Purine Nucleotides Metabolism and Signaling in Huntington’s Disease: Search for a Target for Novel Therapies
Abstract
1. Introduction
1.1. Huntington’s Disease Pathophysiology
1.2. Purine Nucleotides Metabolism and Signaling
2. Purines in Huntington’s Disease
2.1. Cellular Changes Related to Affected Huntingtin Expression
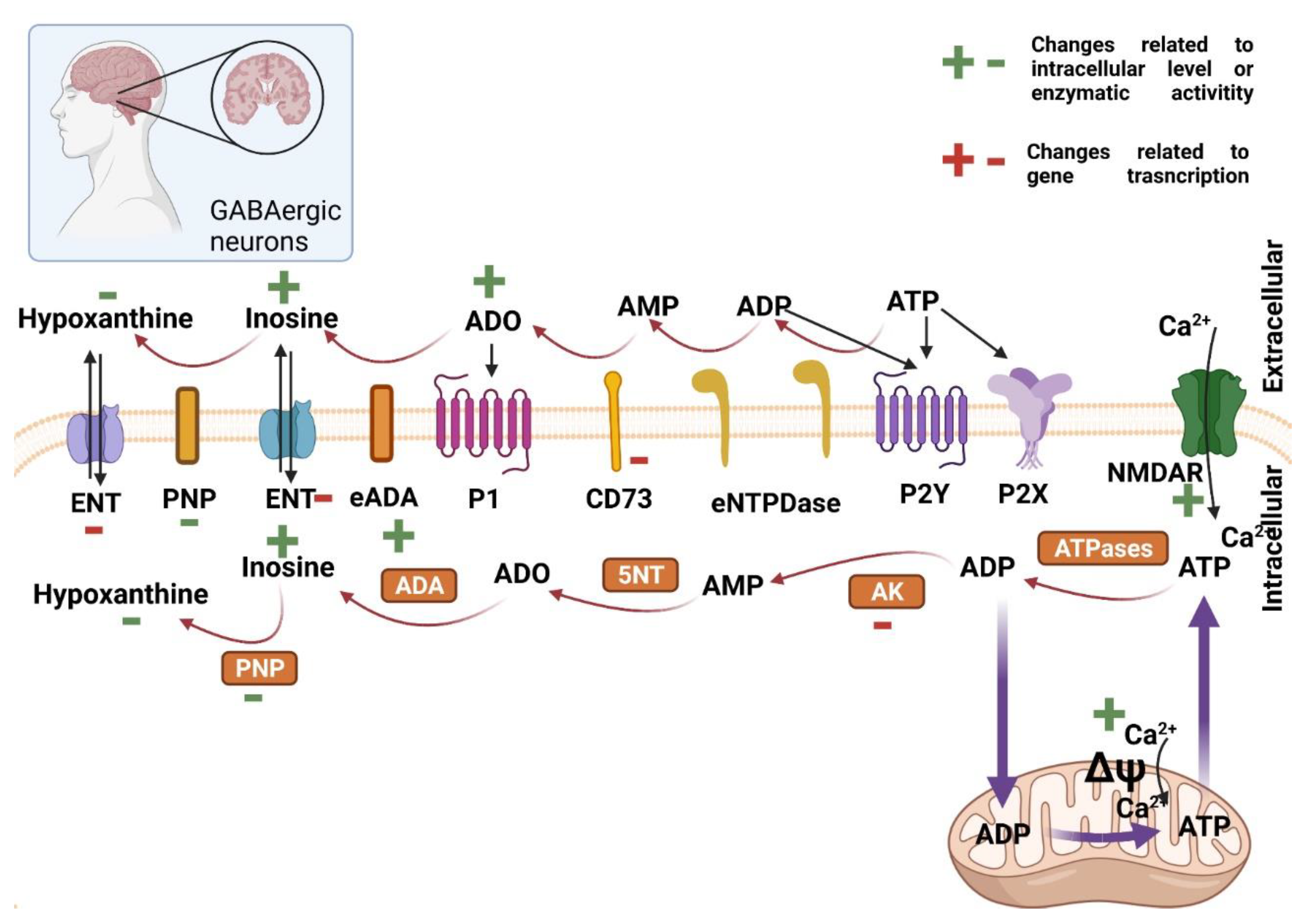
2.2. Purine Nucleotides Metabolism and Signaling in the Central Nervous System in Huntington’s Disease
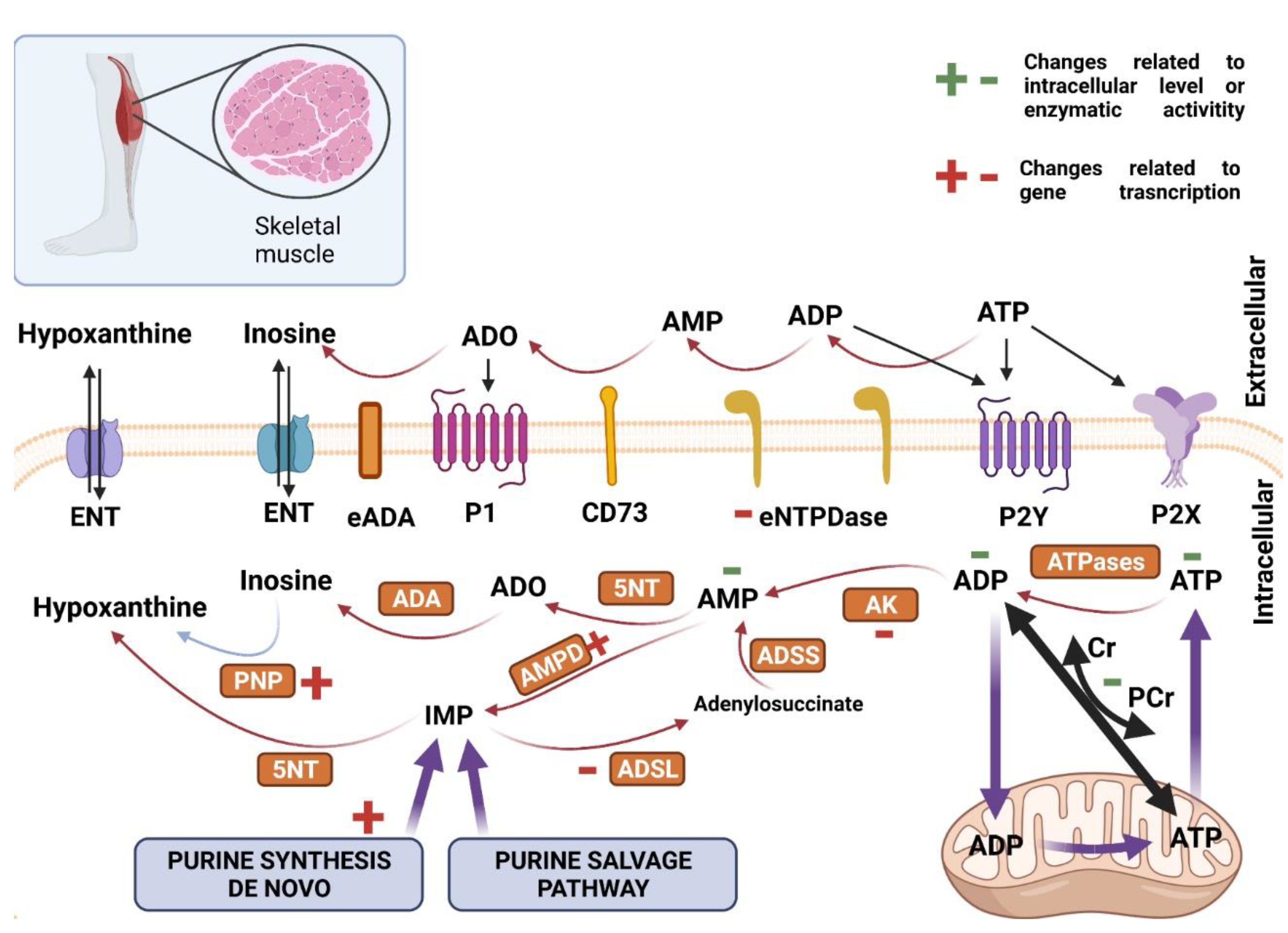
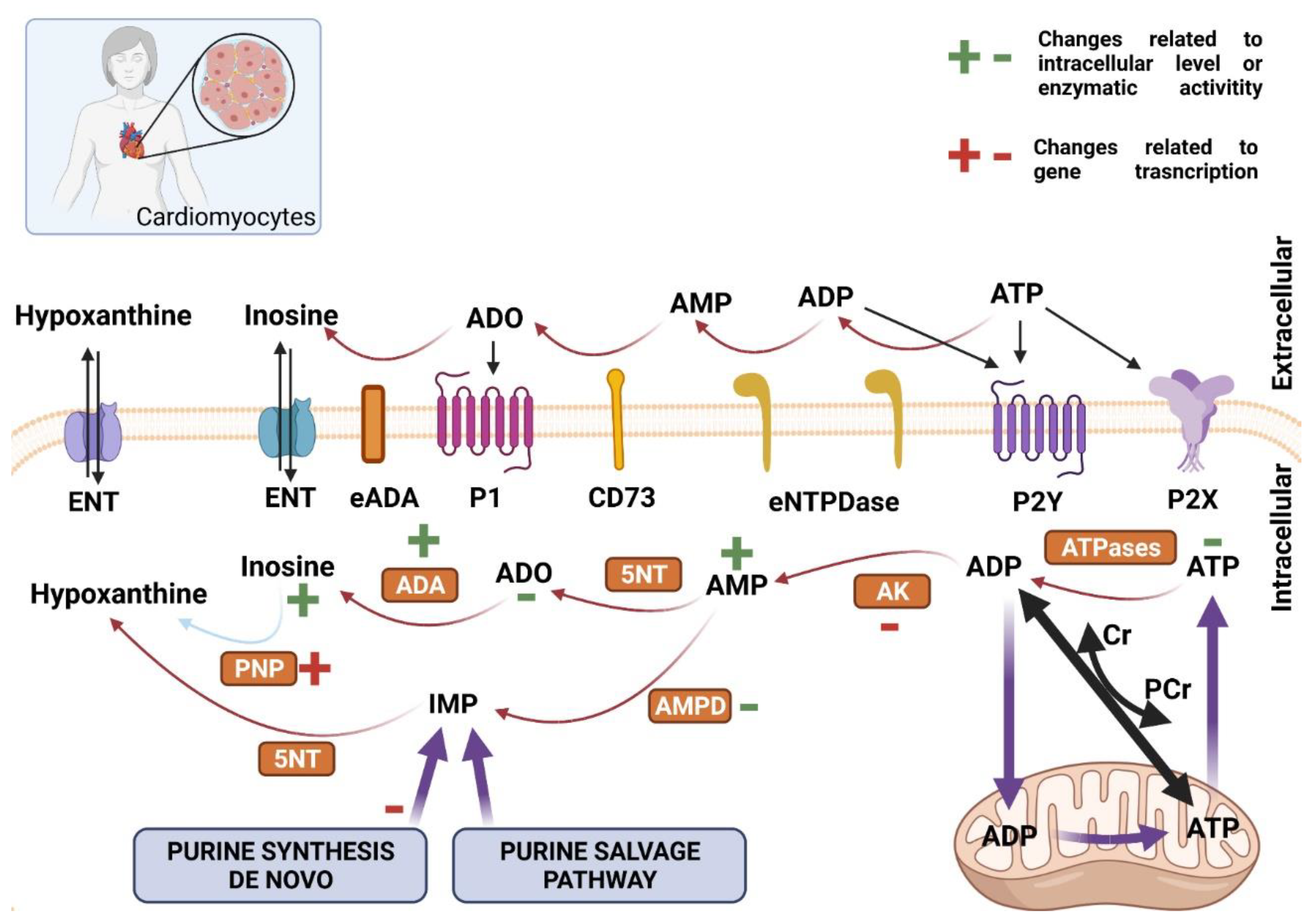
2.3. Purine Nucleotides Metabolism and Signaling in Skeletal Muscle and Heart in Huntington’s Disease
3. Purine Nucleotides Metabolism and Signaling as a Target for Huntington’s Disease Therapies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Walker, F. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Sturrock, A.; Leavitt, B.R. The Clinical and Genetic Features of Huntington Disease. J. Geriatr. Psychiatry Neurol. 2010, 23, 243–259. [Google Scholar] [CrossRef]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef]
- Epping, E.A.; Paulsen, J.S. Depression in the early stages of Huntington disease. Neurodegener. Dis. Manag. 2011, 1, 407–414. [Google Scholar] [CrossRef]
- Ross, C.A.; Tabrizi, S. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011, 10, 83–98. [Google Scholar] [CrossRef]
- Li, S.H.; Schilling, G.; Young, W.S., 3rd; Li, X.J.; Margolis, R.L.; Stine, O.C.; Wagster, M.V.; Abbott, M.H.; Franz, M.L.; Ranen, N.G.; et al. Huntington’s Disease Gene (IT15) Is Widely Expressed in Human and Rat Tissues. Neuron 1993, 11, 985–993. [Google Scholar] [CrossRef]
- Li, S.-H.; Li, X.-J. Huntingtin–protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004, 20, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.W.; Turmaine, M.; Cozens, B.A.; DiFiglia, M.; Sharp, A.H.; Ross, C.A.; Scherzinger, E.; Wanker, E.E.; Mangiarini, L.; Bates, G. Formation of Neuronal Intranuclear Inclusions Underlies the Neurological Dysfunction in Mice Transgenic for the HD Mutation. Cell 1997, 90, 537–548. [Google Scholar] [CrossRef]
- Moffitt, H.; McPhail, G.D.; Woodman, B.; Hobbs, C.; Bates, G.P. Formation of Polyglutamine Inclusions in a Wide Range of Non-CNS Tissues in the HdhQ150 Knock-In Mouse Model of Huntington’s Disease. PLoS ONE 2009, 4, e8025. [Google Scholar] [CrossRef]
- Hoffner, G.; Kahlem, P.; Djian, P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with beta-tubulin: Relevance to Huntington’s disease. J. Cell Sci. 2002, 115 Pt 5, 941–948. [Google Scholar] [CrossRef]
- Godin, J.; Colombo, K.; Molina-Calavita, M.; Keryer, G.; Zala, D.; Charrin, B.C.; Dietrich, P.; Volvert, M.-L.; Guillemot, F.; Dragatsis, I.; et al. Huntingtin Is Required for Mitotic Spindle Orientation and Mammalian Neurogenesis. Neuron 2010, 67, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Veliera, J.; Kima, M.; Schwarza, C.; Kim, T.W.; Sappa, E.; Chaseb, K.; Aroninb, N.; Di Figliaa, M. Wild-Type and Mutant Huntingtins Function in Vesicle Trafficking in the Secretory and Endocytic Pathways. Exp. Neurol. 1998, 152, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Harjes, P.; Wanker, E.E. The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem. Sci. 2003, 28, 425–433. [Google Scholar] [CrossRef]
- Schulte, J.; Littleton, J.T. The biological function of the Huntingtin protein and its relevance to Huntington’s Disease pathology. Curr. Trends Neurol. 2011, 5, 65–78. [Google Scholar] [PubMed]
- Duyao, M.P.; Auerbach, A.B.; Ryan, A.; Persichetti, F.; Barnes, G.T.; McNeil, S.M.; Ge, P.; Vonsattel, J.P.; Gusella, J.F.; Joyner, A.L.; et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 1995, 269, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Robson, S.C.; Sévigny, J.; Zimmermann, H. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal. 2006, 2, 409–430. [Google Scholar] [CrossRef] [PubMed]
- Yegutkin, G.G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: Functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. [Google Scholar] [CrossRef]
- Yegutkin, G.G. Nucleotide- and nucleoside-converting ectoenzymes: Important modulators of purinergic signalling cascade. Biochim. Biophys. Acta 2008, 1783, 673–694. [Google Scholar] [CrossRef]
