Effect of the Electromagnetic Field (EMF) Radiation on Transcriptomic Profile of Pig Myometrium during the Peri-Implantation Period—An In Vitro Study
Abstract
1. Introduction
2. Results
2.1. The Statistics of RNA Sequencing
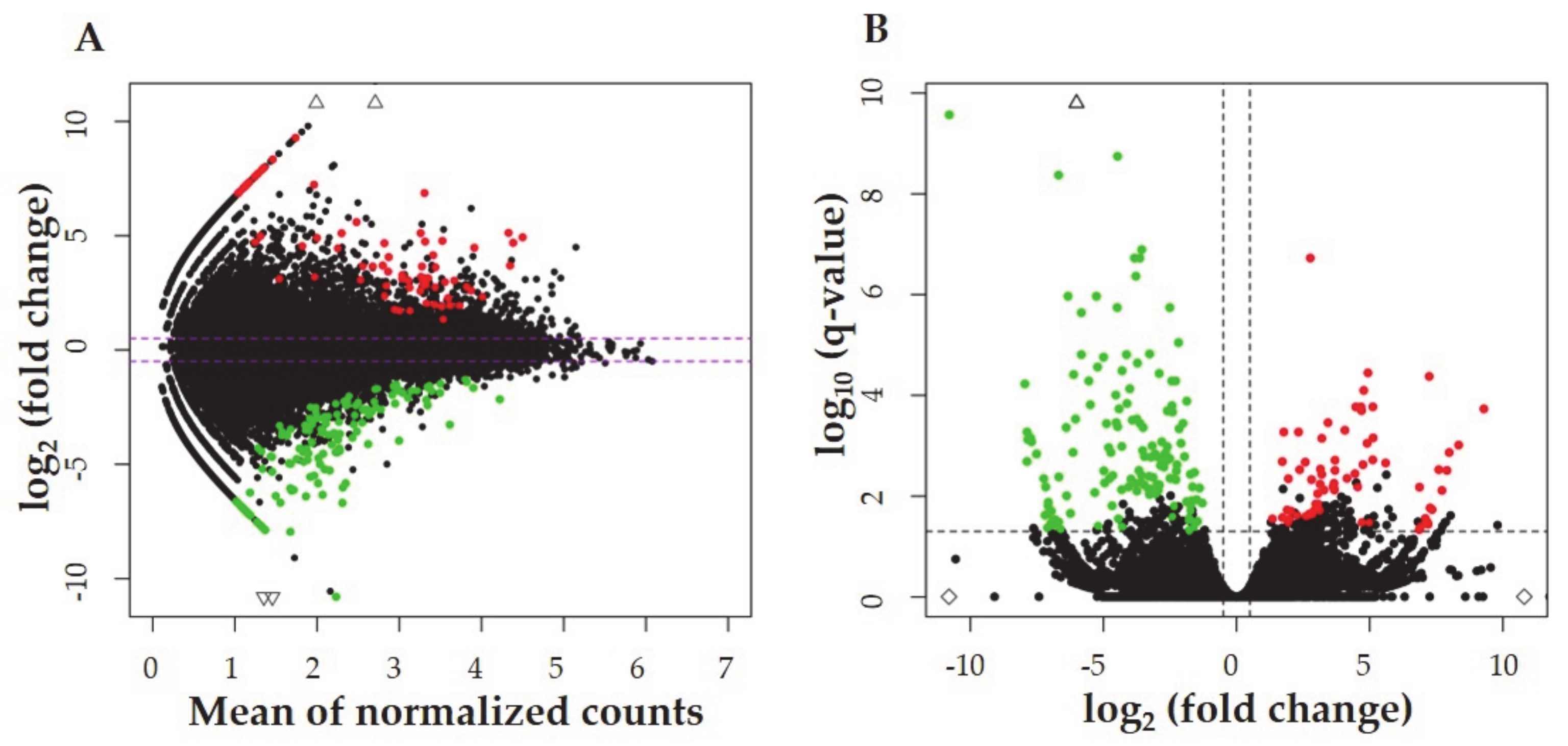
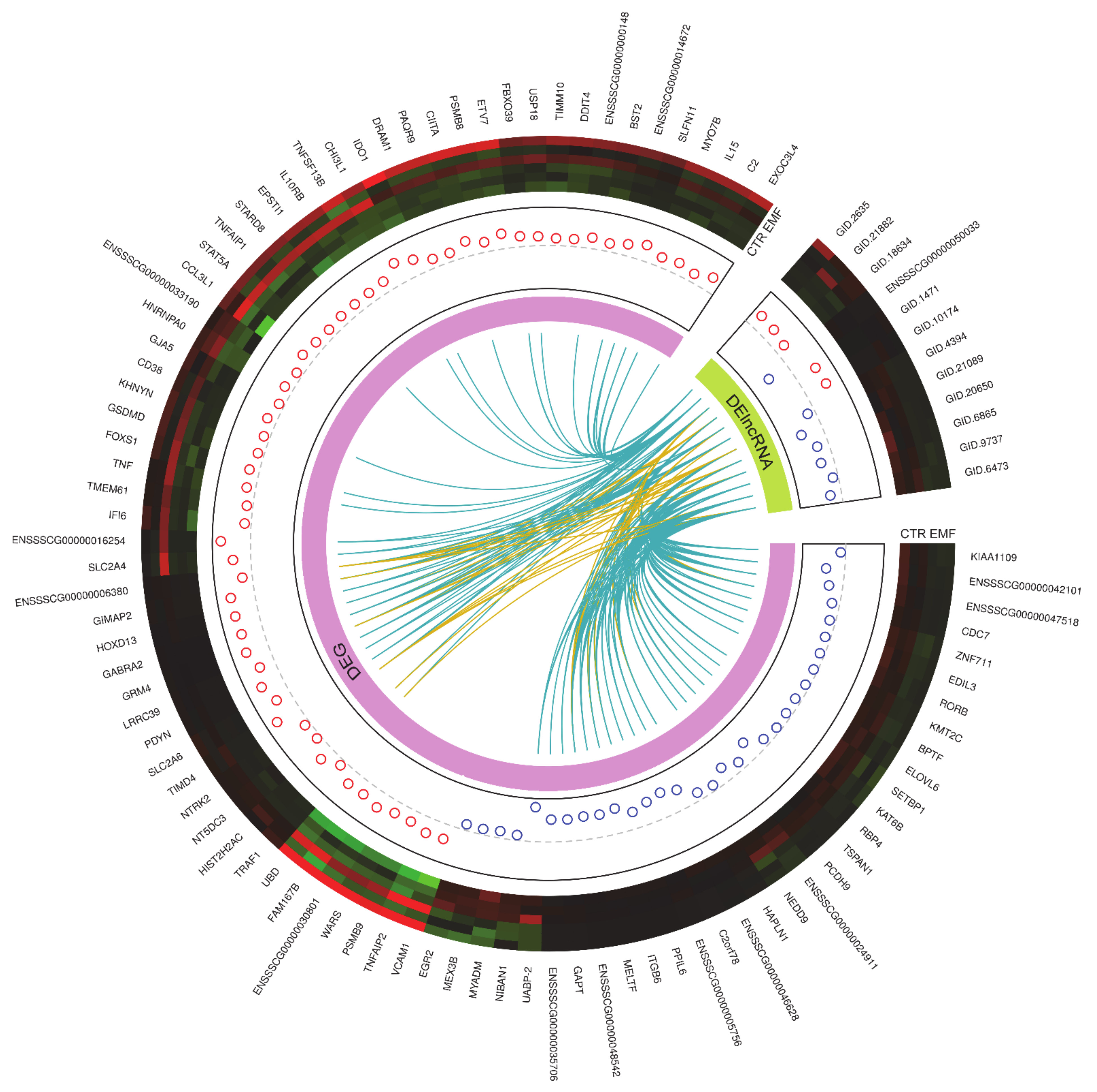
2.2. The Identification of Differentially Expressed Genes (DEGs), and Differentially Expressed Non-Coding RNAs (DE-ncRNAs)
2.3. Functional Ontology Annotations
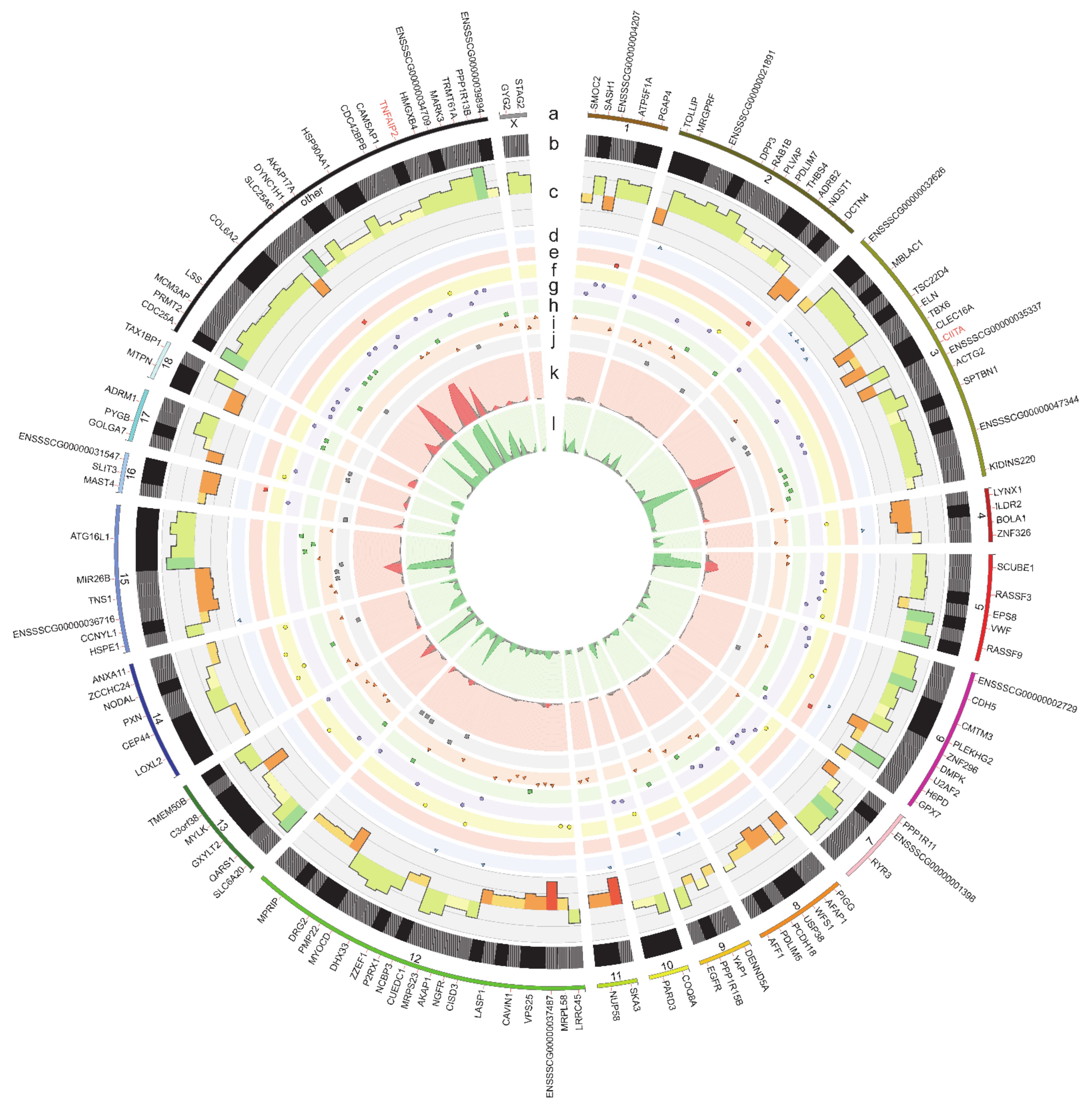
2.4. RNA Editing Site Prediction
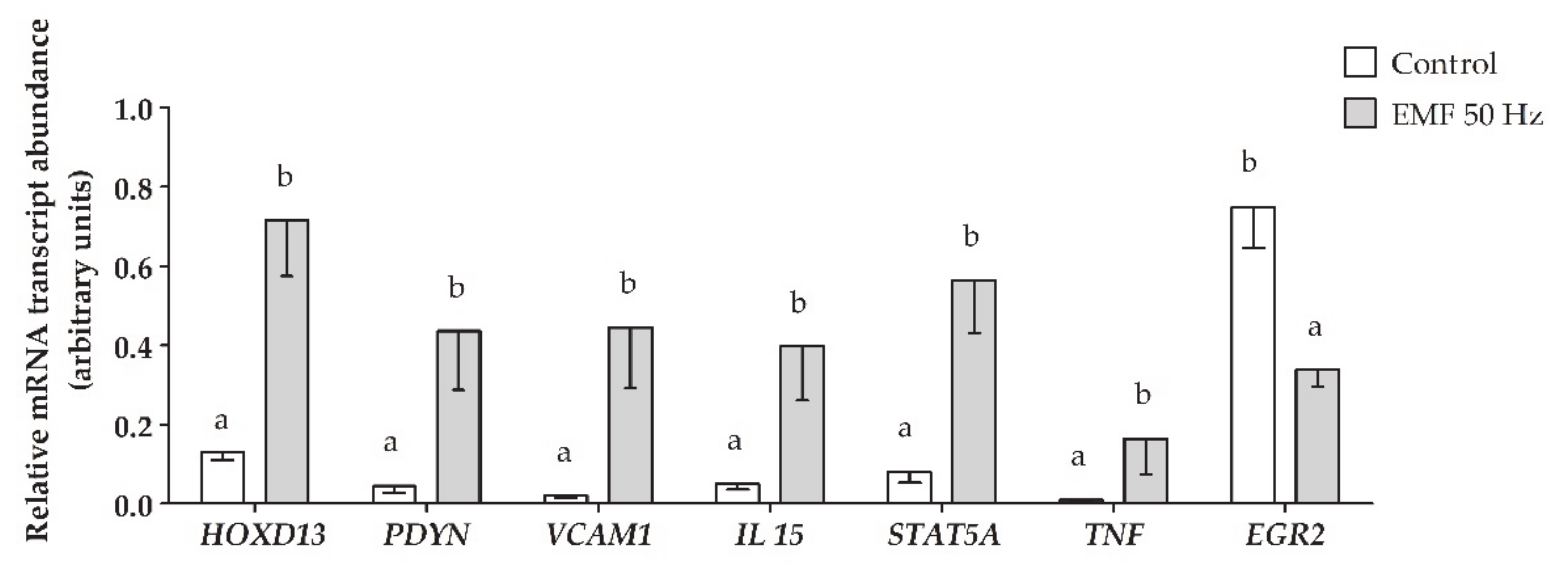
2.5. Validation of NGS Results
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals, Collection of Myometrial Tissue, and EMF Treatment
4.3. RNA Isolation
4.4. Construction and Sequencing of cDNA Libraries—Next-Generation Sequencing
4.5. Transcriptome Profiling
4.6. RNA Editing Sites Prediction
4.7. Validation Procedure
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pritchard, C.; Silk, A.; Hansen, L. Are rises in Electro-Magnetic Field in the human environment, interacting with multiple environmental pollutions, the tripping point for increases in neurological deaths in the Western World? Med. Hypotheses 2019, 127, 76–83. [Google Scholar] [CrossRef]
- Mansuori, E.; Alihemmati, A.; Mesbahi, A. An overview on the effects of power frequency electromagnetic field exposure on the female reproduction system, pregnancy outcome and fetal development. J. Med. Chem. Sci. 2020, 3, 60–70. [Google Scholar]
- Gajšek, P.; Ravazzani, P.; Grellier, J.; Samaras, T.; Bakos, J.; Thuróczy, G. Review of studies concerning electromagnetic field (EMF) exposure assessment in Europe: Low frequency fields (50 Hz–100 kHz). Int. J. Environ. Res. Public Health 2016, 13, 875. [Google Scholar] [CrossRef] [PubMed]
- Gye, M.C.; Park, C.J. Effect of electromagnetic field exposure on the reproductive system. Clin. Exp. Reprod. Med. 2012, 13, 875. [Google Scholar] [CrossRef]
- Behrens, T.; Terschüren, C.; Kaune, W.T.; Hoffmann, W. Quantification of lifetime accumulated ELF-EMF exposure from household appliances in the context of a retrospective epidemiological case-control study. J. Expo. Anal. Environ. Epidemiol. 2004, 14, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Bernabò, N.; Tettamanti, E.; Russo, V.; Martelli, A.; Turriani, M.; Mattoli, M.; Barboni, B. Extremely low frequency electromagnetic field exposure affects fertilization outcome in swine animal model. Theriogenology 2010, 73, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Koziorowska, A.; Waszkiewicz, E.M.; Romerowicz-Misielak, M.; Zglejc-Waszak, K.; Franczak, A. Extremely low-frequency electromagnetic field (EMF) generates alterations in the synthesis and secretion of oestradiol-17β (E2) in uterine tissues: An in vitro study. Theriogenology 2018, 1, 86–95. [Google Scholar] [CrossRef]
