Arabidopsis thaliana G3BP Ortholog Rescues Mammalian Stress Granule Phenotype across Kingdoms
Abstract
1. Introduction
2. Results
2.1. Phylogenetic Relationship of HsG3BPs and the Arabidopsis thaliana G3BP Family
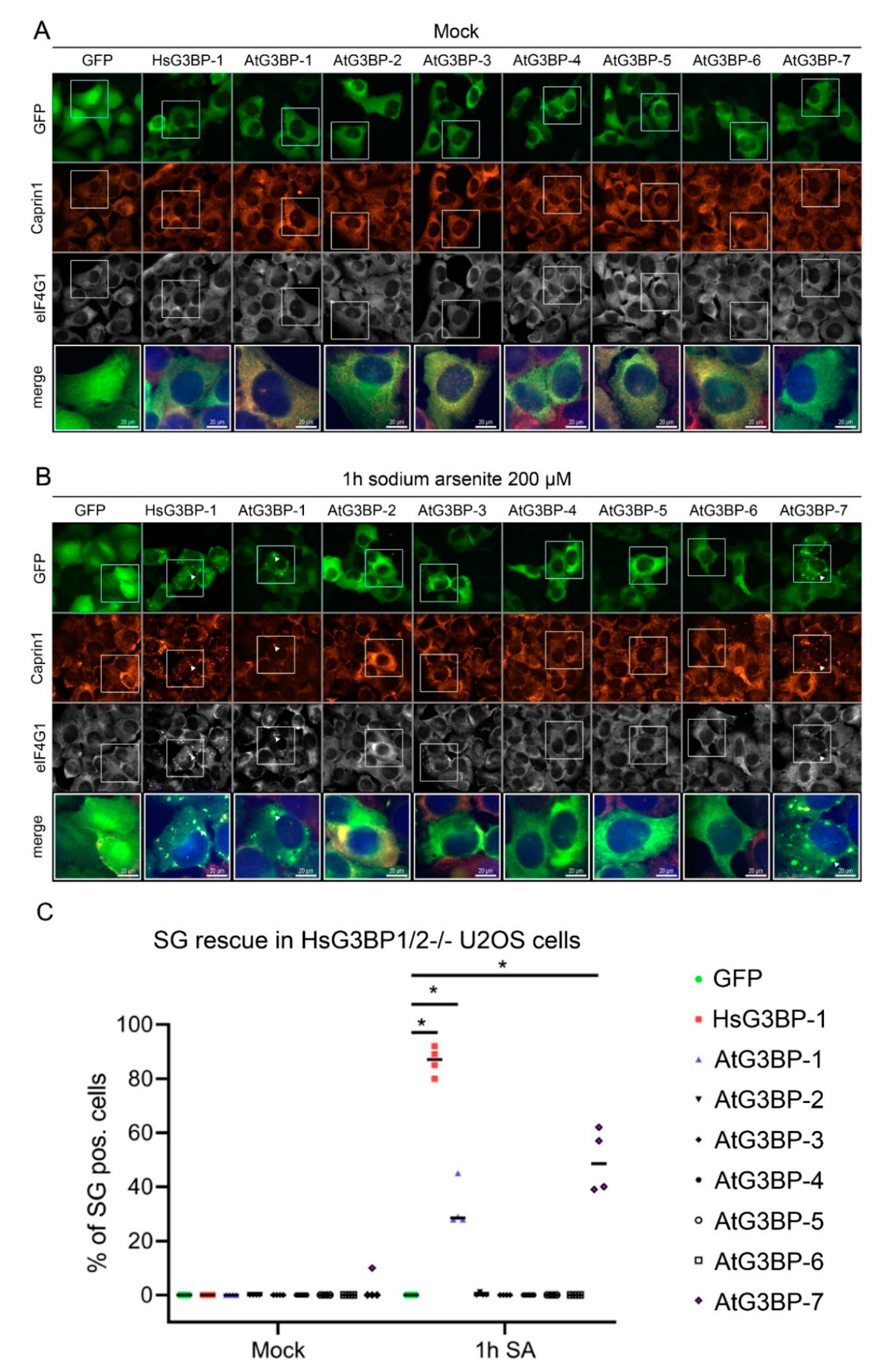
2.2. Expression of EGFP::AtG3BP in HsG3BP1/2−/− U2OS Cells
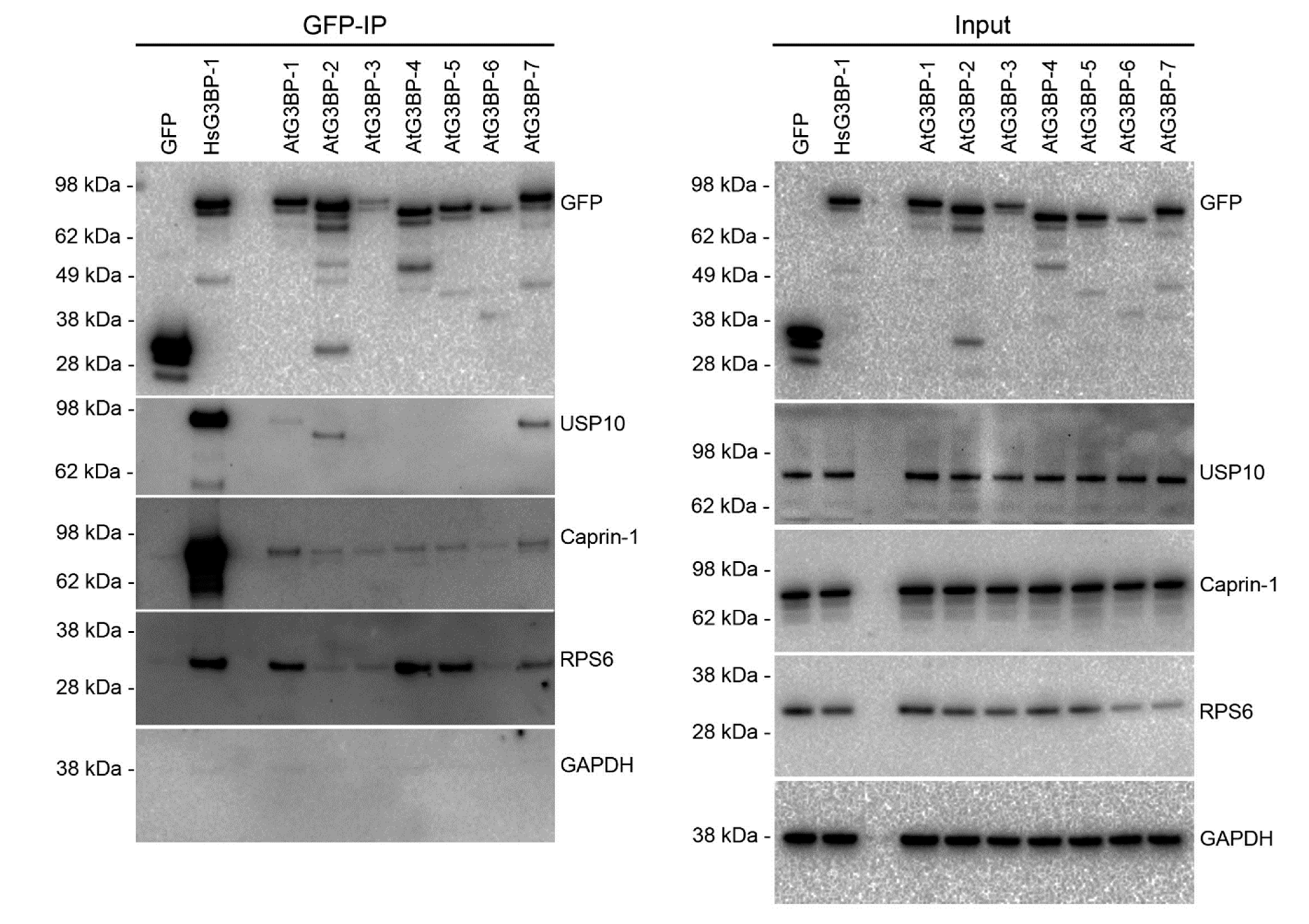
2.3. Interaction Studies of the AtG3BPs with Human Cellular Factors
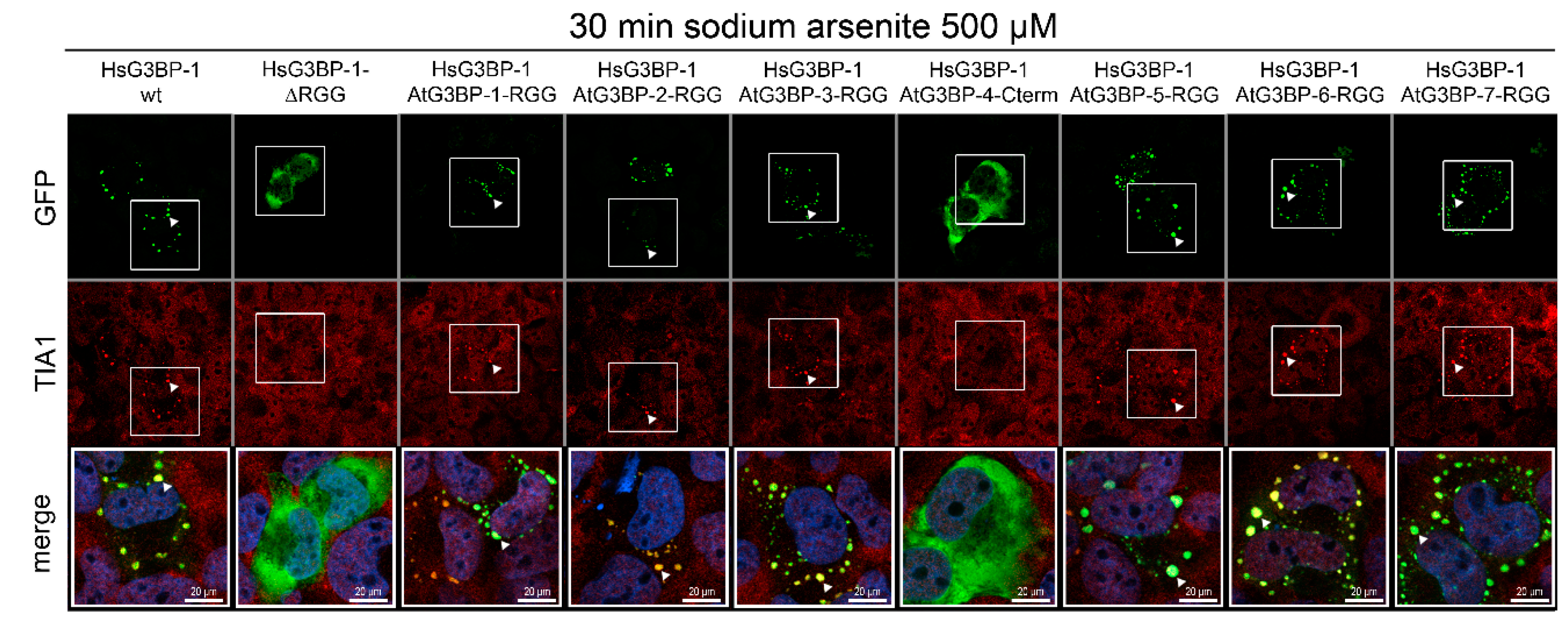
2.4. Exchange of RG/RGG Domains and SG Induction
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunofluorescence and Microscopy
4.3. Plasmids and Transfection
4.4. Immunoprecipitation and Western Blot
4.4.1. Transient Transfection
4.4.2. Stable Expression
4.5. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| (E)GFP | (enhanced) green fluorescent protein |
| BiFC | bimolecular fluorescence complementation |
| CHIKV | chikungunya virus |
| eIF2α | eukaryotic initiation factor 2α |
| eIF4E | eukaryotic initiation factor 4E |
| G3BP | Ras-GAP SH3 domain-binding protein |
| KO | knock-out |
| NTF2-like | nuclear transport factor 2 like |
| RPS6 | 40S ribosomal protein S6 |
| RRM | RNA recognition motif |
| SFV | Semliki Forest virus |
| SG | stress granule |
| TIA-1 | T-cell-restricted intracellular antigen-1 |
| TIAR | TIA-1-related |
| TSN | Tudor staphylococcal nuclease |
| UBP1 | oligouridylate-binding protein 1 |
| UBP-24 | ubiquitin carboxyl-terminal hydrolase 24 |
| USP10 | ubiquitin-specific peptidase 10 |
References
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118876. [Google Scholar] [CrossRef] [PubMed]
- Tourrière, H.H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.R.E.A.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef]
- Kennedy, D.; French, J.; Guitard, E.; Ru, K.; Tocque, B.; Mattick, J. Characterization of G3BPs: Tissue specific expression, chromosomal localisation and rasGAP120 binding studies. J. Cell. Biochem. 2002, 84, 173–187. [Google Scholar] [CrossRef]
