Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways
Abstract
1. Introduction
2. Results
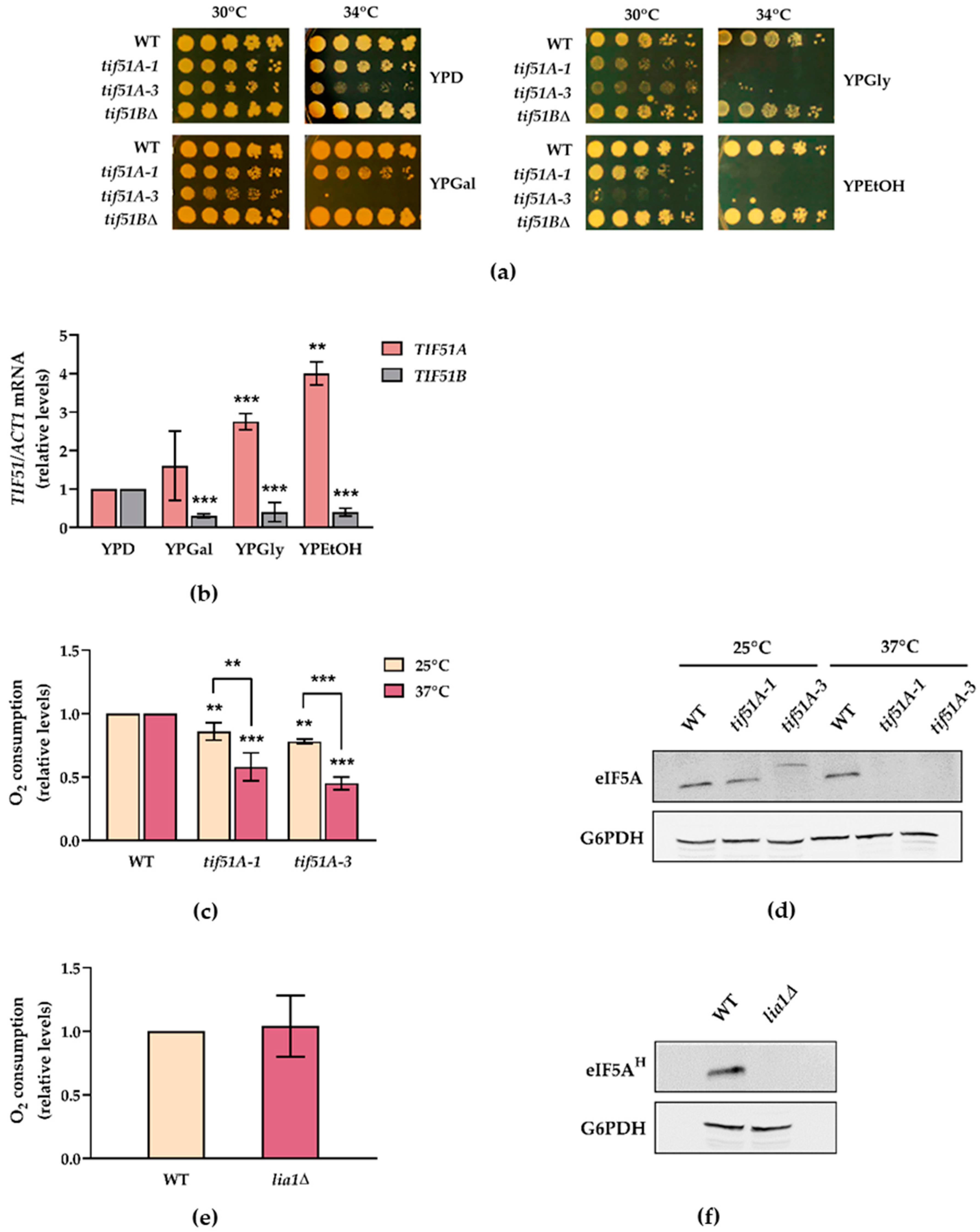
