, B.; Simon, D.V.; Rojas, F.; Young, J.; Robinson, D.R.; Krüger, T.; Engstler, M.; Matthews, K.R. Positional dynamics and glycosomal recruitment of developmental regulators during trypanosome differentiation. MBio 2019, 10, 875–894. [Google Scholar] [CrossRef] [PubMed]
- Michels, P.A.M.; Villafraz, O.; Pineda, E.; Alencar, M.B.; Cáceres, A.J.; Silber, A.M.; Bringaud, F. Carbohydrate metabolism in trypanosomatids: New insight. Revealing novel complexity, diversity and species-unique features. Exp. Parasitol. 2021, 224, 108102. [Google Scholar] [CrossRef] [PubMed]
- Dean, S.; Gould, M.K.; Dewar, C.E.; Schnaufer, A.C. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc. Natl. Acad. Sci. USA 2013, 110, 14741–14746. [Google Scholar] [CrossRef]
- Serricchio, M.; Hierro-Yap, C.; Schädeli, D.; Hamidane, H.B.; Hemphill, A.; Graumann, J.; Zíková, A.; Bütikofer, P. Depletion of cardiolipin induces major changes in energy metabolism in Trypanosoma brucei bloodstream forms. FASEB J. 2021, 35, e21176. [Google Scholar] [CrossRef]