- Kedersha, N.; Panas, M.D.; Achorn, C.A.; Lyons, S.; Tisdale, S.; Hickman, T.; Thomas, M.; Lieberman, J.; McInerney, G.M.; Ivanov, P.; et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016, 212, 845–860. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef]
- Pomeranz, M.C.; Hah, C.; Lin, P.-C.C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.-C.C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010, 152, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.J.; Jang, J.C.; Wu, S.H. Dynamics and functions of stress granules and processing bodies in plants. Plants 2020, 9, 1122. [Google Scholar] [CrossRef]
- Mcinerney, G.M.; Kedersha, N.L.; Kaufman, R.J.; Anderson, P.; Liljestro, P. Importance of eIF2α Phosphorylation and Stress Granule Assembly in Alphavirus Translation Regulation. Mol. Biol. Cell 2005, 16, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Domeradzka, N.E.; Baggen, J.; Geertsema, C.; Flipse, J.; Vlak, J.M.; Pijlman, G.P. Chikungunya Virus nsP3 Blocks Stress Granule Assembly by Recruitment of G3BP into Cytoplasmic Foci. J. Virol. 2012, 86, 10873–10879. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, J.D.; Tsai, W.C.; Lloyd, R.E. Multiple poliovirus proteins repress cytoplasmic RNA granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef] [PubMed]
- Robles-Luna, G.; Furman, N.; Barbarich, M.F.; Carlotto, N.; Attorresi, A.; García, M.L.; Kobayashi, K. Interplay between potato virus X and RNA granules in Nicotiana benthamiana. Virus Res. 2020, 276. [Google Scholar] [CrossRef] [PubMed]
- Götte, B.; Panas, M.D.; Hellström, K.; Liu, L.; Samreen, B.; Larsson, O.; Ahola, T.; McInerney, G.M. Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog. 2019, 15, 1–24. [Google Scholar] [CrossRef]
- Kim, D.Y.; Reynaud, J.M.; Rasalouskaya, A.; Akhrymuk, I.; Mobley, J.A.; Frolov, I.; Frolova, E.I. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog. 2016, 12, 1–31. [Google Scholar] [CrossRef]
- Abulfaraj, A.A.; Mariappan, K.; Bigeard, J.; Manickam, P.; Blilou, I.; Guo, X.; Al-Babili, S.; Pflieger, D.; Hirt, H.; Rayapuram, N. The Arabidopsis homolog of human G3BP1 is a key regulator of stomatal and apoplastic immunity. Life Sci. Alliance 2018, 1, 1–13. [Google Scholar] [CrossRef]
