G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story
Abstract
:1. Introduction
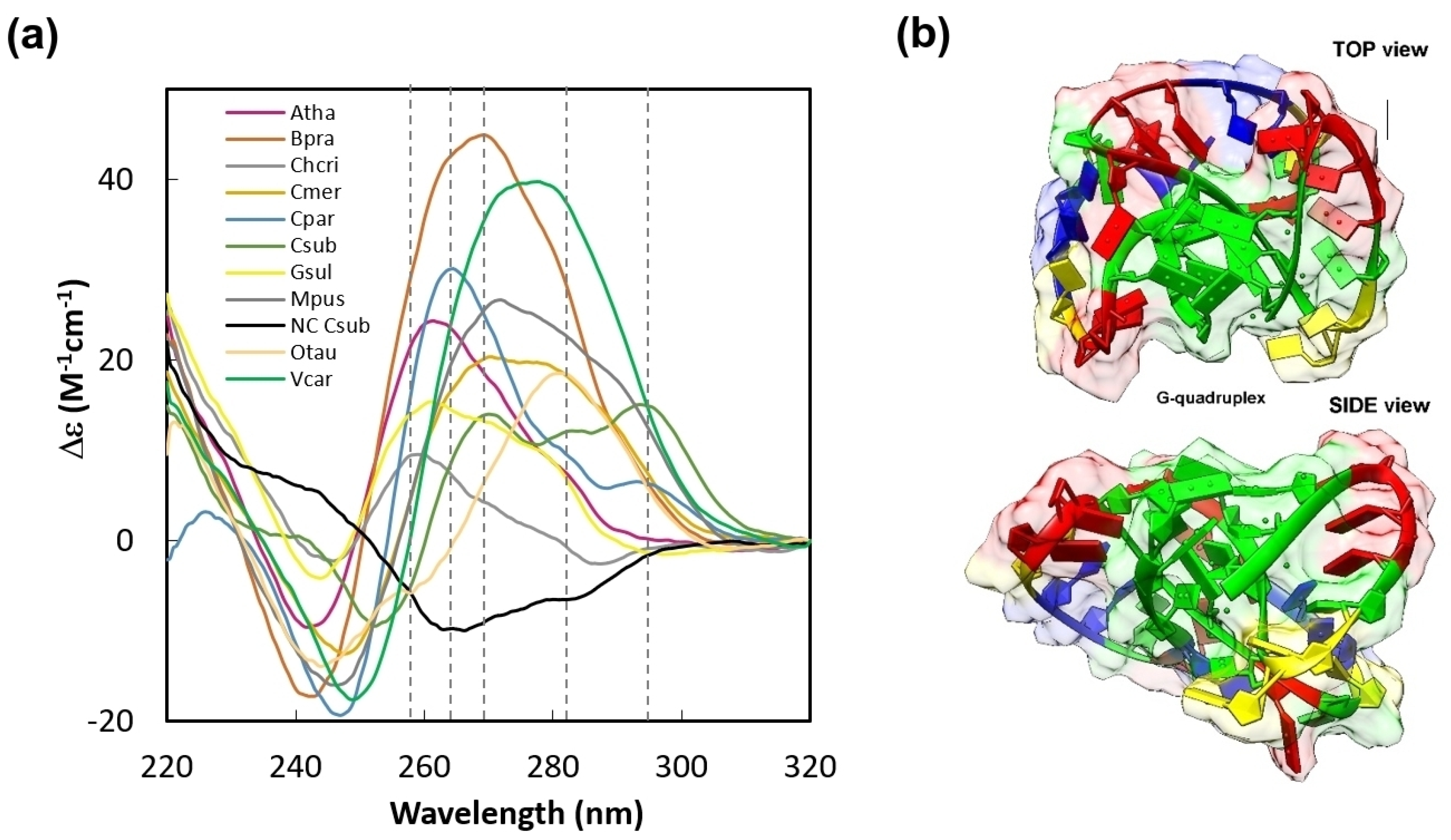
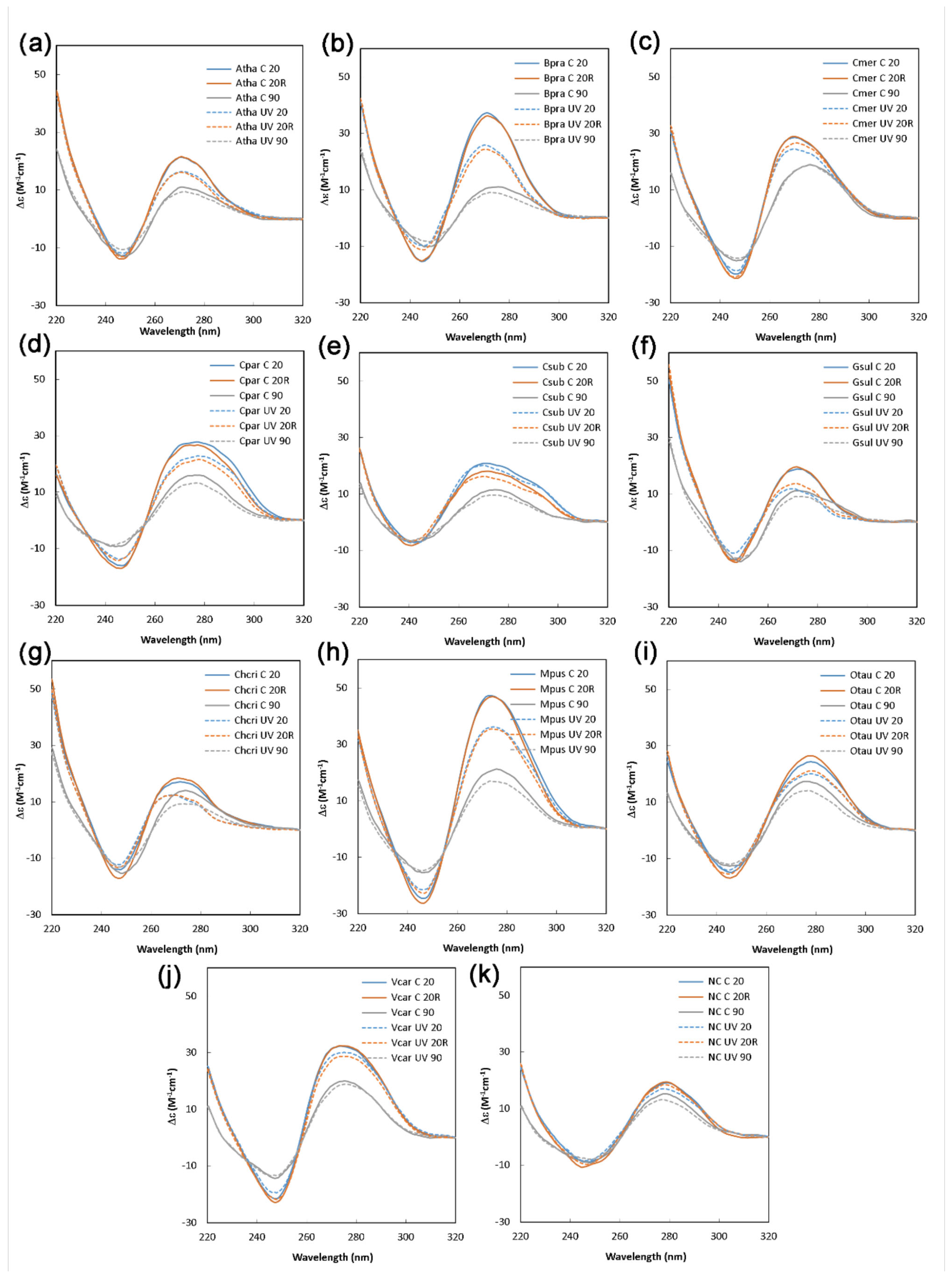
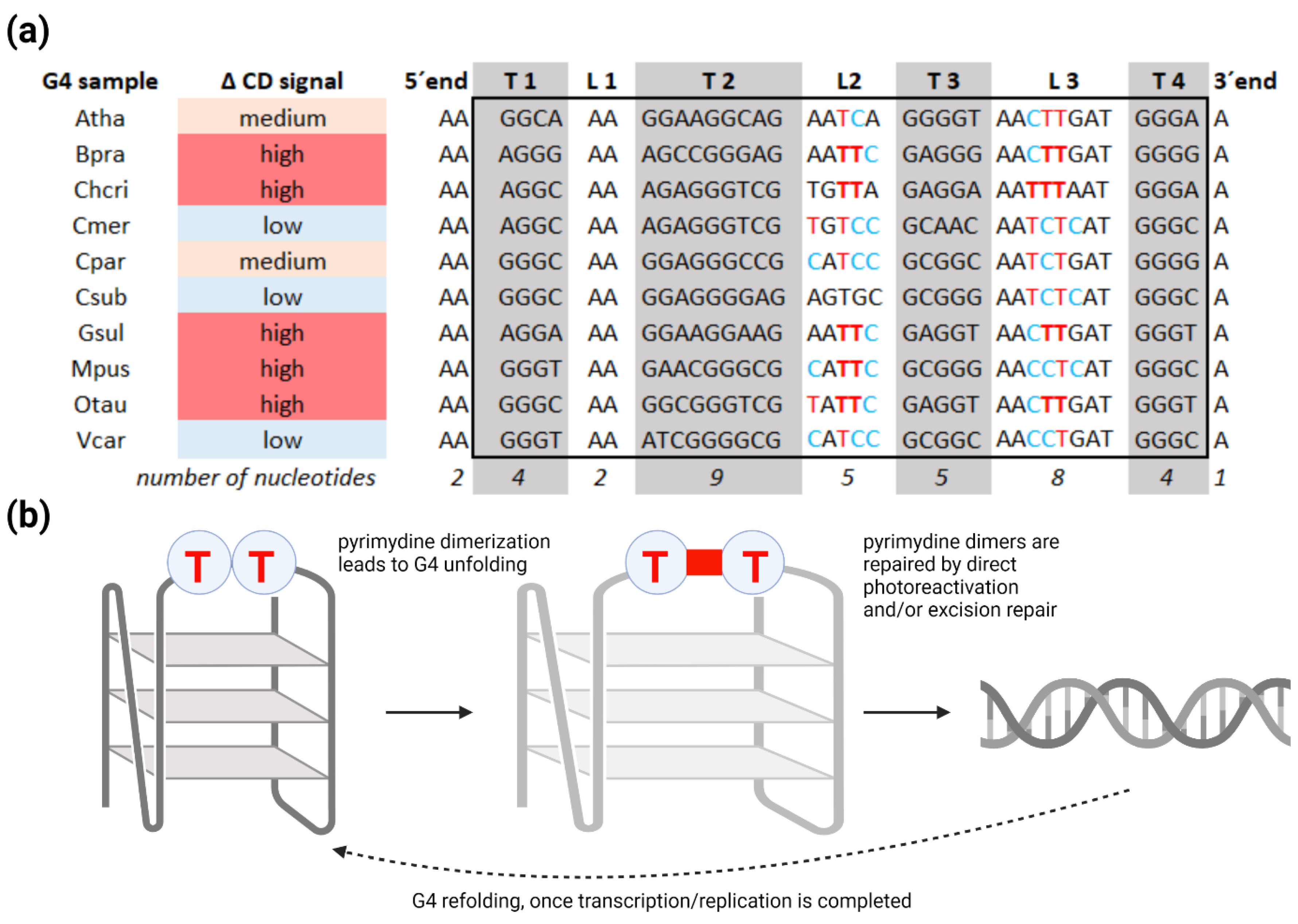
2. Results and Discussion
3. Materials and Methods
3.1. Bioinformatics and Structural Modeling
3.2. Circular Dichroism Measurement
3.3. Gel Electrophoresis and Thioflavin T Staining
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Klug, A. Rosalind Franklin and the Discovery of the Structure of DNA. Nature 1968, 219, 808–810. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix Formation by Guanylic Acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Cheema, J.; Zhang, Y.; Deng, H.; Duncan, S.; Umar, M.I.; Zhao, J.; Liu, Q.; Cao, X.; Kwok, C.K. RNA G-Quadruplex Structures Exist and Function in Vivo in Plants. Genome Biol. 2020, 21, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sánchez-Ferrer, A.; Bagnani, M.; Adamcik, J.; Azzari, P.; Hao, J.; Song, A.; Liu, H.; Mezzenga, R. Metal Ions Confinement Defines the Architecture of G-Quartet, G-Quadruplex Fibrils and Their Assembly into Nematic Tactoids. Proc. Natl. Acad. Sci. USA 2020, 117, 9832–9839. [Google Scholar] [CrossRef]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-Wide Discovery of G-Quadruplex Forming Sequences and Their Functional Relevance in Plants. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yang, D. G-Quadruplex DNA and RNA. In G-Quadruplex Nucleic Acids: Methods and Protocols; Yang, D., Lin, C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 1–24. ISBN 978-1-4939-9666-7. [Google Scholar]
- Warner, E.F.; Bohálová, N.; Brázda, V.; Waller, Z.A.E.; Bidula, S. Analysis of Putative Quadruplex-Forming Sequences in Fungal Genomes: Novel Antifungal Targets? Microb. Genom. 2021, 7, 000570. [Google Scholar] [CrossRef]
- Bartas, M.; Čutová, M.; Brázda, V.; Kaura, P.; Št’astný, J.; Kolomazník, J.; Coufal, J.; Goswami, P.; Červeň, J.; Pečinka, P. The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria. Molecules 2019, 24, 1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brázda, V.; Luo, Y.; Bartas, M.; Kaura, P.; Porubiaková, O.; Št’astný, J.; Pečinka, P.; Verga, D.; Da Cunha, V.; Takahashi, T.S. G-Quadruplexes in the Archaea Domain. Biomolecules 2020, 10, 1349. [Google