ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA
Abstract
1. Introduction
2. Results
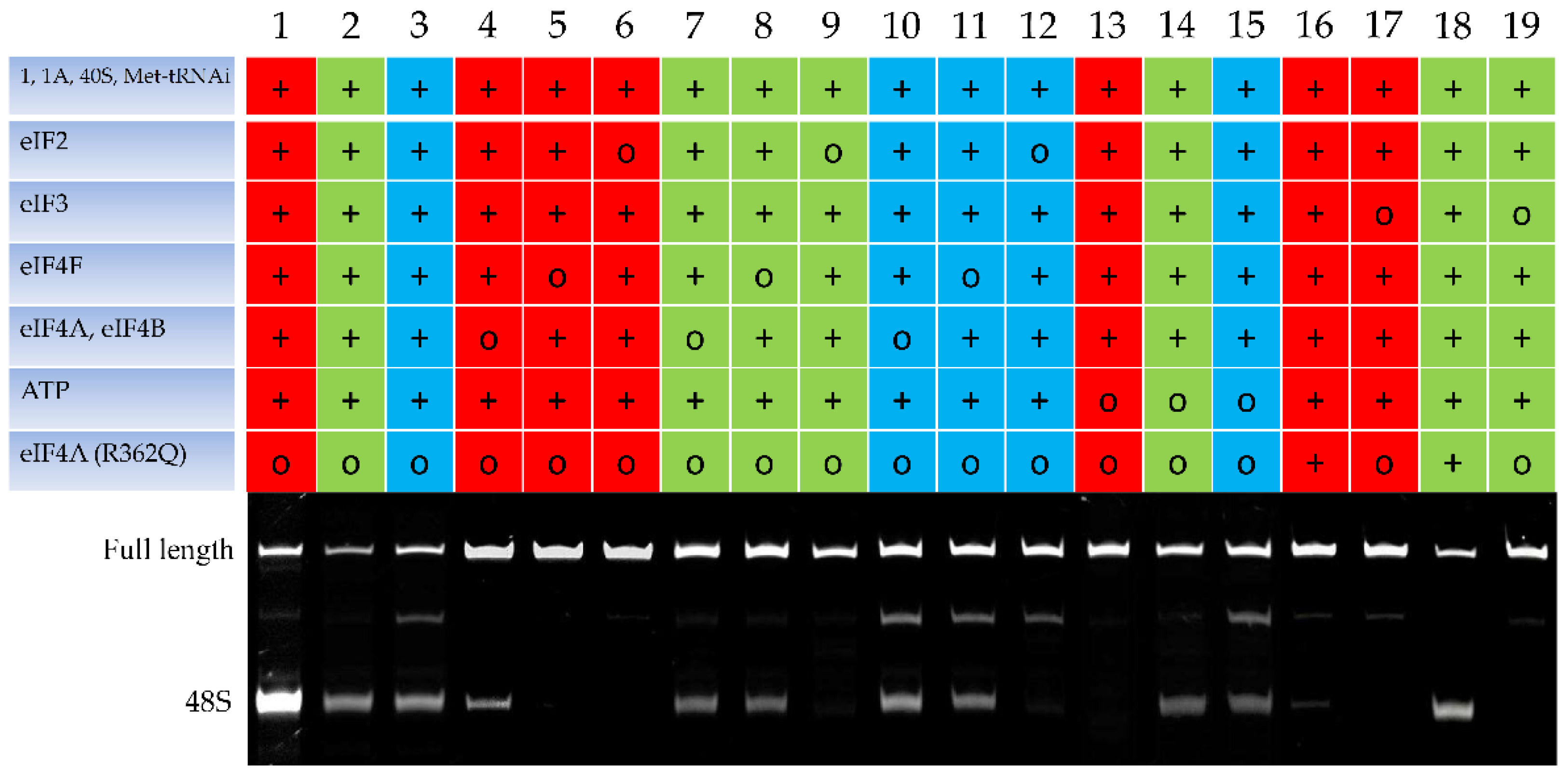
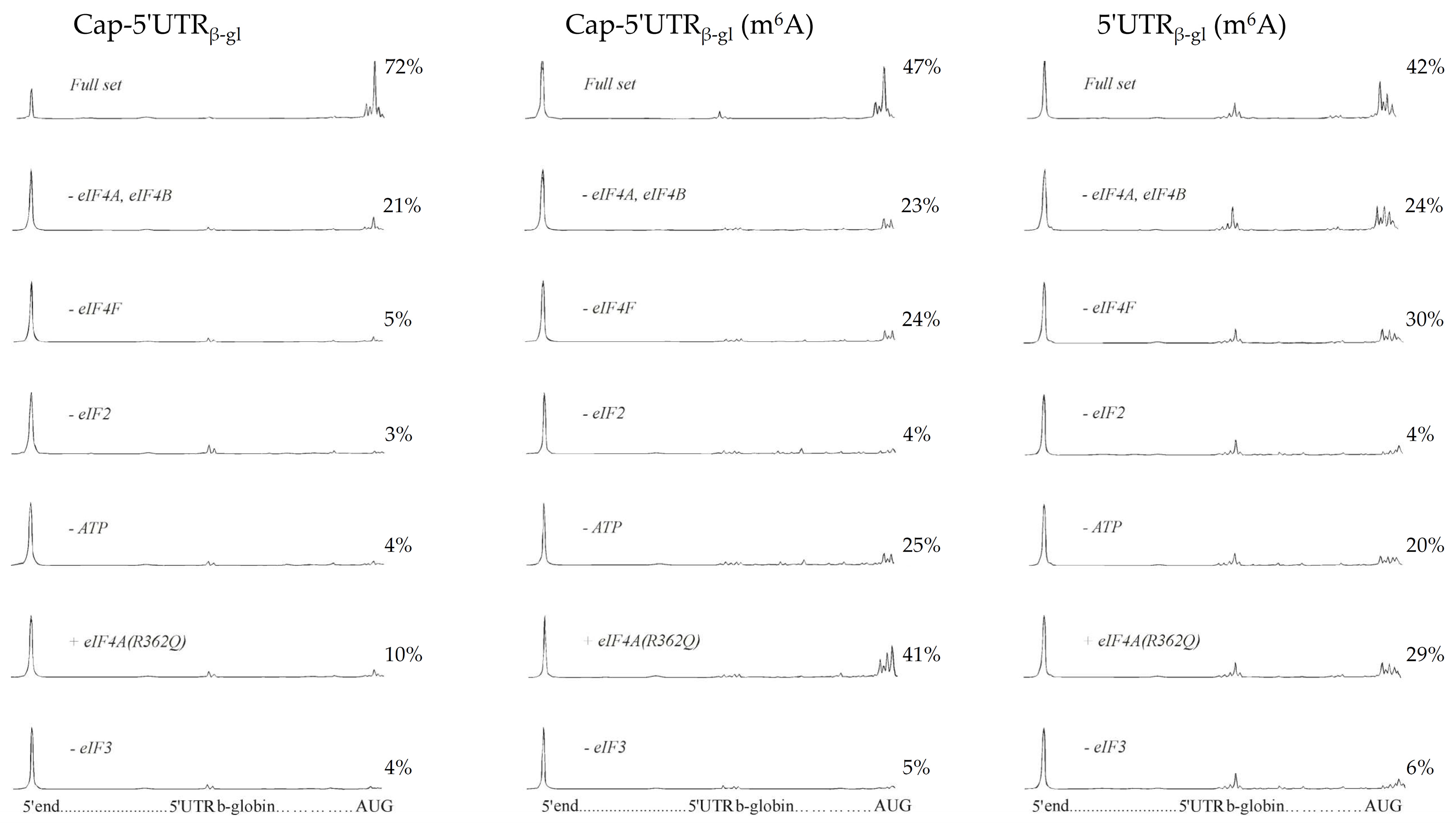
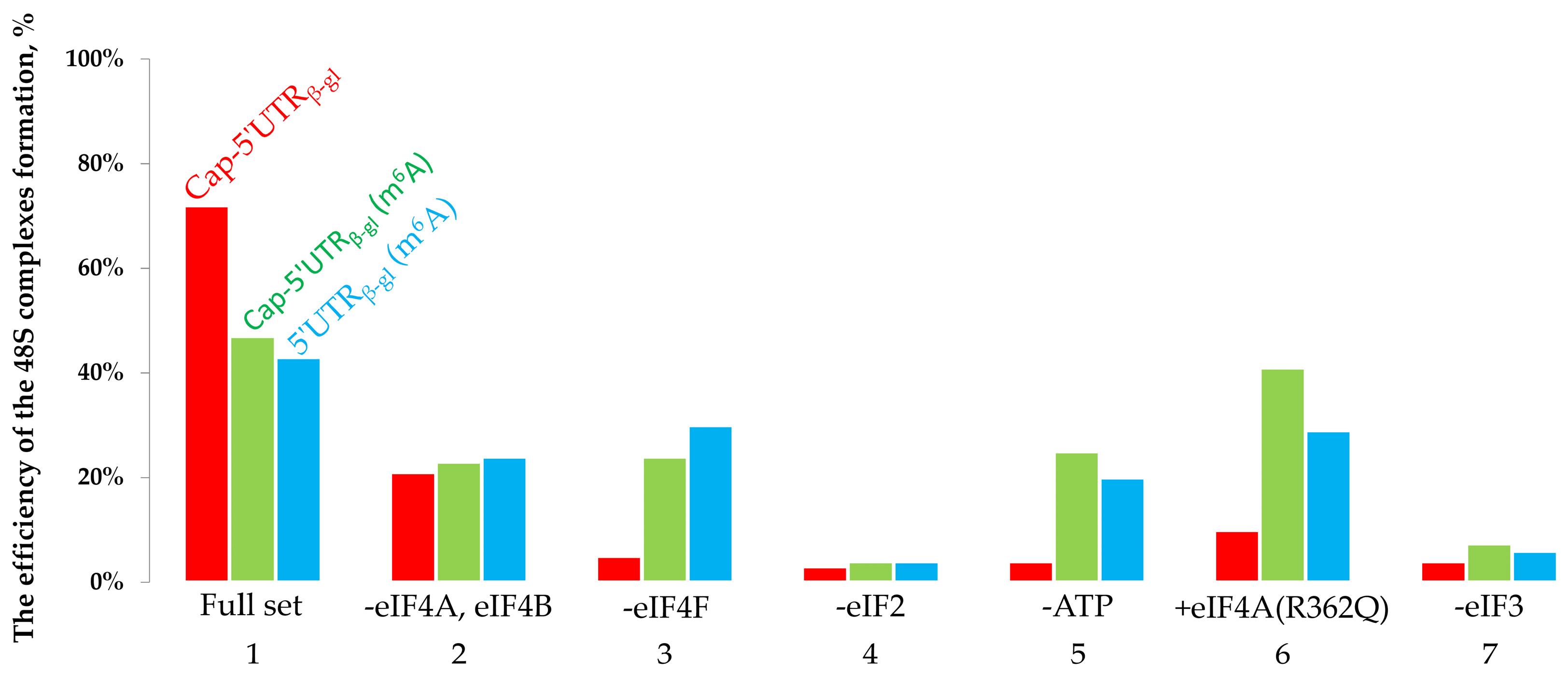
2.1. The Toeprinting Assay of Initiation on m6A-Modified mRNA
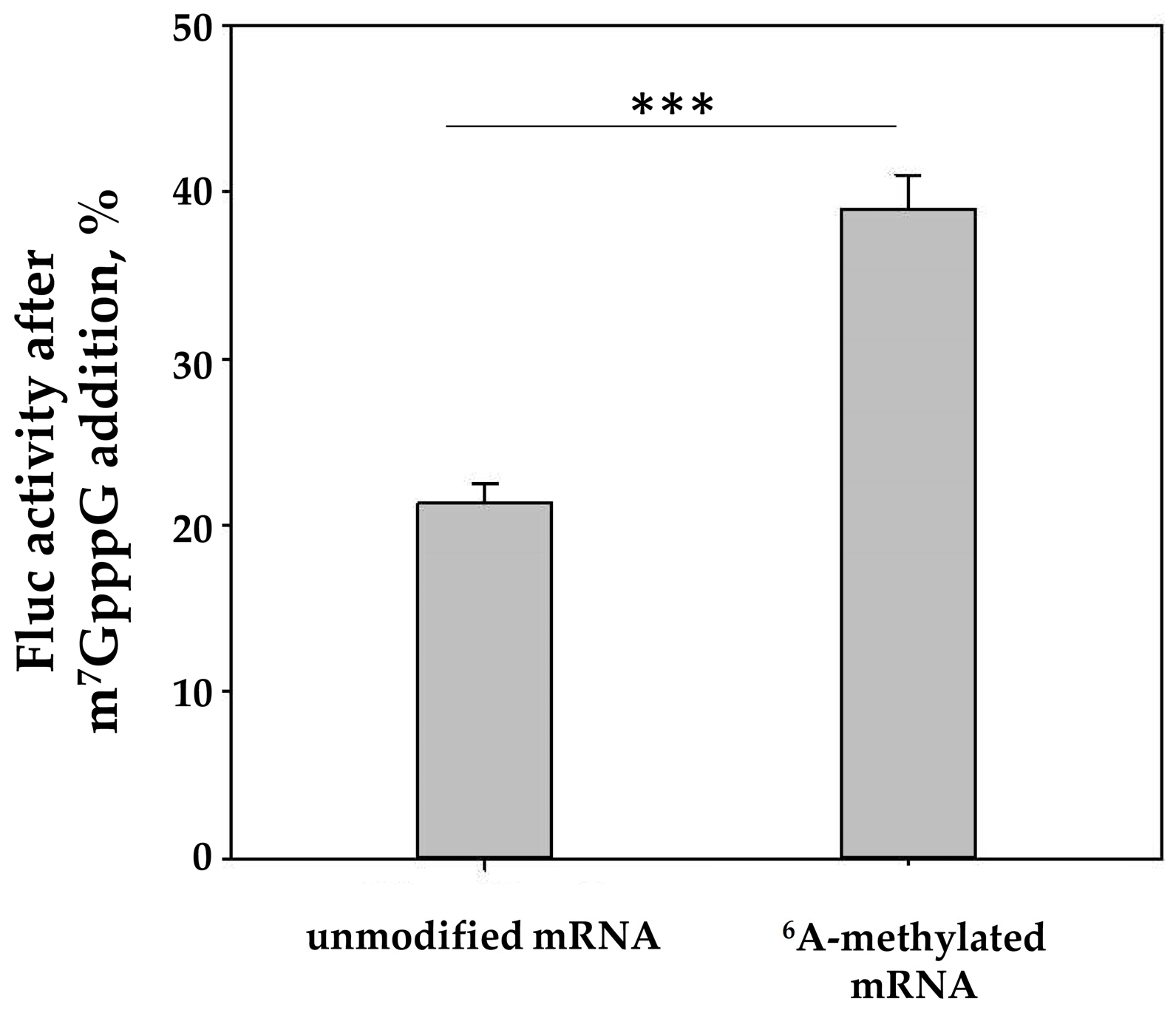
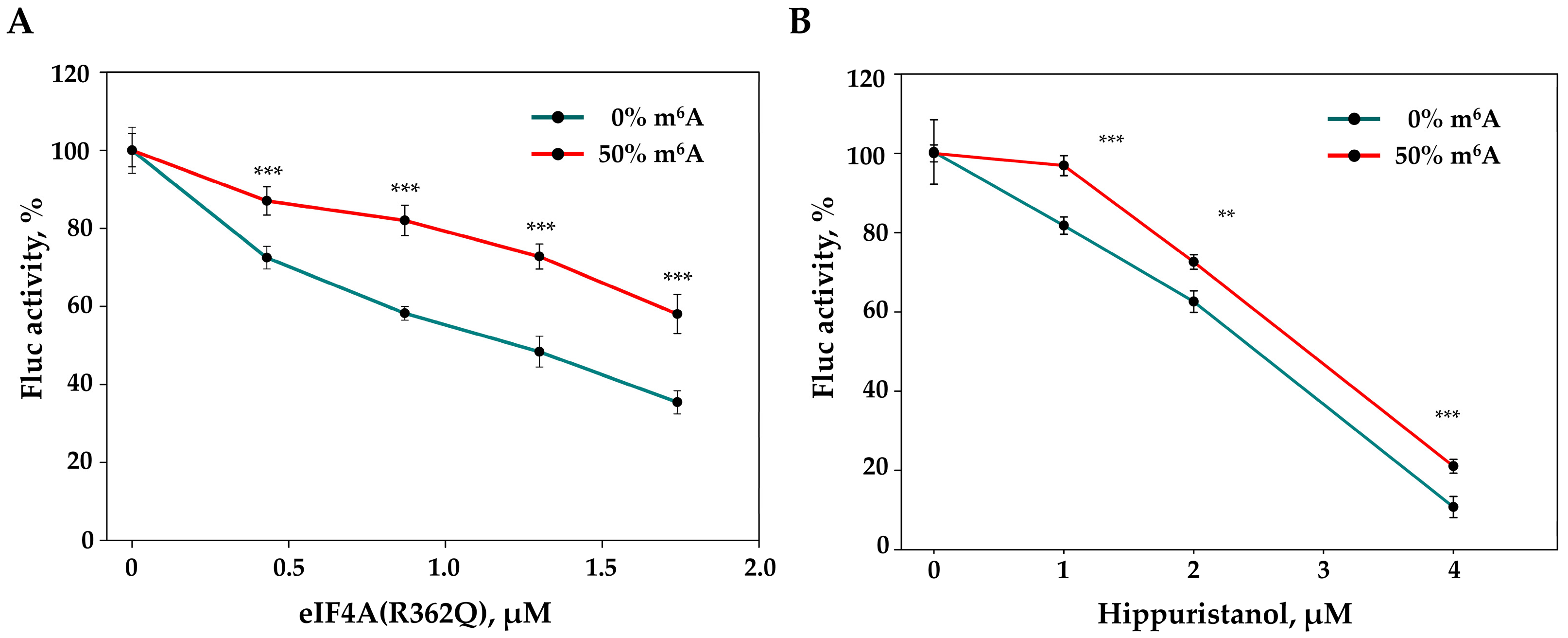
2.2. The In Vitro Translation of m6A-Modified mRNA
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. In Vitro Transcription
4.3. Components for Assembling 48S Initiator Complexes
4.4. Primer Extension Inhibition Assay (Toeprinting)
4.5. In Vitro Translation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Bodi, Z.; Bottley, A.; Archer, N.; May, S.T.; Fray, R.G. Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PLoS ONE 2015, 10, e0132090. [Google Scholar] [CrossRef]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Heck, A.M.; Wilusz, J. The Interplay between the RNA Decay and Translation Machinery in Eukaryotes. Cold Spring Harb. Perspect. Biol 2018, 10. [Google Scholar] [CrossRef]