- Franczak, A.; Waszkiewicz, E.; Kozlowska, W.; Zmijewska, A.; Koziorowska, A. Consequences of electromagnetic field (EMF) radiation during early pregnancy—Androgen synthesis and release from the myometrium of pigs in vitro. Anim. Reprod. Sci. 2020, 218, 106465. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Johnson, G.A. Pig blastocyst-uterine interactions. Differentiation 2014, 87, 52–65. [Google Scholar] [CrossRef]
- Bauersachs, S.; Wolf, E. Uterine responses to the preattachment embryo in domestic ungulates: Recognition of pregnancy and preparation for implantation. Annu. Rev. Anim. Biosci. 2015, 3, 489–511. [Google Scholar] [CrossRef]
- Kozlowska, W.; Drzewiecka, E.; Zmijewska, A.; Koziorowska, A.; Franczak, A. Effects of electromagnetic field (EMF) radiation on androgen synthesis and release from the pig endometrium during the fetal peri-implantation period. Anim. Reprod. Sci. 2021, 226, 106694. [Google Scholar] [CrossRef]
- Drzewiecka, E.M.; Kozlowska, W.; Zmijewska, A.; Franczak, A.; Wydorski, P.J.; Franczak, A. Electromagnetic field (EMF) radiation alters estrogen release from the pig myometrium during the peri-implantation period. Int. J. Mol. Sci. 2021, 22, 2920. [Google Scholar] [CrossRef]
- Franczak, A. Endometrial and myometrial secretion of androgens and estrone during early pregnancy and luteolysis in pigs. Reprod. Biol. 2008, 8, 213–228. [Google Scholar] [CrossRef]
- Makieva, S.; Saunders, P.T.K.; Norman, J.E. Androgens in pregnancy: Roles in parturition. Hum. Reprod. Update 2014, 20, 542–559. [Google Scholar] [CrossRef] [PubMed]
- Chaouat, G.; Dubanchet, S.; Ledée, N. Cytokines: Important for implantation? J. Assist. Reprod. Genet. 2007, 24, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Waclawik, A.; Kaczmarek, M.M.; Blitek, A.; Kaczynski, P.; Ziecik, A.J. Embryo-maternal dialogue during pregnancy establishment and implantation in the pig. Mol. Reprod. Dev. 2017, 84, 842–855. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M. Actions of gonadotrophins on the uterus. Reproduction 2001, 121, 835–842. [Google Scholar] [CrossRef]
- Waszkiewicz, E.M.; Kozlowska, W.; Zmijewska, A.; Franczak, A. Expression of Insulin-Like Growth Factor 1 (IGF-1) and Epidermal Growth Factor (EGF) Receptors and the Effect of IGF-1 and EGF on Androgen and Estrogen Release in the Myometrium of Pigs—In Vitro Study. Animals 2020, 10, 915. [Google Scholar] [CrossRef]
- Franczak, A.; Wojciechowicz, B.; Kolakowska, J.; Kotwica, G. The effect of interleukin-1β, interleukin-6, and tumor necrosis factor-α on estradiol-17β release in the myometrium: The invitro study on the pig model. Theriogenology 2014, 81, 266–274. [Google Scholar] [CrossRef]
- Franczak, A.; Wojciechowicz, B.; Katwica, G. Novel aspects of cytokine action in porcine uterus—Endometrial and myometrial production of estrone (E1) in the presence of interleukin 1β (il1β), interleukin 6 (il6) and tumor necrosis factor (TNFα)—In vitro study. Folia Biol. 2013, 61, 253–261. [Google Scholar] [CrossRef]