Scholar] [CrossRef]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Bartas, M.; Brázda, V.; Bohálová, N.; Cantara, A.; Volná, A.; Stachurová, T.; Malachová, K.; Jagelská, E.B.; Porubiaková, O.; Červeň, J. In-Depth Bioinformatic Analyses of Nidovirales Including Human SARS-CoV-2, SARS-CoV, MERS-CoV Viruses Suggest Important Roles of Non-Canonical Nucleic Acid Structures in Their Lifecycles. Front. Microbiol. 2020, 11, 1583. [Google Scholar] [CrossRef]
- Bohálová, N.; Cantara, A.; Bartas, M.; Kaura, P.; Šťastný, J.; Pečinka, P.; Fojta, M.; Mergny, J.-L.; Brázda, V. Analyses of Viral Genomes for G-Quadruplex Forming Sequences Reveal Their Correlation with the Type of Infection. Biochimie 2021, 186, 13–27. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggiero, E.; Richter, S.N. Viral G-Quadruplexes: New Frontiers in Virus Pathogenesis and Antiviral Therapy. Annu. Rep. Med. Chem. 2020, 54, 101–131. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.-P.; Foiani, M.; Nicolas, A. G-Quadruplex-Induced Instability during Leading-Strand Replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef] [PubMed]
- Lerner, L.K.; Sale, J.E. Replication of G Quadruplex DNA. Genes 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broxson, C.; Beckett, J.; Tornaletti, S. Transcription Arrest by a G Quadruplex Forming-Trinucleotide Repeat Sequence from the Human c-Myb Gene. Biochemistry 2011, 50, 4162–4172. [Google Scholar] [CrossRef]
- Carvalho, J.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-Quadruplex, Friend or Foe: The Role of the G-Quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Szlachta, K.; Thys, R.G.; Atkin, N.D.; Pierce, L.C.T.; Bekiranov, S.; Wang, Y.-H. Alternative DNA Secondary Structure Formation Affects RNA Polymerase II Promoter-Proximal Pausing in Human. Genome Biol. 2018, 19, 89. [Google Scholar] [CrossRef]
- Endoh, T.; Sugimoto, N. Conformational Dynamics of the RNA G-Quadruplex and Its Effect on Translation Efficiency. Molecules 2019, 24, 1613. [Google Scholar] [CrossRef] [Green Version]