2.1. The Tif51A Isoform of Yeast eIF5A Is Required for Respiration and Growth with Non-Fermentative Carbon Sources
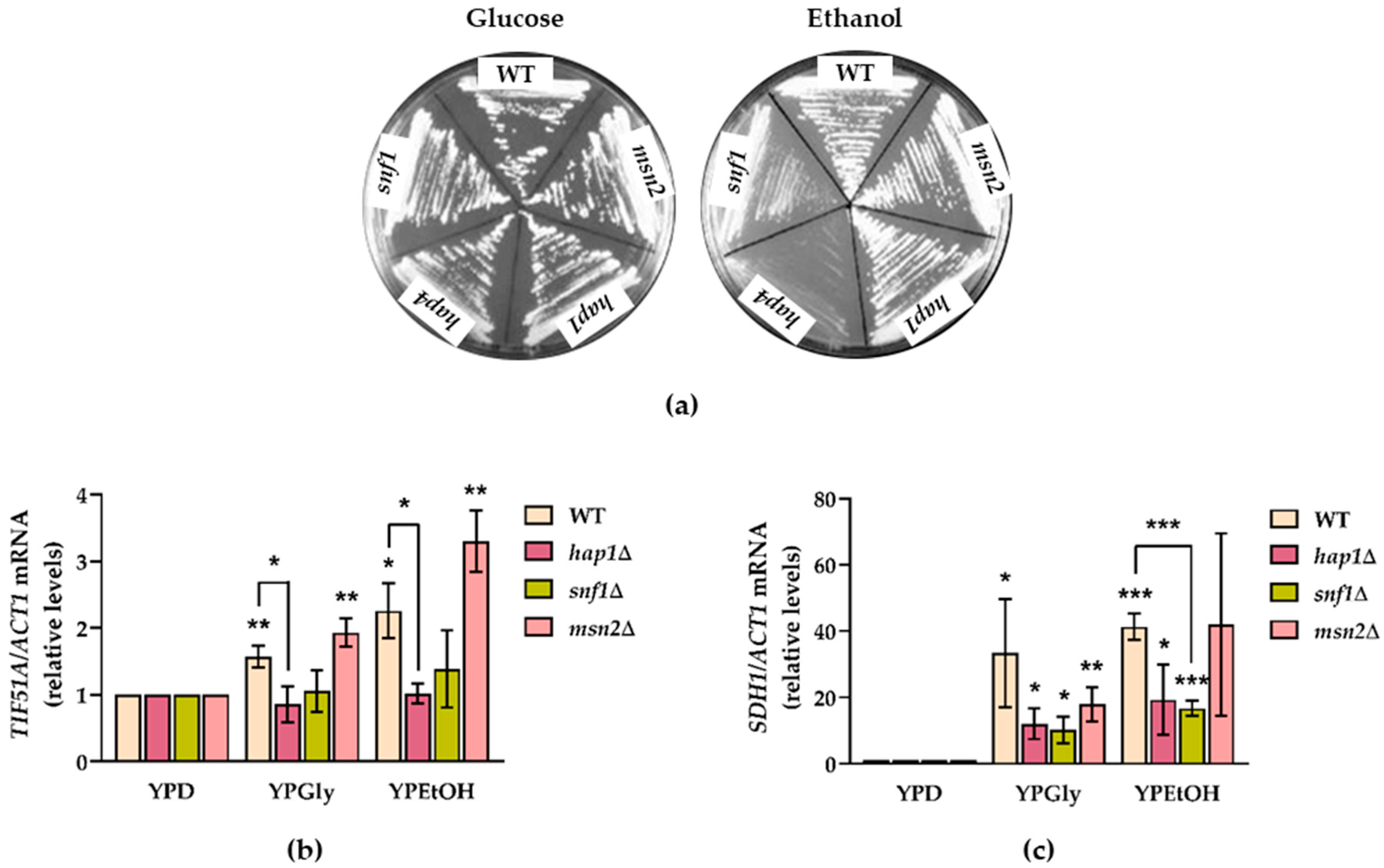