- Reuper, H.; Amari, K.; Krenz, B. Analyzing the G3BP-like gene family of Arabidopsis thaliana in early turnip mosaic virus infection. Sci. Rep. 2021, 11, 2187. [Google Scholar] [CrossRef]
- Krapp, S.; Greiner, E.; Amin, B.; Sonnewald, U.; Krenz, B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res. 2017, 227, 6–14. [Google Scholar] [CrossRef]
- Soncini, C.; Berdo, I.; Draetta, G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 2001, 20, 3869–3879. [Google Scholar] [CrossRef]
- Panas, M.D.; Schulte, T.; Thaa, B.; Sandalova, T.; Kedersha, N.; Achour, A.; McInerney, G.M. Viral and Cellular Proteins Containing FGDF Motifs Bind G3BP to Block Stress Granule Formation. PLoS Pathog. 2015, 11, 1–22. [Google Scholar] [CrossRef]
- Solomon, S.; Xu, Y.; Wang, B.; David, M.D.; Schubert, P.; Kennedy, D.; Schrader, J.W. Distinct Structural Features of Caprin-1 Mediate Its Interaction with G3BP-1 and Its Induction of Phosphorylation of Eukaryotic Translation Initiation Factor 2α, Entry to Cytoplasmic Stress Granules, and Selective Interaction with a Subset of mRNAs. Mol. Cell. Biol. 2007, 27, 2324–2342. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, O.; Vognsen, T.; Runa, I. Crystal structure of the G3BP2 NTF2-like domain in complex with a canonical FGDF motif peptide. Biochem. Biophys. Res. Commun. 2015, 467, 53–57. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- De Castro, E.; Sigrist, C.J.A.; Gattiker, A.; Bulliard, V.; Langendijk-genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic. Acids Res. 2006, 34, 362–365. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Molecules as documents of history. J. Theor. Biol. 1965, 8, 357–366. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Buchan, J.R.; Parker, R. Review Eukaryotic Stress Granules: The Ins and Outs of Translation. Mol. Cell 2009, 36, 932–941. [Google Scholar] [CrossRef]