- Waszkiewicz, E.M.; Zmijewska, A.; Kozlowska, W.; Franczak, A. Effects of LH and FSH on androgen and oestrogen release in the myometrium of pigs during the oestrous cycle and early pregnancy. Reprod. Fertil. Dev. 2020, 32, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Grandolfo, M. Protection of workers from power frequency electric and magnetic fields: A practical guide. In Occupational Safety and Health Series; International Labour Office: Geneva, Switzerland, 1994; p. 69. ISBN 9789221082613. [Google Scholar]
- Cyster, J.C. Chemokines and cell migration in secondary lymphoid organs. Science 1999, 286, 2098–2102. [Google Scholar] [CrossRef] [PubMed]
- Wolpe, S.D.; Cerami, A. Macrophage inflammatory proteins 1 and 2: Members of a novel superfamily of cytokines. FASEB J. 1989, 3, 2565–2573. [Google Scholar] [CrossRef]
- Van Deventer, H.W.; Serody, J.S.; McKinnon, K.P.; Clements, C.; Brickey, W.J.; Ting, J.P.-Y. Transfection of Macrophage Inflammatory Protein 1α into B16 F10 Melanoma Cells Inhibits Growth of Pulmonary Metastases but Not Subcutaneous Tumors. J. Immunol. 2002, 169, 1634–1639. [Google Scholar] [CrossRef]
- Calpe, S.; Wang, N.; Romero, X.; Berger, S.B.; Lanyi, A.; Engel, P.; Terhorst, C. The SLAM and SAP gene families control innate and adaptive immune responses. Adv. Immunol. 2008, 97, 177–250. [Google Scholar]
- Veillette, A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat. Rev. Immunol. 2006, 6, 56–66. [Google Scholar] [CrossRef]
- Engel, P.; Eck, M.J.; Terhorst, C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat. Rev. Immunol. 2003, 3, 813–821. [Google Scholar] [CrossRef]
- Berger, S.B.; Romero, X.; Ma, C.; Wang, G.; Faubion, W.A.; Liao, G.; Compeer, E.; Keszei, M.; Rameh, L.; Wang, N.; et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 2010, 11, 920–927. [Google Scholar] [CrossRef]
- Patidar, M.; Yadav, N.; Dalai, S.K. Interleukin 15: A key cytokine for immunotherapy. Cytokine Growth Factor Rev. 2016, 31, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Goeddel, D.V. TNF-R1 signaling: A beautiful pathway. Science 2002, 296, 1634–1635. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.R. TN-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.K.; Park, L.S. Characterization of interleukin-15 (IL-15) and the IL-15 receptor complex. J. Clin. Immunol. 1996, 16, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Edgar, J.R.; Manna, P.T.; Nishimura, S.; Banting, G.; Robinson, M.S. Tetherin is an exosomal tether. Elife 2016, 5, 17180. [Google Scholar] [CrossRef] [PubMed]
- Tahara, E.; Tahara, H.; Kanno, M.; Naka, K.; Takeda, Y.; Matsuzaki, T.; Yamazaki, R.; Ishihara, H.; Yasui, W.; Barrett, J.C.; et al. G1P3, an interferon inducible gene 6-16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol. Immunother. 2005, 54, 729–740. [Google Scholar] [CrossRef]
- Cheriyath, V.; Glaser, K.B.; Waring, J.F.; Baz, R.; Hussein, M.A.; Borden, E.C. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J. Clin. Investig. 2007, 117, 3107–3117. [Google Scholar] [CrossRef]