- Kukulski, F.; Lévesque, S.A.; Sévigny, J. Impact of Ectoenzymes on P2 and P1 Receptor Signaling. Adv. Pharmacol. 2011, 61, 263–299. [Google Scholar] [CrossRef]
- Lin, J.H.-C.; Takano, T.; Arcuino, G.; Wang, X.; Hu, F.; Darzynkiewicz, Z.; Nunes, M.; Goldman, S.; Nedergaard, M. Purinergic signaling regulates neural progenitor cell expansion and neurogenesis. Dev. Biol. 2007, 302, 356–366. [Google Scholar] [CrossRef]
- Shukla, V.; Zimmermann, H.; Wang, L.; Kettenmann, H.; Raab, S.; Hammer, K.; Sévigny, J.; Robson, S.C.; Braun, N. Functional expression of the ecto-ATPase NTPDase2 and of nucleotide receptors by neuronal progenitor cells in the adult murine hippocampus. J. Neurosci. Res. 2005, 80, 600–610. [Google Scholar] [CrossRef] [PubMed]
- von Kügelgen, I. Pharmacology of P2Y receptors. Brain Res. Bull. 2019, 151, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Knight, G. Purinergic Receptors. In Encyclopedia of Neuroscience; Elsevier BV: Amsterdam, The Netherlands, 2009; pp. 1245–1252. [Google Scholar]
- Ismailoglu, I.; Chen, Q.; Popowski, M.; Yang, L.; Gross, S.S.; Brivanlou, A.H. Huntingtin protein is essential for mitochondrial metabolism, bioenergetics and structure in murine embryonic stem cells. Dev. Biol. 2014, 391, 230–240. [Google Scholar] [CrossRef]
- Glaser, T.; Shimojo, H.; Ribeiro, D.E.; Martins, P.P.L.; Beco, R.P.; Kosinski, M.; Sampaio, V.F.A.; Corrêa-Velloso, J.; Oliveira-Giacomelli, Á.; Lameu, C.; et al. ATP and spontaneous calcium oscillations control neural stem cell fate determination in Huntington’s disease: A novel approach for cell clock research. Mol. Psychiatry 2020, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Tomczyk, M.; Glaser, T.; Ulrich, H.; Slominska, E.M.; Smolenski, R.T. Huntingtin protein maintains balanced energetics in mouse cardiomyocytes. Nucleosides Nucleotides Nucleic Acids 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Irmak, D.; Fatima, A.; Gutiérrez-Garcia, R.; Rinschen, M.M.; Wagle, P.; Altmüller, J.; Arrigoni, L.; Hummel, B.; Klein, C.; Frese, C.K.; et al. Mechanism suppressing H3K9 trimethylation in pluripotent stem cells and its demise by polyQ-expanded huntingtin mutations. Hum. Mol. Genet. 2018, 27, 4117–4134. [Google Scholar] [CrossRef]
- Toczek, M.; Pierzynowska, K.; Kutryb-Zajac, B.-; Gaffke, L.; Slominska, E.M.; Wegrzyn, G.; Smolenski, R. Characterization of adenine nucleotide metabolism in the cellular model of Huntington’s disease. Nucleosides Nucleotides Nucleic Acids 2018, 37, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Coppen, E.M.; Van Der Grond, J.; Roos, R.A.C. Atrophy of the putamen at time of clinical motor onset in Huntington’s disease: A 6-year follow-up study. J. Clin. Mov. Disord. 2018, 5, 2. [Google Scholar] [CrossRef]
- Ipata, P.L. Origin, utilization, and recycling of nucleosides in the central nervous system. Adv. Physiol. Educ. 2011, 35, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Jodeiri Farshbaf, M.; Ghaedi, K. Huntington’s Disease and Mitochondria. Neurotox. Res. 2017, 32, 518–529. [Google Scholar] [CrossRef]
- Herben-Dekker, M.; Van Oostrom, J.C.H.; Roos, R.A.C.; Jurgens, C.K.; Witjes-Ané, M.-N.W.; Kremer, H.P.H.; Leenders, K.L.; Spikman, J.M. Striatal metabolism and psychomotor speed as predictors of motor onset in Huntington’s disease. J. Neurol. 2014, 261, 1387–1397. [Google Scholar] [CrossRef]
- Intihar, T.A.; Martinez, E.A.; Gomez-Pastor, R. Mitochondrial Dysfunction in Huntington’s Disease; Interplay Between HSF1, p53 and PGC-1α Transcription Factors. Front. Cell. Neurosci. 2019, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Gines, S.; Seong, I.S.; Fossale, E.; Ivanova, E.; Trettel, F.; Gusella, J.F.; Wheeler, V.C.; Persichetti, F.; MacDonald, M.E. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Hum. Mol. Genet. 2003, 12, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Mochel, F.; Durant, B.; Meng, X.; O’Callaghan, J.; Yu, H.; Brouillet, E.; Wheeler, V.C.; Humbert, S.; Schiffmann, R.; Durr, A. Early Alterations of Brain Cellular Energy Homeostasis in Huntington Disease Models. J. Biol. Chem. 2012, 287, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Seong, I.S.; Ivanova, E.; Lee, J.-M.; Choo, Y.S.; Fossale, E.; Anderson, M.; Gusella, J.F.; Laramie, J.M.; Myers, R.H.; Lesort, M.; et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005, 14, 2871–2880. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, A.; Rivera-Sánchez, S.; Castro, M.D.R.; Acevedo-Torres, K.; Rane, A.; Torres-Ramos, C.A.; Nicholls, D.G.; Andersen, J.K.; Ayala-Torres, S. Mitochondrial DNA damage Is associated with reduced mitochondrial bioenergetics in Huntington’s disease. Free. Radic. Biol. Med. 2012, 53, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Fuster-Matanzo, A.; Llorens-Martín, M.; De Barreda, E.G.; Avila, J.; Hernandez, F. Different Susceptibility to Neurodegeneration of Dorsal and Ventral Hippocampal Dentate Gyrus: A Study with Transgenic Mice Overexpressing GSK3β. PLoS ONE 2011, 6, e27262. [Google Scholar] [CrossRef]
- Cui, L.; Jeong, H.; Borovecki, F.; Parkhurst, C.N.; Tanese, N.; Krainc, D. Transcriptional Repression of PGC-1α by Mutant Huntingtin Leads to Mitochondrial Dysfunction and Neurodegeneration. Cell 2006, 127, 59–69. [Google Scholar] [CrossRef]
- Yablonska, S.; Ganesan, V.; Ferrando, L.M.; Kim, J.; Pyzel, A.; Baranova, O.V.; Khattar, N.K.; Larkin, T.M.; Baranov, S.V.; Chen, N.; et al. Mutant huntingtin disrupts mitochondrial proteostasis by interacting with TIM. Proc. Natl. Acad. Sci. USA 2019, 116, 16593–16602. [Google Scholar] [CrossRef]
- Orr, A.L.; Li, S.; Wang, C.-E.; Li, H.; Wang, J.; Rong, J.; Xu, X.; Mastroberardino, P.G.; Greenamyre, J.T.; Li, X.-J. N-Terminal Mutant Huntingtin Associates with Mitochondria and Impairs Mitochondrial Trafficking. J. Neurosci. 2008, 28, 2783–2792. [Google Scholar] [CrossRef]
- Mattson, M.P.; Gleichmann, M.; Cheng, A. Mitochondria in Neuroplasticity and Neurological Disorders. Neuron 2008, 60, 748–766. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.E.; Xu, B.; Lu, B.; Hempstead, B.L. New Insights in the Biology of BDNF Synthesis and Release: Implications in CNS Function. J. Neurosci. 2009, 29, 12764–12767. [Google Scholar] [CrossRef] [PubMed]
- Markham, A.; Cameron, I.; Franklin, P.; Spedding, M. BDNF increases rat brain mitochondrial respiratory coupling at complex I, but not complex II. Eur. J. Neurosci. 2004, 20, 1189–1196. [Google Scholar] [CrossRef]
- Zuccato, C.; Ciammola, A.; Rigamonti, D.; Leavitt, B.R.; Goffredo, D.; Conti, L.; MacDonald, M.E.; Friedlander, R.M.; Silani, V.; Hayden, M.; et al. Loss of Huntingtin-Mediated BDNF Gene Transcription in Huntington’s Disease. Science 2001, 293, 493–498. [Google Scholar] [CrossRef]
- Fontán-Lozano, Á.; López-Lluch, G.; Delgado-García, J.M.; Navas, P.; Carrión, Á.M. Molecular Bases of Caloric Restriction Regulation of Neuronal Synaptic Plasticity. Mol. Neurobiol. 2008, 38, 167–177. [Google Scholar] [CrossRef]
- Browne, S.E.; Bowling, A.C.; MacGarvey, U.; Baik, M.J.; Berger, S.C.; Muquit, M.M.K.; Bird, E.D.; Beal, M.F. Oxidative damage and metabolic dysfunction in Huntington’s disease: Selective vulnerability of the basal ganglia. Ann. Neurol. 1997, 41, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Benchoua, A.; Trioulier, Y.; Zala, D.; Gaillard, M.-C.; Lefort, N.; Dufour, N.; Saudou, F.; Elalouf, J.-M.; Hirsch, E.; Hantraye, P.; et al. Involvement of Mitochondrial Complex II Defects in Neuronal Death Produced by N-Terminus Fragment of Mutated Huntingtin. Mol. Biol. Cell 2006, 17, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.K.; Calingasan, N.Y.; Yang, L.; Hennessey, T.; Johri, A.; Beal, M.F. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington’s disease following chronic energy deprivation. Hum. Mol. Genet. 2010, 19, 3190–3205. [Google Scholar] [CrossRef]
- Chaturvedi, R.K.; Adhihetty, P.; Shukla, S.; Hennessy, T.; Calingasan, N.; Yang, L.; Starkov, A.; Kiaei, M.; Cannella, M.; Sassone, J.; et al. Impaired PGC-1α function in muscle in Huntington’s disease. Hum. Mol. Genet. 2009, 18, 3048–3065. [Google Scholar] [CrossRef]
- Lin, J.; Wu, P.-H.; Tarr, P.T.; Lindenberg, K.S.; St-Pierre, J.; Zhang, C.-Y.; Mootha, V.K.; Jager, S.; Vianna, C.R.; Reznick, R.M.; et al. Defects in Adaptive Energy Metabolism with CNS-Linked Hyperactivity in PGC-1α Null Mice. Cell 2004, 119, 121–135. [Google Scholar] [CrossRef]