2.2. Snf1 and Hap1 Signalling Pathways Are Involved in TIF51A Induction under Respiratory Conditions
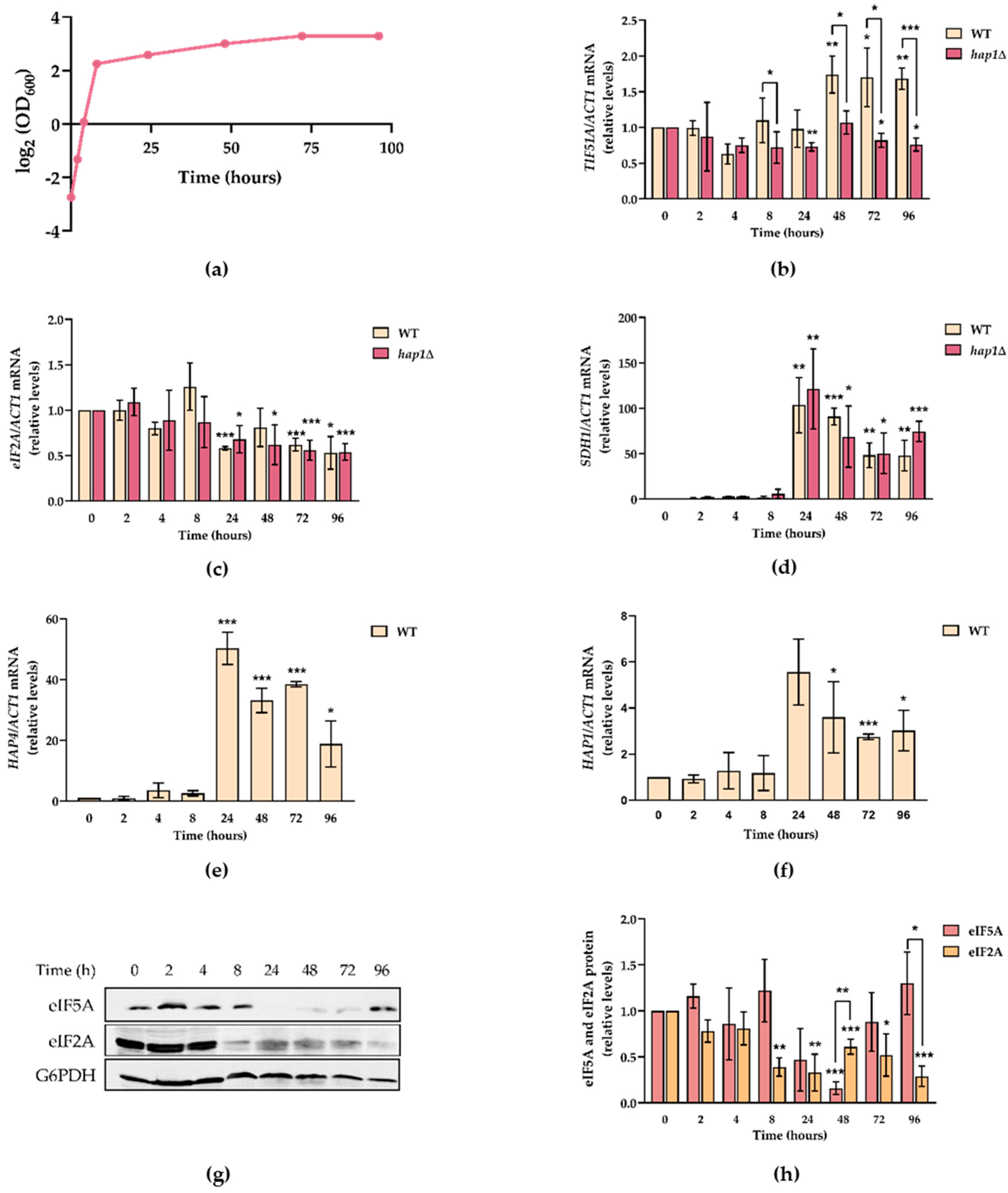
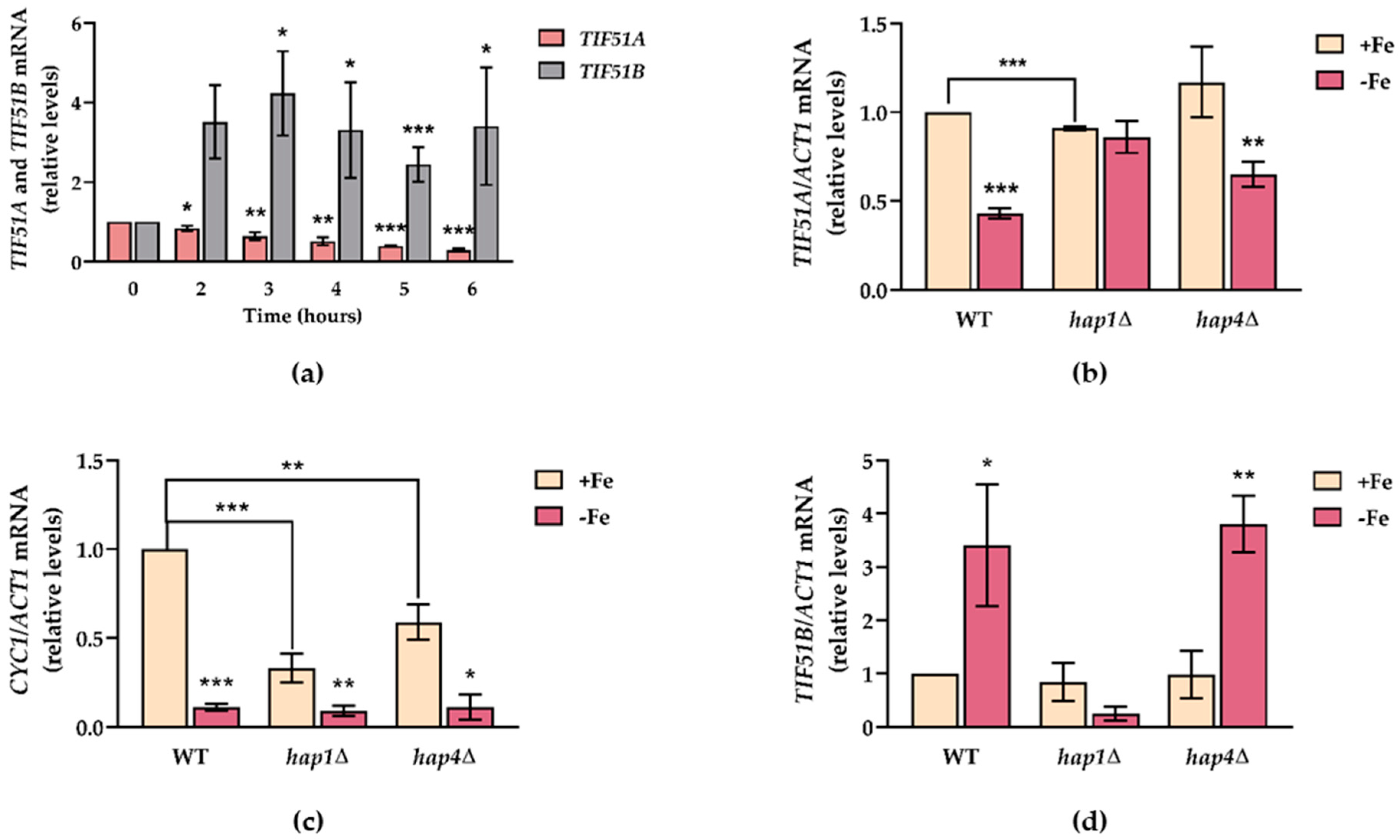
2.3. TIF51A Expression Drops during the Diauxic Shift But Subsequently