- Wolna, A.H.; Fleming, A.M.; Burrows, C.J. Single-Molecule Analysis of Thymine Dimer-Containing G-Quadruplexes Formed from the Human Telomere Sequence. Biochemistry 2014, 53, 7484–7493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, D.G.T.; Fang, H.; Gross, M.L.; Taylor, J.-S.A. Photocrosslinking of Human Telomeric G-Quadruplex Loops by Anti Cyclobutane Thymine Dimer Formation. Proc. Natl. Acad. Sci. USA 2009, 106, 12861–12866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, B.D.; Bass, H.W. Plant G-Quadruplex (G4) Motifs in DNA and RNA.; Abundant, Intriguing Sequences of Unknown Function. Plant Sci. 2018, 269, 143–147. [Google Scholar] [CrossRef]
- Ream, T.S.; Haag, J.R.; Pontvianne, F.; Nicora, C.D.; Norbeck, A.D.; Paša-Tolić, L.; Pikaard, C.S. Subunit Compositions of Arabidopsis RNA Polymerases I and III Reveal Pol I- and Pol III-Specific Forms of the AC40 Subunit and Alternative Forms of the C53 Subunit. Nucleic Acids Res. 2015, 43, 4163–4178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Ma, H. Step-Wise and Lineage-Specific Diversification of Plant RNA Polymerase Genes and Origin of the Largest Plant-Specific Subunits. New Phytol. 2015, 207, 1198–1212. [Google Scholar] [CrossRef] [PubMed]
- Armache, K.-J.; Mitterweger, S.; Meinhart, A.; Cramer, P. Structures of Complete RNA Polymerase II and Its Subcomplex, Rpb4/7. J. Biol. Chem. 2005, 280, 7131–7134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, V.; Weisshart, K. Abundance and Distribution of RNA Polymerase II in Arabidopsis Interphase Nuclei. J. Exp. Bot. 2015, 66, 1687–1698. [Google Scholar] [CrossRef] [Green Version]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA Polymerases IV and V: Purveyors of Non-Coding RNA for Plant Gene Silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef]
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA Polymerase II Allows Controlled Mobilisation of Retrotransposons for Plant Breeding. Genome Biol. 2017, 18, 134. [Google Scholar] [CrossRef]
- Core, L.; Adelman, K. Promoter-Proximal Pausing of RNA Polymerase II: A Nexus of Gene Regulation. Genes Dev. 2019, 33, 960–982. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Liu, M.; Liu, X.; Dong, Z. RNA Polymerase II Activity Revealed by GRO-Seq and PNET-Seq in Arabidopsis. Nat. Plants 2018, 4, 1112–1123. [Google Scholar] [CrossRef]
- Ferrari, C.; Proost, S.; Janowski, M.; Becker, J.; Nikoloski, Z.; Bhattacharya, D.; Price, D.; Tohge, T.; Bar-Even, A.; Fernie, A.; et al. Kingdom-Wide Comparison Reveals the Evolution of Diurnal Gene Expression in Archaeplastida. Nat. Commun. 2019, 10, 737. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Brázda, V.; Kolomazník, J.; Lýsek, J.; Bartas, M.; Fojta, M.; Št’astný, J.; Mergny, J.-L. G4Hunter Web Application: A Web Server for G-Quadruplex Prediction. Bioinformatics 2019, 35, 3493–3495. [Google Scholar] [CrossRef] [Green Version]