- Chou, S.-Y.; Lee, Y.-C.; Chen, H.-M.; Chiang, M.-C.; Lai, H.-L.; Chang, H.-H.; Wu, Y.-C.; Sun, C.-N.; Chien, C.-L.; Lin, Y.-S.; et al. CGS21680 attenuates symptoms of Huntington’s disease in a transgenic mouse model. J. Neurochem. 2005, 93, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Chen, H.-M.; Lin, J.-T.; Chang, C.-P.; Chang, W.-C.; Kang, J.-J.; Sun, C.-P.; Tao, M.-H.; Tu, P.-H.; Chang, C.; et al. Nuclear translocation of AMPK-α1 potentiates striatal neurodegeneration in Huntington’s disease. J. Cell Biol. 2011, 194, 209–227. [Google Scholar] [CrossRef]
- Glaser, T.; Andrejew, R.; Oliveira-Giacomelli, Á.; Ribeiro, D.E.; Marques, L.B.; Ye, Q.; Ren, W.-J.; Semyanov, A.; Illes, P.; Tang, Y.; et al. Purinergic Receptors in Basal Ganglia Diseases: Shared Molecular Mechanisms between Huntington’s and Parkinson’s Disease. Neurosci. Bull. 2020, 36, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Dayalu, P.; Albin, R.L. Huntington Disease: Pathogenesis and Treatment. Neurol. Clin. 2015, 33, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Patassini, S.; Begley, P.; Xu, J.; Church, S.J.; Reid, S.J.; Kim, E.H.; Curtis, M.; Dragunow, M.; Waldvogel, H.J.; Snell, R.G.; et al. Metabolite mapping reveals severe widespread perturbation of multiple metabolic processes in Huntington’s disease human brain. Biochim. Biophys. Acta 2016, 1862, 1650–1662. [Google Scholar] [CrossRef]
- Nambron, R.; Silajdzic, E.; Kalliolia, E.; Ottolenghi, C.; Hindmarsh, P.; Hill, N.R.; Costelloe, S.J.; Martin, N.G.; Positano, V.; Watt, H.C.; et al. A Metabolic Study of Huntington’s Disease. PLoS ONE 2016, 11, e014648. [Google Scholar] [CrossRef]
- Gianfriddo, M.; Melani, A.; Turchi, D.; Giovannini, M.; Pedata, F. Adenosine and glutamate extracellular concentrations and mitogen-activated protein kinases in the striatum of Huntington transgenic mice. Selective antagonism of adenosine A2A receptors reduces transmitter outflow. Neurobiol. Dis. 2004, 17, 77–88. [Google Scholar] [CrossRef]
- Lee, C.-F.; Chern, Y. Adenosine Receptors and Huntington’s Disease. Int. Rev. Neurobiol. 2014, 119, 195–232. [Google Scholar] [CrossRef]
- Glass, M.; Dragunow, M.; Faull, R. The pattern of neurodegeneration in Huntington’s disease: A comparative study of cannabinoid, dopamine, adenosine and GABAA receptor alterations in the human basal ganglia in Huntington’s disease. Neuroscience 2000, 97, 505–519. [Google Scholar] [CrossRef]
- Bauer, A.; Zilles, K.; Matusch, A.; Holzmann, C.; Riess, O.; Von Hörsten, S. Regional and subtype selective changes of neurotransmitter receptor density in a rat transgenic for the Huntington’s disease mutation. J. Neurochem. 2005, 94, 639–650. [Google Scholar] [CrossRef]
- Cha, J.-H.J.; Frey, A.S.; Alsdorf, S.A.; Kerner, J.A.; Kosinski, C.M.; Mangiarini, L.; Penney, J.B.; Davies, S.W.; Bates, G.; Young, A.B. Altered neurotransmitter receptor expression in transgenic mouse models of Huntington’s disease. Philos. Trans. R. Soc. B Biol. Sci. 1999, 354, 981–989. [Google Scholar] [CrossRef]
- Mievis, S.; Blum, D.; Ledent, C. A2A receptor knockout worsens survival and motor behaviour in a transgenic mouse model of Huntington’s disease. Neurobiol. Dis. 2011, 41, 570–576. [Google Scholar] [CrossRef]
- Villar-Menéndez, I.; Blanch, M.; Tyebji, S.; Pereira-Veiga, T.; Albasanz, J.L.; Martín, M.; Ferrer, I.; Pérez-Navarro, E.; Barrachina, M. Increased 5-Methylcytosine and Decreased 5-Hydroxymethylcytosine Levels are Associated with Reduced Striatal A2AR Levels in Huntington’s Disease. NeuroMolecular Med. 2013, 15, 295–309. [Google Scholar] [CrossRef]
- Domenici, M.; Scattoni, M.L.; Martire, A.; Lastoria, G.; Potenza, R.; Borioni, A.; Venerosi, A.; Calamandrei, G.; Popoli, P. Behavioral and electrophysiological effects of the adenosine A2A receptor antagonist SCH 58261 in R6/2 Huntington’s disease mice. Neurobiol. Dis. 2007, 28, 197–205. [Google Scholar] [CrossRef]
- Li, W.; Silva, H.; Real, J.I.; Wang, Y.-M.; Rial, D.; Li, P.; Payen, M.-P.; Zhou, Y.; Muller, C.E.; Tomé, A.R.; et al. Inactivation of adenosine A2A receptors reverses working memory deficits at early stages of Huntington’s disease models. Neurobiol. Dis. 2015, 79, 70–80. [Google Scholar] [CrossRef]
- Díaz-Hernández, M.; Díez-Zaera, M.; Sánchez-Nogueiro, J.; Gómez-Villafuertes, R.; Canals, J.M.; Alberch, J.; Miras-Portugal, M.T.; Lucas, J.J. Altered P2X7-receptor level and function in mouse models of Huntington’s disease and therapeutic efficacy of antagonist administration. FASEB J. 2009, 23, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