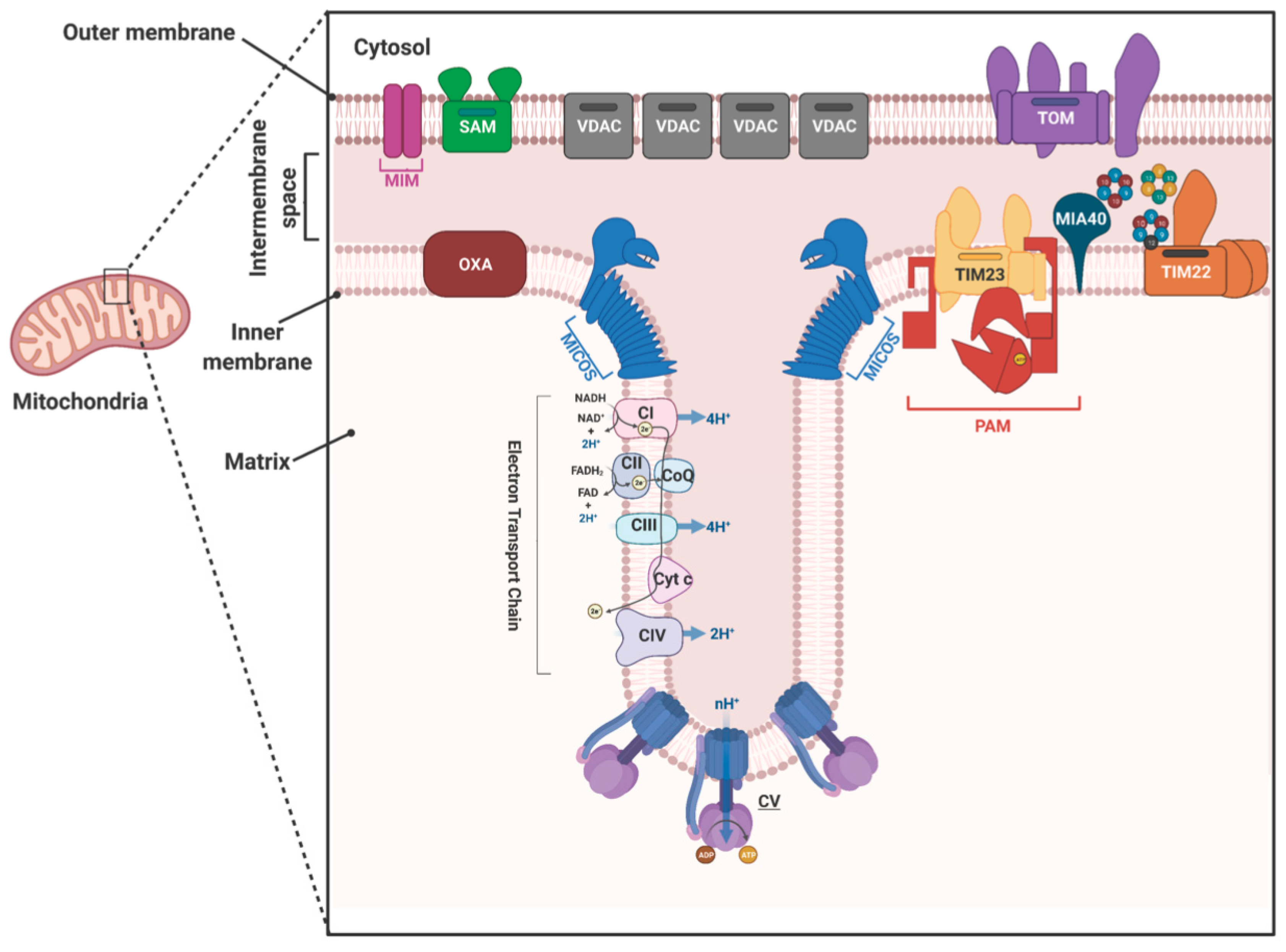
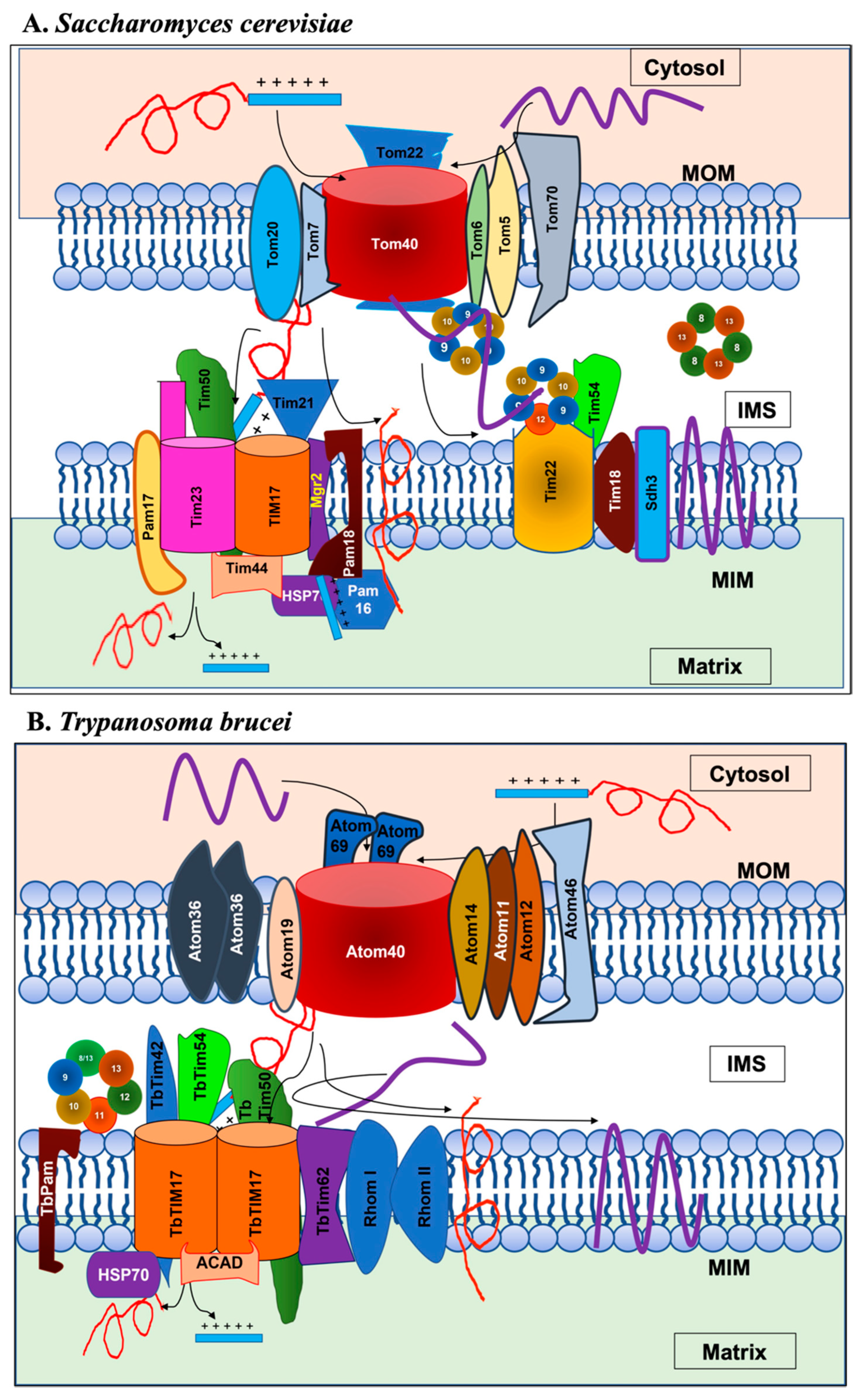

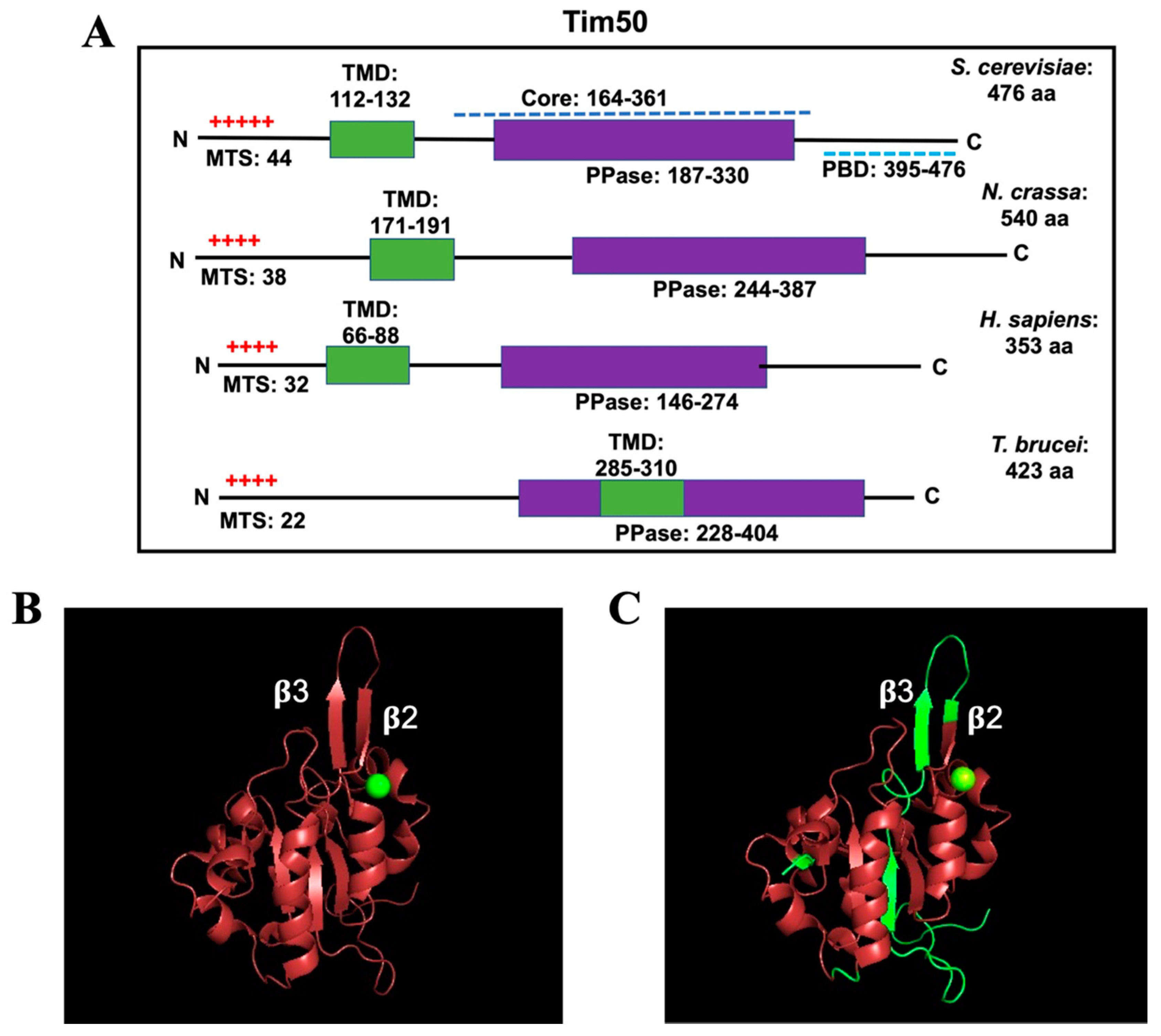
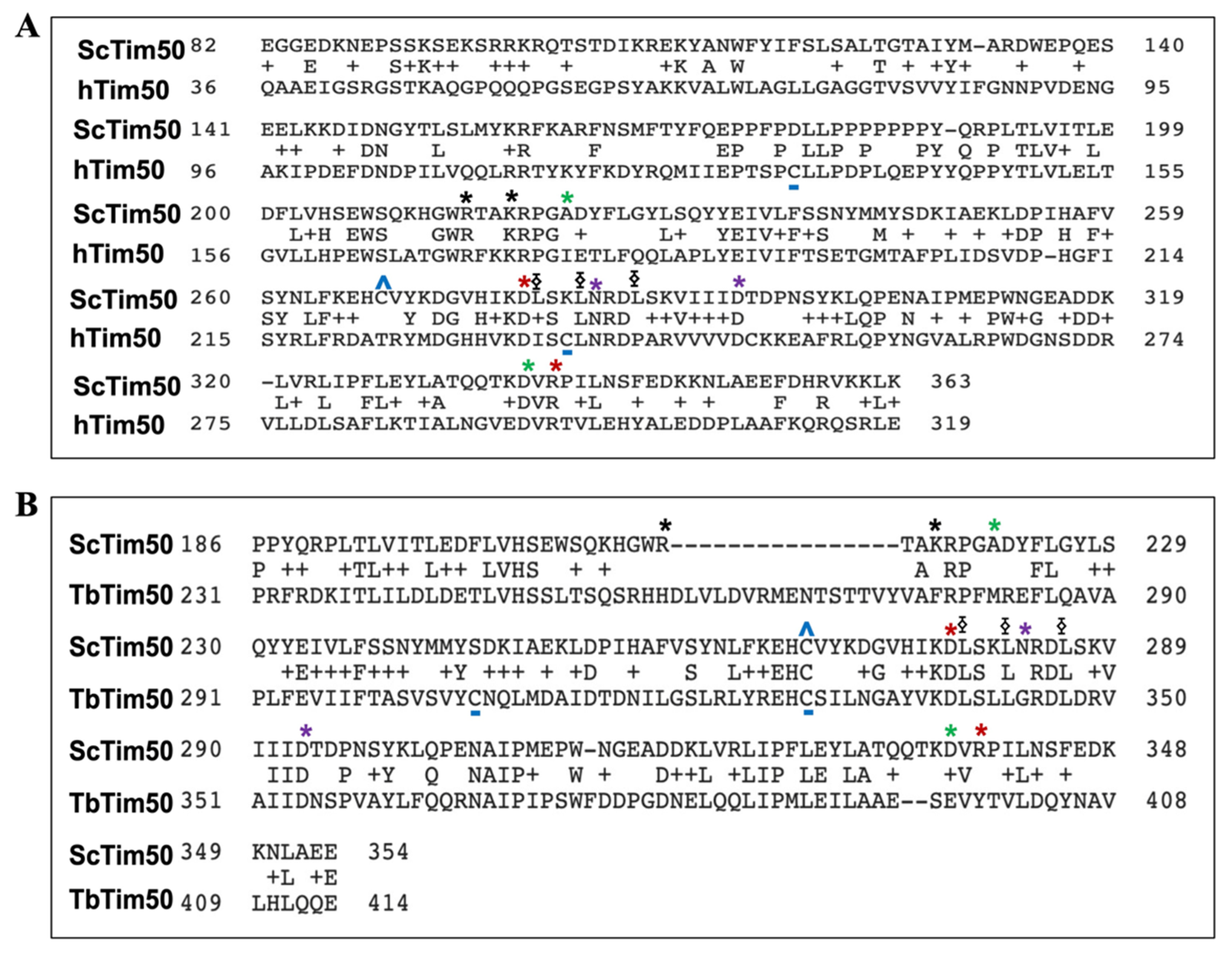
| Complex | Yeast/Fungi | Human | Plant | Trypanosomatids |
|---|---|---|---|---|
| SAM/TOB | Sam50/Tob55, Sam35/Tob38, Sam37/Mas37, Mdm10 | Sam50/Tob55, Metaxin 1, Metaxin 2 | Sam50/Tob55, Sam37(Metaxin), Mdm10 | Sam50/Tob55 |
| TOM | Tom40, Tom22, Tom5, Tom6, Tom7, Tom20, Tom70 | Tom40, Tom22, Tom5, Tom6, Tom7, Tom20, Tom70 | Tom40, Tom9(Tom22), Tom5, Tom6, Tom7, Tom20, OM64 e | Atom40 f, Atom46 f, Atom69 f Atom19 g, Atom14 g, Atom11 g, Atom12 g, |
| MIM | Mim1, Mim2 | Atom36 h | ||
| MIA | Mia40, Erv1 | Mia40, Erv1 | Erv1 | |
| Small Tims | Tim9, Tim10, Tim8, Tim13, Tim12 | Tim9, Tim10a, Tim10b, Tim8, Tim13 | Tim9, Tim10, Tim8, Tim13 | TbTim9, Tbim10, TbTim11 g, TbTim12 g, TbTim13 g, TbTim8/13 g |