- Garant, J.-M.; Perreault, J.-P.; Scott, M.S. G4RNA Screener Web Server: User Focused Interface for RNA G-Quadruplex Prediction. Biochimie 2018, 151, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.-D.; Jodoin, R.; Perreault, J.-P. New Scoring System to Identify RNA G-Quadruplex Folding. Nucleic Acids Res. 2014, 42, 1209–1223. [Google Scholar] [CrossRef] [Green Version]
- Garant, J.-M.; Perreault, J.-P.; Scott, M.S. Motif Independent Identification of Potential RNA G-Quadruplexes by G4RNA Screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef] [Green Version]
- Sabouri, N.; Capra, J.A.; Zakian, V.A. The Essential Schizosaccharomyces Pombe Pfh1 DNA Helicase Promotes Fork Movement Past G-Quadruplex Motifs to Prevent DNA Damage. BMC Biol. 2014, 12, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig Lombardi, E.; Holmes, A.; Verga, D.; Teulade-Fichou, M.-P.; Nicolas, A.; Londoño-Vallejo, A. Thermodynamically Stable and Genetically Unstable G-Quadruplexes Are Depleted in Genomes across Species. Nucleic Acids Res. 2019, 47, 6098–6113. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P.; Bushnell, D.A.; Fu, J.; Gnatt, A.L.; Maier-Davis, B.; Thompson, N.E.; Burgess, R.R.; Edwards, A.M.; David, P.R.; Kornberg, R.D. Architecture of RNA Polymerase II and Implications for the Transcription Mechanism. Science 2000, 288, 640–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cogoi, S.; Xodo, L.E. G-Quadruplex Formation within the Promoter of the KRAS Proto-Oncogene and Its Effect on Transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Yadav, V.; Kim, N.; Tuteja, N.; Yadav, P. G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole Genome Experimental Maps of DNA G-Quadruplexes in Multiple Species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [Green Version]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-Evaluation of G-Quadruplex Propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Thomas, Q.A.; Ard, R.; Liu, J.; Li, B.; Wang, J.; Pelechano, V.; Marquardt, S. Transcript Isoform Sequencing Reveals Widespread Promoter-Proximal Transcriptional Termination in Arabidopsis. Nat. Commun. 2020, 11, 2589. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, K.; Xiao, S.; Hao, Y.; Tan, Z. Mechanism and Manipulation of DNA: RNA Hybrid G-Quadruplex Formation in Transcription of G-Rich DNA. J. Am. Chem. Soc. 2014, 136, 1381–1390. [Google Scholar] [CrossRef]