- Collier, N.C.; Heuser, J.; Levy, M.A.; Schlesinger, M.J. Ultrastructural and biochemical analysis of the stress granule in chicken embryo fibroblasts. J. Cell Biol. 1988, 106, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Scharf, K.D.; Neumann, D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 1983, 3, 1648–1655. [Google Scholar] [CrossRef]
- Hoyle, N.P.; Castelli, L.M.; Campbell, S.G.; Holmes, L.E.A.; Ashe, M.P. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007, 179, 65–74. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef]
- Kosmacz, M.; Gorka Michałand Schmidt, S.; Luzarowski, M.; Moreno, J.C.; Szlachetko, J.; Leniak, E.; Sokolowska, E.M.; Sofroni, K.; Schnittger, A.; Skirycz, A.; et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 2019, 222, 1420–1433. [Google Scholar] [CrossRef]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar] [CrossRef]
- Gao, X.; Fu, X.; Song, J.; Zhang, Y.; Cui, X.; Su, C.; Ge, L.; Shao, J.; Xin, L.; Saarikettu, J.; et al. Poly(A)+ mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics. FEBS J. 2015, 282, 874–890. [Google Scholar] [CrossRef]
- Lokdarshi, A.; Craig Conner, W.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 2016, 170, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor Staphylococcal Nuclease Links Formation of Stress Granules and Processing Bodies with mRNA Catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yan, Z.; Wang, Y.; Yan, X.; Han, Y. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 2014, 65, 5933–5944. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.C.; Nakaminami, K.; Matsui, A.; Watanabe, S.; Kanno, Y.; Seo, M.; Seki, M. Overexpression of oligouridylate binding protein 1b results in ABA hypersensitivity. Plant Signal. Behav. 2017, 12, e1282591. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.C.; Nakaminami, K.; Matsui, A.; Kobayashi, S.; Kurihara, Y.; Toyooka, K.; Tanaka, M.; Seki, M. Oligouridylate Binding Protein 1b Plays an Integral Role in Plant Heat Stress Tolerance. Front. Plant Sci. 2016, 7, 853. [Google Scholar] [CrossRef]