- Cheriyath, V.; Kaur, J.; Davenport, A.; Khalel, A.; Chowdhury, N.; Gaddipati, L. G1P3 (IFI6), a mitochondrial localised antiapoptotic protein, promotes metastatic potential of breast cancer cells through mtROS. Br. J. Cancer 2018, 119, 52–64. [Google Scholar] [CrossRef]
- Mahaki, H.; Tanzadehpanah, H.; Jabarivasal, N.; Sardanian, K.; Zamani, A. A review on the effects of extremely low frequency electromagnetic field (ELF-EMF) on cytokines of innate and adaptive immunity. Electromagn. Biol. Med. 2019, 38, 84–95. [Google Scholar] [CrossRef]
- Guerriero, F.; Ricevuti, G. Extremely low frequency electromagnetic fields stimulation modulates autoimmunity and immune responses: A possible immuno-modulatory therapeutic effect in neurodegenerative diseases. Neural Regen. Res. 2016, 11, 1888–1895. [Google Scholar] [CrossRef]
- Rosado, M.M.; Simkó, M.; Mattsson, M.O.; Pioli, C. Immune-modulating perspectives for low frequency electromagnetic fields in innate immunity. Front. Public Health 2018, 6, 1. [Google Scholar] [CrossRef]
- Markov, M.S. Expanding use of pulsed electromagnetic field therapies. Electromagn. Biol. Med. 2007, 26, 257–274. [Google Scholar] [CrossRef]
- Imada, K.; Leonard, W.J. The Jak-STAT pathway. Mol. Immunol. 2000, 37, 1–11. [Google Scholar] [CrossRef]
- Jang, Y.W.; Gil, K.C.; Lee, J.S.; Kang, W.K.; Park, S.Y.; Hwang, K.W. T-Cell differentiation to T helper 9 phenotype is elevated by extremely low-frequency electromagnetic fields via induction of IL-2 signaling. Bioelectromagnetics 2019, 40, 588–601. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.J.; Croy, B.A. A Map of Relationships Between Uterine Natural Killer Cells and Progesterone Receptor Expressing Cells During Mouse Pregnancy. Placenta 2008, 29, 317–323. [Google Scholar] [CrossRef]
- Jana, B.; Koszykowska, M.; Andronowska, A. The influence of interleukin (IL)-1β and IL-6 and tumour necrosis factor-α on prostaglandin secretion from porcine myometrium during the first third of pregnancy. Acta Vet. Brno 2010, 79, 559–569. [Google Scholar] [CrossRef][Green Version]
- Sarma, V.; Wolf, F.W.; Marks, R.M.; Shows, T.B.; Dixit, V.M. Cloning of a novel tumor necrosis factor-α-inducible primary response gene that is differentially expressed in development and capillary tube-like formation in vitro. J. Immunol. 1992, 148, 3302–3312. [Google Scholar]
- Jia, L.; Shi, Y.; Wen, Y.; Li, W.; Feng, J.; Chen, C. The roles of TNFAIP2 in cancers and infectious diseases. J. Cell. Mol. Med. 2018, 22, 5188–5195. [Google Scholar] [CrossRef]
- Ciarmela, P.; Bloise, E.; Gray, P.C.; Carrarelli, P.; Islam, M.S.; De Pascalis, F.; Severi, F.M.; Vale, W.; Castellucci, M.; Petraglia, F. Activin-A and myostatin response and steroid regulation in human myometrium: Disruption of their signalling in uterine fibroid. J. Clin. Endocrinol. Metab. 2011, 96, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Ciarmela, P.; Carrarelli, P.; Islam, M.S.; Janjusevic, M.; Zupi, E.; Tosti, C.; Castellucci, M.; Petraglia, F. Ulipristal acetate modulates the expression and functions of activin A in leiomyoma cells. Reprod. Sci. 2014, 21, 1120–1125. [Google Scholar] [CrossRef]