- Busse, M.E.; Hughes, G.; Wiles, C.M.; Rosser, A.E. Use of hand-held dynamometry in the evaluation of lower limb muscle strength in people with Huntington’s disease. J. Neurol. 2008, 255, 1534–1540. [Google Scholar] [CrossRef]
- Ribchester, R.R.; Thomson, D.; Wood, N.I.; Hinks, T.; Gillingwater, T.H.; Wishart, T.M.; Court, F.A.; Morton, A.J. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington’s disease mutation. Eur. J. Neurosci. 2004, 20, 3092–3114. [Google Scholar] [CrossRef] [PubMed]
- Romer, S.H.; Metzger, S.; Peraza, K.; Wright, M.C.; Jobe, D.S.; Song, L.-S.; Rich, M.M.; Foy, B.D.; Talmadge, R.J.; Voss, A.A. A mouse model of Huntington’s disease shows altered ultrastructure of transverse tubules in skeletal muscle fibers. J. Gen. Physiol. 2021, 153, 153. [Google Scholar] [CrossRef]
- Orth, M.; Cooper, J.M.; Bates, G.; Schapira, A.H.V. Inclusion formation in Huntington’s disease R6/2 mouse muscle cultures. J. Neurochem. 2003, 87, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Valadão, P.A.C.; De Aragão, B.C.; Andrade, J.N.; Magalhães-Gomes, M.P.S.; Foureaux, G.; Joviano-Santos, J.V.; Nogueira, J.C.; Machado, T.; De Jesus, I.C.G.; Nogueira, J.M.; et al. Abnormalities in the Motor Unit of a Fast-Twitch Lower Limb Skeletal Muscle in Huntington’s Disease. ASN Neuro 2019, 11, 11. [Google Scholar] [CrossRef]
- Valadão, P.A.C.; De Aragão, B.C.; Andrade, J.N.; Magalhães-Gomes, M.P.S.; Foureaux, G.; Joviano-Santos, J.V.; Nogueira, J.C.; Ribeiro, F.; Tapia, J.C.; Guatimosim, C. Muscle atrophy is associated with cervical spinal motoneuron loss in BACHD mouse model for Huntington’s disease. Eur. J. Neurosci. 2016, 45, 785–796. [Google Scholar] [CrossRef]
- Strand, A.D.; Aragaki, A.K.; Shaw, D.; Bird, T.; Holton, J.; Turner, C.; Tapscott, S.J.; Tabrizi, S.; Schapira, A.H.; Kooperberg, C.; et al. Gene expression in Huntington’s disease skeletal muscle: A potential biomarker. Hum. Mol. Genet. 2005, 14, 1863–1876. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Toczek, M.; Smeets, C.J.L.M.; Franklin, S.A.; Bondulich, M.K.; Jolinon, N.; Muller, T.; Ahmed, M.; Dick, J.R.T.; Piotrowska, I.; et al. HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy. PLoS Genet. 2015, 11, e1005021. [Google Scholar] [CrossRef]
- Ezielonka, D.; Epiotrowska, I.; Marcinkowski, J.T.; Emielcarek, M. Skeletal muscle pathology in Huntington’s disease. Front. Physiol. 2014, 5, 380. [Google Scholar] [CrossRef]
- Saft, C.; Zange, J.; Andrich, J.; Müller, K.; Lindenberg, K.; Landwehrmeyer, B.; Vorgerd, M.; Kraus, P.H.; Przuntek, H.; Schöls, L.; et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington’s disease. Mov. Disord. 2005, 20, 674–679. [Google Scholar] [CrossRef]
- Kojer, K.; Hering, T.; Bazenet, C.; Weiss, A.; Herrmann, F.; Taanman, J.-W.; Orth, M. Huntingtin Aggregates and Mitochondrial Pathology in Skeletal Muscle but not Heart of Late-Stage R6/2 Mice. J. Huntington’s Dis. 2019, 8, 145–159. [Google Scholar] [CrossRef]
- Tsang, T.M.; Woodman, B.; McLoughlin, G.A.; Griffin, J.L.; Tabrizi, S.J.; Bates, G.P.; Holmes, E. Metabolic Characterization of the R6/2 Transgenic Mouse Model of Huntington’s Disease by High-Resolution MAS1H NMR Spectroscopy. J. Proteome Res. 2006, 5, 483–492. [Google Scholar] [CrossRef]
- Ciammola, A.; Sassone, J.; Alberti, L.; Meola, G.; Mancinelli, E.; Russo, M.A.; Squitieri, F.; Silani, V. Increased apoptosis, huntingtin inclusions and altered differentiation in muscle cell cultures from Huntington’s disease subjects. Cell Death Differ. 2006, 13, 2068–2078. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.; Calingasan, N.Y.; Hennessey, T.M.; Sharma, A.; Yang, L.; Wille, E.; Chandra, A.; Beal, M.F. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2011, 21, 1124–1137. [Google Scholar] [CrossRef]
- Miller, S.G.; Hafen, P.S.; Brault, J.J. Increased Adenine Nucleotide Degradation in Skeletal Muscle Atrophy. Int. J. Mol. Sci. 2019, 21, 88. [Google Scholar] [CrossRef]
- Lanska, D.J.; LaVine, L.; Schoenberg, B.S. Huntington’s disease mortality in the United States. Neurology 1988, 38, 769. [Google Scholar] [CrossRef] [PubMed]