- Frey, K.; Pucker, B. Animal, Fungi, and Plant Genome Sequences Harbor Different Non-Canonical Splice Sites. Cells 2020, 9, 458. [Google Scholar] [CrossRef] [Green Version]
- Vinnarasi, S.; Radhika, R.; Vijayakumar, S.; Shankar, R. Structural Insights into the Anti-Cancer Activity of Quercetin on G-Tetrad, Mixed G-Tetrad, and G-Quadruplex DNA Using Quantum Chemical and Molecular Dynamics Simulations. J. Biomol. Struct. Dyn. 2020, 38, 317–339. [Google Scholar] [CrossRef]
- Brown, R.V.; Wang, T.; Chapetta, V.R.; Wu, G.; Onel, B.; Chawla, R.; Quijada, H.; Camp, S.M.; Chiang, E.T.; Lassiter, Q.R.; et al. The Consequences of Overlapping G-Quadruplexes and i-Motifs in the Platelet-Derived Growth Factor Receptor β Core Promoter Nuclease Hypersensitive Element Can Explain the Unexpected Effects of Mutations and Provide Opportunities for Selective Targeting of Both Structures by Small Molecules To Downregulate Gene Expression. J. Am. Chem. Soc. 2017, 2017, 7456–7475. [Google Scholar] [CrossRef]
- Megger, D.A.; Lax, P.M.; Paauwe, J.; Fonseca Guerra, C.; Lippert, B. Mixed Guanine, Adenine Base Quartets: Possible Roles of Protons and Metal Ions in Their Stabilization. J. Biol. Inorg. Chem. 2018, 23, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Patro, L.P.P.; Kumar, A.; Kolimi, N.; Rathinavelan, T. 3D-NuS: A Web Server for Automated Modeling and Visualization of Non-Canonical 3-Dimensional Nucleic Acid Structures. J. Mol. Biol. 2017, 429, 2438–2448. [Google Scholar] [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.-L. How Long Is Too Long? Effects of Loop Size on G-Quadruplex Stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisa, M.-U.; Huang, Y.; Benhamed, M.; Raynaud, C. The Plant DNA Damage Response: Signaling Pathways Leading to Growth Inhibition and Putative Role in Response to Stress Conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balanikas, E.; Banyasz, A.; Baldacchino, G.; Markovitsi, D. Guanine Radicals Generated in Telomeric G-Quadruplexes by Direct Absorption of Low-Energy UV Photons: Effect of Potassium Ions. Molecules 2020, 25, 2094. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M. DNA Repair. In The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000; ISBN 0-87893-106-6. [Google Scholar]