- Islam, M.S.; Catherino, W.H.; Protic, O.; Janjusevic, M.; Gray, P.C.; Giannubilo, S.R.; Ciavattini, A.; Lamanna, P.; Tranquilli, A.L.; Petraglia, F.; et al. Role of activin-A and myostatin and their signaling pathway in human myometrial and leiomyoma cell function. J. Clin. Endocrinol. Metab. 2014, 99, 775–785. [Google Scholar] [CrossRef]
- Protic, O.; Toti, P.; Islam, M.S.; Occhini, R.; Giannubilo, S.R.; Catherino, W.H.; Cinti, S.; Petraglia, F.; Ciavattini, A.; Castellucci, M.; et al. Possible involvement of inflammatory/reparative processes in the development of uterine fibroids. Cell Tissue Res. 2016, 364, 415–427. [Google Scholar] [CrossRef]
- Patruno, A.; Ferrone, A.; Costantini, E.; Franceschelli, S.; Pesce, M.; Speranza, L.; Amerio, P.; D’Angelo, C.; Felaco, M.; Grilli, A.; et al. Extremely low-frequency electromagnetic fields accelerates wound healing modulating MMP-9 and inflammatory cytokines. Cell Prolif. 2018, 51, e12432. [Google Scholar] [CrossRef] [PubMed]
- Cook-Mills, J.M.; Marchese, M.E.; Abdala-Valencia, H. Vascular cell adhesion molecule-1 expression and signaling during disease: Regulation by reactive oxygen species and antioxidants. Antioxid. Redox Signal. 2011, 15, 1607–1638. [Google Scholar] [CrossRef]
- Kong, D.H.; Kim, Y.K.; Kim, M.R.; Jang, J.H.; Lee, S. Emerging roles of vascular cell adhesion molecule-1 (VCAM-1) in immunological disorders and cancer. Int. J. Mol. Sci. 2018, 19, 1057. [Google Scholar] [CrossRef]
- Williams, C.D.; Markov, M.S.; Hardman, W.E.; Cameron, I.T. Therapeutic electromagnetic field effects on angiogenesis and tumor growth - PubMed. Aniticancer Res. 2001, 21, 3887–3891. [Google Scholar]
- Chen, J.; Tu, C.; Tang, X.; Li, H.; Yan, J.; Ma, Y.; Wu, H.; Liu, C. The combinatory effect of sinusoidal electromagnetic field and VEGF promotes osteogenesis and angiogenesis of mesenchymal stem cell-laden PCL/HA implants in a rat subcritical cranial defect. Stem Cell Res. Ther. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, I.; Lovett, M.; Rice, W.; Kaplan, D.L.; Balduini, A. Bone marrow osteoblastic niche: A new model to study physiological regulation of megakaryopoiesis. PLoS ONE 2009, 4, e8359. [Google Scholar] [CrossRef]
- Salsi, V.; Ferrari, S.; Ferraresi, R.; Cossarizza, A.; Grande, A.; Zappavigna, V. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol. Cell. Biol. 2009, 29, 5775–5788. [Google Scholar] [CrossRef]
- Cheng, K.; Zou, C. Electromagnetic field effect on separation of nucleotide sequences and unwinding of a double helix during DNA replication. Med. Hypotheses 2006, 66, 148–153. [Google Scholar] [CrossRef]
- Lee, H.M.; Kwon, U.H.; Kim, H.; Kim, H.J.; Kim, B.; Park, J.O.; Moon, E.S.; Moon, S.H. Pulsed electromagnetic field stimulates cellular proliferation in human intervertebral disc cells. Yonsei Med. J. 2010, 51, 954–959. [Google Scholar] [CrossRef]