- Chiu, E.; Alexander, L. Causes of death in Huntington’s Disease. Med. J. Aust. 1982, 1, 153. [Google Scholar] [CrossRef] [PubMed]
- Andrich, J.; Schmitz, T.; Saft, C.; Postert, T.; Kraus, P.; Epplen, J.T.; Przuntek, H.; Agelink, M.W. Autonomic nervous system function in Huntington’s disease. J. Neurol. Neurosurg. Psychiatry 2002, 72, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Hasselbalch, S.G.; Oberg, G.; Sorensen, S.A.; Andersen, A.R.; Waldemar, G.; Schmidt, J.F.; Fenger, K.; Paulson, O.B. Reduced regional cerebral blood flow in Huntington’s disease studied by SPECT. J. Neurol. Neurosurg. Psychiatry 1992, 55, 1018–1023. [Google Scholar] [CrossRef]
- Melik, Z.; Kobal, J.; Cankar, K.; Strucl, M. Microcirculation response to local cooling in patients with Huntington’s disease. J. Neurol. 2012, 259, 921–928. [Google Scholar] [CrossRef]
- Sharma, K.R.; Romano, J.G.; Ayyar, D.R.; Rotta, F.T.; Facca, A.; Sanchez-Ramos, J. Sympathetic Skin Response and Heart Rate Variability in Patients With Huntington Disease. Arch. Neurol. 1999, 56, 1248–1252. [Google Scholar] [CrossRef]
- Terroba-Chambi, C.; Bruno, V.; Vigo, D.E.; Merello, M. Heart rate variability and falls in Huntington’s disease. Clin. Auton. Res. 2021, 31, 281–292. [Google Scholar] [CrossRef]
- Mielcarek, M.; Inuabasi, L.; Bondulich, M.K.; Muller, T.; Osborne, G.; Franklin, S.A.; Smith, D.L.; Neueder, A.; Rosinski, J.; Rattray, I.; et al. Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington’s Disease. PLoS Genet. 2014, 10, e1004550. [Google Scholar] [CrossRef] [PubMed]
- Wood, N.I.; Sawiak, S.J.; Buonincontri, G.; Niu, Y.; Kane, A.D.; Carpenter, T.A.; Giussani, D.; Morton, A.J. Direct Evidence of Progressive Cardiac Dysfunction in a Transgenic Mouse Model of Huntington’s Disease. J. Huntington’s Dis. 2012, 1, 57–64. [Google Scholar] [CrossRef]
- Kiriazis, H.; Jennings, N.L.; Davern, P.; Lambert, G.; Su, Y.; Pang, T.; Du, X.; La Greca, L.; Head, G.; Hannan, A.J.; et al. Neurocardiac dysregulation and neurogenic arrhythmias in a transgenic mouse model of Huntington’s disease. J. Physiol. 2012, 590, 5845–5860. [Google Scholar] [CrossRef]
- Mielcarek, M.; Bondulich, M.K.; Inuabasi, L.; Franklin, S.A.; Muller, T.; Bates, G.P. The Huntington’s Disease-Related Cardiomyopathy Prevents a Hypertrophic Response in the R6/2 Mouse Model. PLoS ONE 2014, 9, e108961. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-T.; Chiang, M.-C.; Tasi, C.-Y.; Kuo, C.-H.; Shyu, W.-C.; Kao, C.-L.; Huang, C.-Y.; Lee, S.-D. Cardiac Fas-Dependent and Mitochondria-Dependent Apoptotic Pathways in a Transgenic Mouse Model of Huntington’s Disease. Cardiovasc. Toxicol. 2016, 16, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Joviano-Santos, J.V.; Santos-Miranda, A.; Botelho, A.F.M.; De Jesus, I.C.G.; Andrade, J.N.; Barreto, T.D.O.; Magalhães-Gomes, M.P.S.; Valadão, P.A.C.; Cruz, J.; Melo, M.M.; et al. Increased oxidative stress and CaMKIIactivity contribute to electro-mechanical defects in cardiomyocytes from a murine model of Huntington’s disease. FEBS J. 2019, 286, 110–123. [Google Scholar] [CrossRef]
- Dridi, H.; Liu, X.; Yuan, Q.; Reiken, S.; Yehia, M.; Sittenfeld, L.; Apostolou, P.; Buron, J.; Sicard, P.; Matecki, S.; et al. Role of defective calcium regulation in cardiorespiratory dysfunction in Huntington’s disease. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Mihm, M.J.; Amann, D.M.; Schanbacher, B.L.; Altschuld, R.A.; Bauer, J.A.; Hoyt, K.R. Cardiac dysfunction in the R6/2 mouse model of Huntington’s disease. Neurobiol. Dis. 2007, 25, 297–308. [Google Scholar] [CrossRef]
- Child, D.; Lee, J.H.; Pascua, C.J.; Chen, Y.H.; Monteys, A.M.; Davidson, B.L. Cardiac mTORC1 Dysregulation Impacts Stress Adaptation and Survival in Huntington’s Disease. Cell Rep. 2018, 23, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Critchley, B.J.; Isalan, M.; Mielcarek, M. Neuro-Cardio Mechanisms in Huntington’s Disease and Other Neurodegenerative Disorders. Front. Physiol. 2018, 9, 559. [Google Scholar] [CrossRef]
- Lodi, R.; Schapira, A.H.V.; Manners, D.; Styles, P.; Wood, N.W.; Taylor, D.J.; Warner, T.T. Abnormal in Vivo Skeletal Muscle Energy Metabolism in Huntington’s Disease and Dentatorubropallidoluysian Atrophy. Ann. Neurol. 2000. [Google Scholar] [CrossRef]