| TIM23 | Tim23, Tim17, Tim50, Tim21 | TIMM23, TIMM17, TIMM50 | Tim23, Tim17, Tim50, Tim21 | TbTim17, TbTim62 g, TbTim54 g, TbTim42 g, TbTim50, ACAD g, Rhomboid I g, Rhomboid II g, small TbTims h |
| PAM | Pam18, Pam16, Pam17, Tim44, Hsp70, Mge-1 | DnaJC19 a, DnaJC15 a, Magma b, Mortalin/HSPA9 c | Pam18, Pam16, Tim14, Tim44, Hsp70, Mge1 | TbPam27 g, Hsp70 |
| TIM22 | Tim22, Tim54, Tim18, Sdh3, Tim12 | TIMM22, TIMM29 d, AGK d | Tim22 | |
| OXA | Oxa1 | Oxa1 | Oxa1 | Oxa1 |
| HAD Family | Examples | Substrate | Function |
|---|---|---|---|
| RNA polymerase | 1 FCP, 2 SCP1-3 | p-Serine | Gene expression |
| C-terminal domain | Dullard | in proteins | regulation |
| Phosphatase | 3 UBLCP1 | ||
| Polynucleotide | PNKP | 3′-Phosphorylated | DNA repair |
| Kinase phosphatase | DNA termini | ||
| Epoxide hydrolase | sEH2 | Dihydroxylipid | Lipid metabolism |
| phosphate, iso- | cholesterol | ||
| prenoid phosphate | biosynthesis | ||
| Intracellular | 4 cN-I-III | AMP, GMP | Nucleotide/ |