- Kimura, S.; Tahira, Y.; Ishibashi, T.; Mori, Y.; Mori, T.; Hashimoto, J.; Sakaguchi, K. DNA Repair in Higher Plants; Photoreactivation Is the Major DNA Repair Pathway in Non-Proliferating Cells While Excision Repair (Nucleotide Excision Repair and Base Excision Repair) Is Active in Proliferating Cells. Nucleic Acids Res. 2004, 32, 2760–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spampinato, C.P. Protecting DNA from Errors and Damage: An Overview of DNA Repair Mechanisms in Plants Compared to Mammals. Cell. Mol. Life Sci. 2017, 74, 1693–1709. [Google Scholar] [CrossRef]
- Marquevielle, J.; Robert, C.; Lagrabette, O.; Wahid, M.; Bourdoncle, A.; Xodo, L.E.; Mergny, J.-L.; Salgado, G.F. Structure of Two G-Quadruplexes in Equilibrium in the KRAS Promoter. Nucleic Acids Res. 2020, 48, 9336–9345. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Wild, A.T.; Wu, W.H.; Shah, R.; Danussi, C.; Riggins, G.J.; Kannan, K.; Sulman, E.P.; Chan, T.A. G-Quadruplex DNA Drives Genomic Instability and Represents a Targetable Molecular Abnormality in ATRX-Deficient Malignant Glioma. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Shin-Ya, K.; Brosh, R.M., Jr. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA to Defend Genomic Stability. Mol. Cell. Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [Green Version]
- Armas, P.; David, A.; Calcaterra, N.B. Transcriptional Control by G-Quadruplexes: In Vivo Roles and Perspectives for Specific Intervention. Transcription 2017, 8, 21–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Varshney, D.; Simeone, A.; Zhang, X.; Adhikari, S.; Tannahill, D.; Balasubramanian, S. Promoter G-Quadruplex Folding Precedes Transcription and Is Controlled by Chromatin. Genome Biol. 2021, 22, 143. [Google Scholar] [CrossRef]
- Bolduc, F.; Garant, J.-M.; Allard, F.; Perreault, J.-P. Irregular G-Quadruplexes Found in the Untranslated Regions of Human MRNAs Influence Translation. J. Biol. Chem. 2016, 291, 21751–21760. [Google Scholar] [CrossRef] [Green Version]