- Toczek, M.; Zielonka, D.; Zukowska, P.; Marcinkowski, J.T.; Slominska, E.; Isalan, M.; Smolenski, R.T.; Mielcarek, M. An impaired metabolism of nucleotides underpins a novel mechanism of cardiac remodeling leading to Huntington’s disease related cardiomyopathy. Biochim. Biophys. Acta 2016, 1862, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Smolenski, R.; Isalan, M. Transcriptional Signature of an Altered Purine Metabolism in the Skeletal Muscle of a Huntington’s Disease Mouse Model. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Toczek, M.; Kutryb-Zajac, B.; Zukowska, P.; Slominska, E.M.; Isalan, M.; Mielcarek, M.; Smolenski, R. Changes in cardiac nucleotide metabolism in Huntington’s disease. Nucleosides Nucleotides Nucleic Acids 2016, 35, 707–712. [Google Scholar] [CrossRef]
- Fortuin, F.D.; Morisaki, T.; Holmes, E.W. Subunit composition of AMPD varies in response to changes in AMPD1 and AMPD3 gene expression in skeletal muscle. Proc. Assoc. Am. Physicians 1996, 108, 329–333. [Google Scholar] [PubMed]
- Ferrando, B.; Gomez-Cabrera, M.C.; Salvador-Pascual, A.; Puchades, C.; Derbré, F.; Gratas-Delamarche, A.; Laparre, L.; Olaso-González, G.; Cerda, M.; Viosca, E.; et al. Allopurinol partially prevents disuse muscle atrophy in mice and humans. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Krasowska, E.; Róg, J.; Sinadinos, A.; Young, C.N.J.; Górecki, D.C.; Zabłocki, K. Purinergic receptors in skeletal muscles in health and in muscular dystrophy. Postępy Biochemii 2014, 60, 483–489. [Google Scholar] [PubMed]
- Górecki, D.C. P2X7 purinoceptor as a therapeutic target in muscular dystrophies. Curr. Opin. Pharmacol. 2019, 47, 40–45. [Google Scholar] [CrossRef]
- Ryten, M.; Yang, S.Y.; Dunn, P.M.; Goldspink, G.; Burnstock, G. Purinoceptor expression in regenerating skeletal muscle in the mdx mouse model of muscular dystrophy and in satellite cell cultures. FASEB J. 2004, 18, 1404–1406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Berry, D.A.; Barden, J.A.; Balcar, V.J.; Keogh, A.; Dos Remedios, C.G. Increase in expression of P2X1 receptors in the atria of patients suffering from dilated cardiomyopathy. Electrophoresis 1999, 20, 2059–2064. [Google Scholar] [CrossRef]
- Yang, A.; Sonin, D.; Jones, L.; Barry, W.H.; Liang, B.T. A beneficial role of cardiac P2X4 receptors in heart failure: Rescue of the calsequestrin overexpression model of cardiomyopathy. Am. J. Physiol. Circ. Physiol. 2004, 287, H1096–H1103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, J.-B.; Cronin, C.; Sonin, D.; Joshi, B.V.; Nieto, M.G.; Harrison, D.; Jacobson, K.A.; Liang, B.T. P2X purinergic receptor-mediated ionic current in cardiac myocytes of calsequestrin model of cardiomyopathy: Implications for the treatment of heart failure. Am. J. Physiol. Circ. Physiol. 2007, 292, H1077–H1084. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.G.; Zamith-Miranda, D.; Da Silva, M.G.; Ribeiro, K.C.; Brandão, I.T.; Silva, C.L.; Diaz, B.L.; Bellio, M.; Persechini, P.M.; Kurtenbach, E. P2×7 purinergic signaling in dilated cardiomyopathy induced by auto-immunity against muscarinic M2 receptors: Autoantibody levels, heart functionality and cytokine expression. Sci. Rep. 2015, 5, 16940. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Jiang, M.; Jin, J. Metabolism in HD: Still a relevant mechanism? Mov. Disord. 2014, 29, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Dickey, A.S.; Pineda, V.V.; Tsunemi, T.; Liu, P.P.; Miranda, H.C.; Gilmore-Hall, S.K.; Lomas, N.; Sampat, K.R.; Buttgereit, A.; Torres, M.-J.M.; et al. PPAR-δ is repressed in Huntington’s disease, is required for normal neuronal function and can be targeted therapeutically. Nat. Med. 2016, 22, 37–45. [Google Scholar] [CrossRef]
- Smolenski, R.T.; Raisky, O.; Slominska, E.M.; Abunasra, H.; Kalsi, K.K.; Jayakumar, J.; Suzuki, K.; Yacoub, M.H. Protection from reperfusion injury after cardiac transplantation by inhibition of adenosine metabolism and nucleotide precursor supply. Circulation 2001, 104, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, R.; Kalsi, K.K.; Zych, M.; Kochan, Z.; Yacoub, M.H. Adenine/Ribose Supply Increases Adenosine Production and Protects ATP Pool in Adenosine Kinase-inhibited Cardiac Cells. J. Mol. Cell. Cardiol. 1998, 30, 673–683. [Google Scholar] [CrossRef]