- Meyer, K.D. m(6)A-mediated translation regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 301–309. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef] [PubMed]
- Roost, C.; Lynch, S.R.; Batista, P.J.; Qu, K.; Chang, H.Y.; Kool, E.T. Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J. Am. Chem. Soc. 2015, 137, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.; Thompson, S.R. Noncanonical Translation Initiation in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, N.; Hsu, B.E.; Istomine, R.; Alvarez, F.; Blagih, J.; Ma, E.H.; Morales, S.V.; Dai, D.L.; Li, G.; Souleimanova, M.; et al. Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc. Natl. Acad. Sci. USA 2018, 115, E2202–E2209. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Coots, R.A.; Liu, X.M.; Mao, Y.; Dong, L.; Zhou, J.; Wan, J.; Zhang, X.; Qian, S.B. m(6)A Facilitates eIF4F-Independent mRNA Translation. Mol. Cell 2017, 68, 504–514 e507. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef] [PubMed]
- Hartz, D.; McPheeters, D.S.; Traut, R.; Gold, L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988, 164, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Shirokikh, N.E.; Spirin, A.S. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl. Acad. Sci. USA 2008, 105, 10738–10743. [Google Scholar] [CrossRef]
- Sakharov, P.A.; Agalarov, S.C. Free Initiation Factors eIF4A and eIF4B Are Dispensable for Translation Initiation on Uncapped mRNAs. Biochemistry 2016, 81, 1198–1204. [Google Scholar] [CrossRef]
- Pause, A.; Methot, N.; Sonenberg, N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell Biol. 1993, 13, 6789–6798. [Google Scholar] [CrossRef][Green Version]