- Fan, W.; Qian, F.; Ma, Q.; Zhang, P.; Chen, T.; Chen, C.; Zhang, Y.; Deng, P.; Zhou, Z.; Yu, Z. 50 Hz electromagnetic field exposure promotes proliferation and cytokine production of bone marrow mesenchymal stem cells. Int. J. Clin. Exp. Med. 2015, 8, 7394–7404. [Google Scholar]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Makiyama, K.; Hamada, J.I.; Takada, M.; Murakawa, K.; Takahashi, Y.; Tada, M.; Tamoto, E.; Shindo, G.; Matsunaga, A.; Teramoto, K.I.; et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol. Rep. 2005, 13, 673–679. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cantile, M.; Scognamiglio, G.; La Sala, L.; La Mantia, E.; Scaramuzza, V.; Valentino, E.; Tatangelo, F.; Losito, S.; Pezzullo, L.; Chiofalo, M.G.; et al. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int. J. Mol. Sci. 2013, 14, 21727–21740. [Google Scholar] [CrossRef] [PubMed]
- Masai, H.; Arai, K.I. Cdc7 kinase complex: A key regulator in the initiation of DNA replication. J. Cell. Physiol. 2002, 190, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.C.; Kato, H.; Shindo, M.; Masai, H. Cdc7 activates replication checkpoint by phosphorylating the chk1 binding domain of claspin in human cells. Elife 2019, 8, 50796. [Google Scholar] [CrossRef]
- Bonte, D.; Lindvall, C.; Liu, H.; Dykema, K.; Furge, K.; Weinreich, M. Cdc7-Dbf4 kinase overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia 2008, 10, 920–931. [Google Scholar] [CrossRef]
- Montagnoli, A.; Moll, J.; Colotta, F. Targeting cell division cycle 7 kinase: A new approach for cancer therapy. Clin. Cancer Res. 2010, 16, 4503–4508. [Google Scholar] [CrossRef]
- Bortkiewicz, A. Health effects of radiofrequency electromagnetic fields (RF EMF). Ind. Health 2019, 57, 403–405. [Google Scholar] [CrossRef]
- Sharma, S.; Wu, S.Y.; Jimenez, H.; Xing, F.; Zhu, D.; Liu, Y.; Wu, K.; Tyagi, A.; Zhao, D.; Lo, H.W.; et al. Ca2+ and CACNA1H mediate targeted suppression of breast cancer brain metastasis by AM RF EMF. EBioMedicine 2019, 44, 194–208. [Google Scholar] [CrossRef]
- Xu, A.; Wang, Q.; Lin, T. Low-frequency magnetic fields (Lf-mfs) inhibit proliferation by triggering apoptosis and altering cell cycle distribution in breast cancer cells. Int. J. Mol. Sci. 2020, 21, 2952. [Google Scholar] [CrossRef]
- Yin, P.; Navarro, A.; Fang, F.; Xie, A.; Coon, J.S.; Richardson, C.; Bulun, S.E. Early growth response-2 expression in uterine leiomyoma cells: Regulation and function. Fertil. Steril. 2011, 96, 439–444. [Google Scholar] [CrossRef]
- Li, H.; Cao, G.; Zhang, N.; Lou, T.; Wang, Q.; Zhang, Z.; Liu, C. RBP4 regulates trophoblastic cell proliferation and invasion via the PI3K/AKT signaling pathway. Mol. Med. Rep. 2018, 18, 2873–2879. [Google Scholar] [CrossRef]