- Kanoh, Y.; Matsumoto, S.; Fukatsu, R.; Kakusho, N.; Kono, N.; Renard-Guillet, C.; Masuda, K.; Iida, K.; Nagasawa, K.; Shirahige, K.; et al. Rif1 Binds to G Quadruplexes and Suppresses Replication over Long Distances. Nat. Struct. Mol. Biol. 2015, 22, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Poggi, L.; Richard, G.-F. Alternative DNA Structures In Vivo: Molecular Evidence and Remaining Questions. Microbiol. Mol. Biol. Rev. 2020, 85, e00110-20. [Google Scholar] [CrossRef] [PubMed]
- Schiavone, D.; Guilbaud, G.; Murat, P.; Papadopoulou, C.; Sarkies, P.; Prioleau, M.-N.; Balasubramanian, S.; Sale, J.E. Determinants of G Quadruplex-Induced Epigenetic Instability in REV1-Deficient Cells. EMBO J. 2014, 33, 2507–2520. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-Molecule Visualization of DNA G-Quadruplex Formation in Live Cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Buma, A.G.J.; Engelen, A.H.; Gieskes, W.W.C. Wavelength-Dependent Induction of Thymine Dimers and Growth Rate Reduction in the Marine Diatom Cyclotella Sp. Exposed to Ultraviolet Radiation. Mar. Ecol. Prog. Ser. 1997, 153, 91–97. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Bartas, M.; Brázda, V.; Karlický, V.; Červeň, J.; Pečinka, P. Bioinformatics Analyses and in Vitro Evidence for Five and Six Stacked G-Quadruplex Forming Sequences. Biochimie 2018, 150, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Brazda, V.; Kolomaznik, J.; Mergny, J.-L.; Stastny, J. G4Killer Web Application: A Tool to Design G-Quadruplex Mutations. Bioinformatics 2020, 36, 3246–3247. [Google Scholar] [CrossRef] [PubMed]
- Renaud de la Faverie, A.; Guédin, A.; Bedrat, A.; Yatsunyk, L.A.; Mergny, J.-L. Thioflavin T as a Fluorescence Light-up Probe for G4 Formation. Nucleic Acids Res. 2014, 42, e65. [Google Scholar] [CrossRef] [Green Version]

| Latin Name/Common Name | Higher Taxonomy Unit | Lower Taxonomy Unit | RPB1 CDS NCBI ID | Predicted Conserved G4 Site 1 |
|---|---|---|---|---|
| Amborella trichopoda/ amborella | Viridiplantae | Amborella | XM_006828781.3 | Yes |
| Arabidopsis thaliana/ thale cress | Viridiplantae | Brassicales | NM_119746.4 | Yes |
| Bathycoccus prasinos | Viridiplantae | Chlorophyta | XM_007510177.1 | Yes |
| Chondrus crispus | Rhodophyta | Gigartinales | XM_005718456.1 | Yes |
| Cyanidioschyzon merolae | Rhodophyta | Cyanidiales | XM_005538436.1 | No |