- Choi, J.; Ieong, K.W.; Demirci, H.; Chen, J.; Petrov, A.; Prabhakar, A.; O’Leary, S.E.; Dominissini, D.; Rechavi, G.; Soltis, S.M.; et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016, 23, 110–115. [Google Scholar] [CrossRef]
- Slobodin, B.; Han, R.; Calderone, V.; Vrielink, J.; Loayza-Puch, F.; Elkon, R.; Agami, R. Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell 2017, 169, 326–337 e312. [Google Scholar] [CrossRef] [PubMed]
- Agalarov, S.; Sakharov, P.A.; Fattakhova, D.; Sogorin, E.A.; Spirin, A.S. Internal translation initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic ribosomal particles. Sci. Rep. 2014, 4, 4438. [Google Scholar] [CrossRef]
- Spirin, A.S. How does a scanning ribosomal particle move along the 5’-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry 2009, 48, 10688–10692. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Boye, E.; Grallert, B. eIF2alpha phosphorylation and the regulation of translation. Curr. Genet. 2020, 66, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Shu, X.E.; Mao, Y.; Liu, X.M.; Yuan, X.; Zhang, X.; Hess, M.E.; Bruning, J.C.; Qian, S.B. N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol. Cell 2018, 69, 636–647 e637. [Google Scholar] [CrossRef]
- Alkalaeva, E.Z.; Pisarev, A.V.; Frolova, L.Y.; Kisselev, L.L.; Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 2006, 125, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Wakiyama, M.; Futami, T.; Miura, K. Poly(A) dependent translation in rabbit reticulocyte lysate. Biochimie 1997, 79, 781–785. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Unbehaun, A.; Hellen, C.U.; Pestova, T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007, 430, 147–177. [Google Scholar] [CrossRef]
- Terenin, I.M.; Andreev, D.E.; Dmitriev, S.E.; Shatsky, I.N. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic. Acids Res. 2013, 41, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakharov, P.A.; Smolin, E.A.; Lyabin, D.N.; Agalarov, S.C. ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA. Int. J. Mol. Sci. 2021, 22, 3662. https://doi.org/10.3390/ijms22073662
Sakharov PA, Smolin EA, Lyabin DN, Agalarov SC. ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA. International Journal of Molecular Sciences. 2021; 22(7):3662. https://doi.org/10.3390/ijms22073662
Chicago/Turabian StyleSakharov, Pavel A., Egor A. Smolin, Dmitry N. Lyabin, and Sultan C. Agalarov. 2021. "ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA" International Journal of Molecular Sciences 22, no. 7: 3662. https://doi.org/10.3390/ijms22073662
APA StyleSakharov, P. A., Smolin, E. A., Lyabin, D. N., & Agalarov, S. C. (2021). ATP-Independent Initiation during Cap-Independent Translation of m6A-Modified mRNA. International Journal of Molecular Sciences, 22(7), 3662. https://doi.org/10.3390/ijms22073662