- Plaideau, C.; Lai, Y.-C.; Kviklyte, S.; Zanou, N.; Löfgren, L.; Andersén, H.; Vertommen, D.; Gailly, P.; Hue, L.; Bohlooly, Y.M.; et al. Effects of Pharmacological AMP Deaminase Inhibition and Ampd1 Deletion on Nucleotide Levels and AMPK Activation in Contracting Skeletal Muscle. Chem. Biol. 2014, 21, 1497–1510. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, R.T.; Rybakowska, I.; Turyn, J.; Romaszko, P.; Zabielska, M.; Taegtmeyer, A.; Słomińska, E.M.; Kaletha, K.K.; Barton, P.J.R. AMP deaminase 1 gene polymorphism and heart disease-a genetic association that highlights new treatment. Cardiovasc. Drugs Ther. 2014, 28, 183–189. [Google Scholar] [CrossRef]
- Zabielska, M.A.; Borkowski, T.; Slominska, E.M.; Smolenski, R.T. Inhibition of AMP deaminase as therapeutic target in cardiovascular pathology. Pharmacol. Rep. 2015, 67, 682–688. [Google Scholar] [CrossRef]
- Guieu, R.; Deharo, J.-C.; Maille, B.; Crotti, L.; Torresani, E.; Brignole, M.; Parati, G. Adenosine and the Cardiovascular System: The Good and the Bad. J. Clin. Med. 2020, 9, 1366. [Google Scholar] [CrossRef]
- Boison, D. Role of adenosine in status epilepticus: A potential new target? Epilepsia 2013, 54 (Suppl. 6) (Suppl. 6), 20–22. [Google Scholar] [CrossRef]
- Boison, D. Adenosine dysfunction in epilepsy. Glia 2012, 60, 1234–1243. [Google Scholar] [CrossRef]
- Kutryb-Zajac, B.; Mierzejewska, P.; Slominska, E.M.; Smolenski, R.T. Therapeutic Perspectives of Adenosine Deaminase Inhibition in Cardiovascular Diseases. Molecules 2020, 25, 4652. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-H.; Lin, M.-S.; Chen, C.-M.; Wu, Y.-R.; Chen, H.-M.; Lai, H.-L.; Chern, Y.; Lin, C.-J. Targeting ENT1 and adenosine tone for the treatment of Huntington’s disease. Hum. Mol. Genet. 2016, 26, 467–478. [Google Scholar] [CrossRef]
- Mezzaroma, E.; Toldo, S.; Farkas, D.; Seropian, I.M.; Van Tassell, B.W.; Salloum, F.; Kannan, H.R.; Menna, A.C.; Voelkel, N.F.; Abbate, A. The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc. Natl. Acad. Sci. USA 2011, 108, 19725–19730. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yin, J.; Shi, Y.; Hu, H.; Li, X.; Xue, M.; Cheng, W.; Wang, Y.; Li, X.; Li, Y.; et al. Targeted P2X7R shRNA delivery attenuates sympathetic nerve sprouting and ameliorates cardiac dysfunction in rats with myocardial infarction. Cardiovasc. Ther. 2016, 35, e12245. [Google Scholar] [CrossRef] [PubMed]
- Bracey, N.A.; Beck, P.L.; Muruve, D.A.; Hirota, S.A.; Guo, J.; Jabagi, H.; Jr, J.R.W.; Macdonald, J.A.; Lees-Miller, J.P.; Roach, D.; et al. The Nlrp3 inflammasome promotes myocardial dysfunction in structural cardiomyopathy through interleukin-1β. Exp. Physiol. 2013, 98, 462–472. [Google Scholar] [CrossRef]
- Frantz, S.; Ducharme, A.; Sawyer, D.; Rohde, L.E.; Kobzik, L.; Fukazawa, R.; Tracey, D.; Allen, H.; Lee, R.T.; Kelly, R.A. Targeted deletion of caspase-1 reduces early mortality and left ventricular dilatation following myocardial infarction. J. Mol. Cell. Cardiol. 2003, 35, 685–694. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomczyk, M.; Glaser, T.; Slominska, E.M.; Ulrich, H.; Smolenski, R.T. Purine Nucleotides Metabolism and Signaling in Huntington’s Disease: Search for a Target for Novel Therapies. Int. J. Mol. Sci. 2021, 22, 6545. https://doi.org/10.3390/ijms22126545
Tomczyk M, Glaser T, Slominska EM, Ulrich H, Smolenski RT. Purine Nucleotides Metabolism and Signaling in Huntington’s Disease: Search for a Target for Novel Therapies. International Journal of Molecular Sciences. 2021; 22(12):6545. https://doi.org/10.3390/ijms22126545
Chicago/Turabian StyleTomczyk, Marta, Talita Glaser, Ewa M. Slominska, Henning Ulrich, and Ryszard T. Smolenski. 2021. "Purine Nucleotides Metabolism and Signaling in Huntington’s Disease: Search for a Target for Novel Therapies" International Journal of Molecular Sciences 22, no. 12: 6545. https://doi.org/10.3390/ijms22126545
APA StyleTomczyk, M., Glaser, T., Slominska, E. M., Ulrich, H., & Smolenski, R. T. (2021). Purine Nucleotides Metabolism and Signaling in Huntington’s Disease: Search for a Target for Novel Therapies. International Journal of Molecular Sciences, 22(12), 6545. https://doi.org/10.3390/ijms22126545