- Safari, M.; Jadidi, M.; Baghian, A.; Hasanzadeh, H. Proliferation and differentiation of rat bone marrow stem cells by 400 μT electromagnetic field. Neurosci. Lett. 2016, 612, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi-Gahrouei, D.; Hashemi-Beni, B.; Moradi, A.; Aliakbari, M.; Shahbazi-Gahrouei, S. Exposure to global system for mobile communication 900 mhz cellular phone radiofrequency alters growth, proliferation and morphology of michigan cancer foundation-7 cells and mesenchymal stem cells. Int. J. Prev. Med. 2018, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Buckner, C.A.; Buckner, A.L.; Koren, S.A.; Persinger, M.A.; Lafrenie, R.M. Exposure to a specific time-varying electromagnetic field inhibits cell proliferation via cAMP and ERK signaling in cancer cells. Bioelectromagnetics 2018, 39, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Im, S.H.; Yoon, Y.J.; Kim, H.M.; Lee, H.J.; Park, G.S. A 60 Hz uniform electromagnetic field promotes human cell proliferation by decreasing intracellular reactive oxygen species levels. PLoS ONE 2018, 13, e0199753. [Google Scholar] [CrossRef]
- Wojciechowicz, B.; Kotwica, G.; Kołakowska, J.; Zglejc, K.; Martyniak, M.; Franczak, A. The alterations in endometrial and myometrial transcriptome at the time of maternal recognition of pregnancy in pigs. Agri Gene 2016, 2, 5–10. [Google Scholar] [CrossRef]
- Fang, Z.; Rajewsky, N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS ONE 2011, 6, 18067. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Singer, D.S. CIITA and its dual roles in MHC gene transcription. Front. Immunol. 2013, 4, 476. [Google Scholar] [CrossRef]
- Simon, E.J. Opioid receptors and endogenous opioid peptides. Med. Res. Rev. 1991, 11, 357–374. [Google Scholar] [CrossRef]
- Kovatsi, L.; Nikolaou, K. Opioids and the hormone oxytocin. Vitam. Horm. 2019, 111, 195–225. [Google Scholar]
- Rosenblum, A.; Marsch, L.A.; Joseph, H.; Portenoy, R.K. Opioids and the treatment of chronic pain: Controversies, current status, and future directions. Exp. Clin. Psychopharmacol. 2008, 16, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Dziekoński, M.; Zmijewska, A.; Franczak, A.; Kotwica, G.; Czelejewska, W.; Okrasa, S. The expression of mRNAs for opioid precursors in endometrium of cyclic and early pregnant pigs; effects of IL-1β, IL-6 and TNFα. J. Anim. Feed Sci. 2015, 24, 118–126. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinforma. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 January 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 30 June 2021).
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler—A web server for functional interpretation of gene lists. Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, 498–503. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, J.; Pan, Y.; Shen, S.; Lin, L.; Xing, Y. rMATS-DVR: rMATS discovery of differential variants in RNA. Bioinformatics 2017, 33, 2216–2217. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
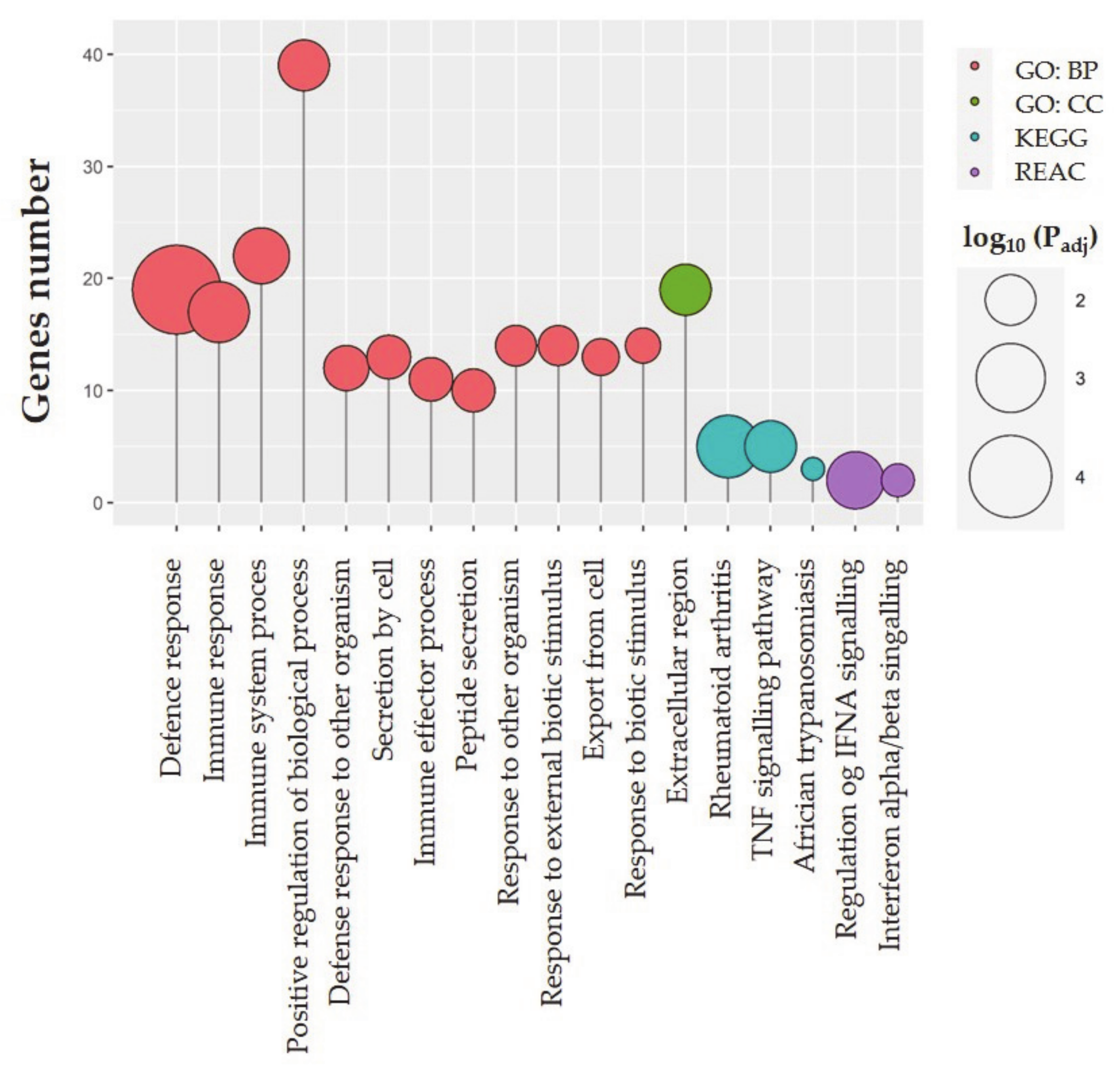
| Term Name | Term ID | Adjusted p-Value |
|---|---|---|
| GO BP | ||
| Defense response | GO:0006952 | 0.00002344438413677965 |
| Immune response | GO:0006955 | 0.0031852702186542744 |
| Immune system process | GO:0002376 | 0.005703042047974045 |
| Positive regulation of biological process | GO:0048518 | 0.009437770426041985 |