| Cyanophora paradoxa | Glaucophyta | Cyanophoraceae | DQ223186.1 | Yes |
| Coccomyxa subellipsoidea | Viridiplantae | Chlorophyta | XM_005651715.1 | Yes |
| Galdieria sulphuraria | Rhodophyta | Cyanidiales | XM_005707370.1 | Yes |
| Helianthus annuus/ common sunflower | Viridiplantae | Asterales | XM_035988906.1 | Yes |
| Juglans regia/ English walnut | Viridiplantae | Fagales | XM_035687192.1 | Yes |
| Micromonas pusilla | Viridiplantae | Chlorophyta | XM_003055558.1 | Yes |
| Ostreococcus tauri | Viridiplantae | Chlorophyta | XM_022983138.1 | Yes |
| Papaver somniferum/ opium poppy | Viridiplantae | Ranunculales | XM_026541050.1 | Yes |
| Populus alba/ white poplar | Viridiplantae | Malpighiales | XM_035033875.1 | Yes |
| Prunus dulcis/ almond | Viridiplantae | Rosales | XM_034368569.1 | Yes |
| Setaria viridis/ bristlegrass | Viridiplantae | Poales | XM_034744839.1 | No |
| Solanum tuberosum/ potato | Viridiplantae | Solanales | XM_006340725.2 | Yes |
| Vitis riparia/ riverbank grape | Viridiplantae | Vitales | XM_034819617.1 | Yes |
| Volvox carteri | Viridiplantae | Chlorophyta | XM_002949413.1 | Yes |
| Zea mays/ maize | Viridiplantae | Poales | NM_001305817.1 | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volná, A.; Bartas, M.; Karlický, V.; Nezval, J.; Kundrátová, K.; Pečinka, P.; Špunda, V.; Červeň, J. G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story. Int. J. Mol. Sci. 2021, 22, 7381. https://doi.org/10.3390/ijms22147381
Volná A, Bartas M, Karlický V, Nezval J, Kundrátová K, Pečinka P, Špunda V, Červeň J. G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story. International Journal of Molecular Sciences. 2021; 22(14):7381. https://doi.org/10.3390/ijms22147381
Chicago/Turabian StyleVolná, Adriana, Martin Bartas, Václav Karlický, Jakub Nezval, Kristýna Kundrátová, Petr Pečinka, Vladimír Špunda, and Jiří Červeň. 2021. "G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story" International Journal of Molecular Sciences 22, no. 14: 7381. https://doi.org/10.3390/ijms22147381
APA StyleVolná, A., Bartas, M., Karlický, V., Nezval, J., Kundrátová, K., Pečinka, P., Špunda, V., & Červeň, J. (2021). G-Quadruplex in Gene Encoding Large Subunit of Plant RNA Polymerase II: A Billion-Year-Old Story. International Journal of Molecular Sciences, 22(14), 7381. https://doi.org/10.3390/ijms22147381