- Brocca, S.; Grandori, R.; Longhi, S.; Uversky, V. Liquid–liquid phase separation by intrinsically disordered protein regions of viruses: Roles in viral life cycle and control of virus–host interactions. Int. J. Mol. Sci. 2020, 21, 9045. [Google Scholar] [CrossRef] [PubMed]
- Malm, M.; Krohn, K.; Blazevic, V. Immunization with dendritic cells transfected in vivo with HIV-1 plasmid DNA induces HIV-1-specific immune responses. Arch. Virol. 2011, 156, 1607–1610. [Google Scholar] [CrossRef] [PubMed]
- Utt, A.; Quirin, T.; Saul, S.; Hellström, K.; Ahola, T.; Merits, A. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS ONE 2016, 11, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.D.; Kedersha, N.; Schulte, T.; Branca, R.M.; Ivanov, P.; Anderson, P. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J. Cell Biol. 2019, 218, 2425–2432. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reuper, H.; Götte, B.; Williams, L.; Tan, T.J.C.; McInerney, G.M.; Panas, M.D.; Krenz, B. Arabidopsis thaliana G3BP Ortholog Rescues Mammalian Stress Granule Phenotype across Kingdoms. Int. J. Mol. Sci. 2021, 22, 6287. https://doi.org/10.3390/ijms22126287
Reuper H, Götte B, Williams L, Tan TJC, McInerney GM, Panas MD, Krenz B. Arabidopsis thaliana G3BP Ortholog Rescues Mammalian Stress Granule Phenotype across Kingdoms. International Journal of Molecular Sciences. 2021; 22(12):6287. https://doi.org/10.3390/ijms22126287
Chicago/Turabian StyleReuper, Hendrik, Benjamin Götte, Lucy Williams, Timothy J. C. Tan, Gerald M. McInerney, Marc D. Panas, and Björn Krenz. 2021. "Arabidopsis thaliana G3BP Ortholog Rescues Mammalian Stress Granule Phenotype across Kingdoms" International Journal of Molecular Sciences 22, no. 12: 6287. https://doi.org/10.3390/ijms22126287
APA StyleReuper, H., Götte, B., Williams, L., Tan, T. J. C., McInerney, G. M., Panas, M. D., & Krenz, B. (2021). Arabidopsis thaliana G3BP Ortholog Rescues Mammalian Stress Granule Phenotype across Kingdoms. International Journal of Molecular Sciences, 22(12), 6287. https://doi.org/10.3390/ijms22126287