| Defense response to other organism | GO:0098542 | 0.015461210039798225 |
| Secretion by cell | GO:0032940 | 0.017106824946529414 |
| Immune effector process | GO:0002252 | 0.0175096188187713 |
| Peptide secretion | GO:0002790 | 0.018160684956297413 |
| Response to other organism | GO:0051707 | 0.020710132468959617 |
| Response to external biotic stimulus | GO:0043207 | 0.02169456662389253 |
| Export from cell | GO:0140352 | 0.025696623178163416 |
| Response to biotic stimulus | GO:0009607 | 0.027680534899551905 |
| GO CC | ||
| Extracellular region | GO:0005576 | 0.009251891504629992 |
| KEGG pathways | ||
| Rheumatoid arthritis | KEGG:05323 | 0.0025090688431046978 |
| TNF signaling pathway | KEGG:04668 | 0.008835243157774076 |
| African trypanosomiasis | KEGG:05143 | 0.03556479600406204 |
| REACTOME | ||
| Regulation of IFNA signaling | REAC: R-SSC-912694 | 0.005030993232569516 |
| Interferon-alpha/beta signaling | REAC: R-SSC-909733 | 0.03000607786831512 |
| Fold Change 1 | Gene Symbol | Assay ID |
|---|---|---|
| Differentially expressed genes | ||
| 7.339974418 | HOXD13 | Ss06916655_m1 |
| 7.084714455 | PDYN | Ss03391715_m1 |
| 4.690302179 | VCAM1 | Ss03390912_m1 |
| 3.659258776 | IL15 | Ss03394854_m1 |
| 2.971483779 | STAT5A | Ss03394621_m1 |
| 2.81226065 | TNF | Ss03391318_g1 |
| −1.878442068 | EGR2 | Ss03388929_m1 |
| Reference genes | ||
| - | ACTB | Ss03374854_g1 |
| - | GAPDH | Ss03376081_u1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drzewiecka, E.M.; Kozlowska, W.; Paukszto, L.; Zmijewska, A.; Wydorski, P.J.; Jastrzebski, J.P.; Franczak, A. Effect of the Electromagnetic Field (EMF) Radiation on Transcriptomic Profile of Pig Myometrium during the Peri-Implantation Period—An In Vitro Study. Int. J. Mol. Sci. 2021, 22, 7322. https://doi.org/10.3390/ijms22147322
Drzewiecka EM, Kozlowska W, Paukszto L, Zmijewska A, Wydorski PJ, Jastrzebski JP, Franczak A. Effect of the Electromagnetic Field (EMF) Radiation on Transcriptomic Profile of Pig Myometrium during the Peri-Implantation Period—An In Vitro Study. International Journal of Molecular Sciences. 2021; 22(14):7322. https://doi.org/10.3390/ijms22147322
Chicago/Turabian StyleDrzewiecka, Ewa Monika, Wiktoria Kozlowska, Lukasz Paukszto, Agata Zmijewska, Pawel Jozef Wydorski, Jan Pawel Jastrzebski, and Anita Franczak. 2021. "Effect of the Electromagnetic Field (EMF) Radiation on Transcriptomic Profile of Pig Myometrium during the Peri-Implantation Period—An In Vitro Study" International Journal of Molecular Sciences 22, no. 14: 7322. https://doi.org/10.3390/ijms22147322
APA StyleDrzewiecka, E. M., Kozlowska, W., Paukszto, L., Zmijewska, A., Wydorski, P. J., Jastrzebski, J. P., & Franczak, A. (2021). Effect of the Electromagnetic Field (EMF) Radiation on Transcriptomic Profile of Pig Myometrium during the Peri-Implantation Period—An In Vitro Study. International Journal of Molecular Sciences, 22(14), 7322. https://doi.org/10.3390/ijms22147322