| 5′-Nucleotidase | 5 cdN, 6 mdN | IMP | nucleoside balance |
| Phosphoserine | PSPH | L-3-Phosphoserine | Serine biosynthesis |
| Phospho hydrolase | |||
| Eyes absent | EYA1-4 | p-Tyrosine | Organ development |
| in proteins | DNA repair | ||
| Cell proliferation | |||
| HAD-like | pseudouridine 5′ | Pseudouridine-P | Pseudouridine |
| Hydrolase | monophosphatase, | secretion | |
| pyridoxal 5′-phosphate | Pyridoxal-P | Vitamin B6 | |
| phosphatase | metabolism | ||
| Lipins | Lipin1-3, 7 PAP | Phosphatidic acid | Lipid metabolism |
| Organism | Functions | References |
|---|---|---|
| Yeast/Fungi |
| [35,36,65,66] |
| [84,85,97] | |
| [90] | |
| Human |
| [15,83] |
| [68,69] | |
| [69] | |
| [98,99,100] | |
| [101] | |
| [95,102,103] | |
| [104,105,106,107] | |
| Zebra fish |
| [69] |
| [69] | |
| [69] | |
| Drosophila |
| [68] |
| [68] | |
| [68] | |
| Plant |
| [16,96] |
| [96] | |
| Trypanosoma |
| [58] |
| [58,93,94] | |
| [58,74] | |
| [93,94] | |
| [93,94] | |
| [74] | |
| [74,93,94] | |
| [74] | |
| [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhuri, M.; Tripathi, A.; Gonzalez, F.S. Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase. Int. J. Mol. Sci. 2021, 22, 7779. https://doi.org/10.3390/ijms22157779
Chaudhuri M, Tripathi A, Gonzalez FS. Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase. International Journal of Molecular Sciences. 2021; 22(15):7779. https://doi.org/10.3390/ijms22157779
Chicago/Turabian StyleChaudhuri, Minu, Anuj Tripathi, and Fidel Soto Gonzalez. 2021. "Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase" International Journal of Molecular Sciences 22, no. 15: 7779. https://doi.org/10.3390/ijms22157779
APA StyleChaudhuri, M., Tripathi, A., & Gonzalez, F. S. (2021). Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase. International Journal of Molecular Sciences, 22(15), 7779. https://doi.org/10.3390/ijms22157779