Increases in a Hap1-Dependent Manner
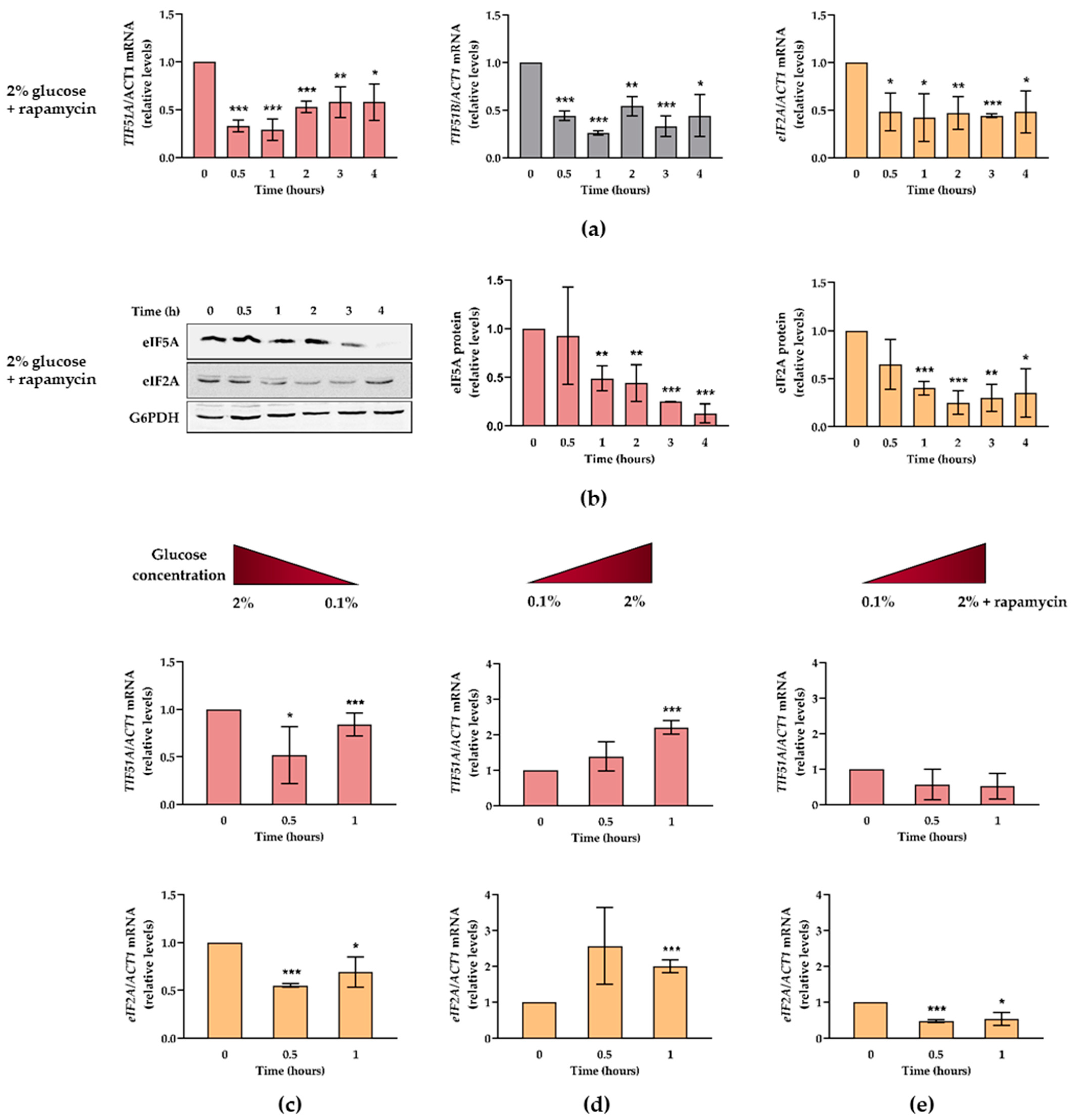
2.4. Glucose Availability and TORC1 Regulate eIF5A Expression
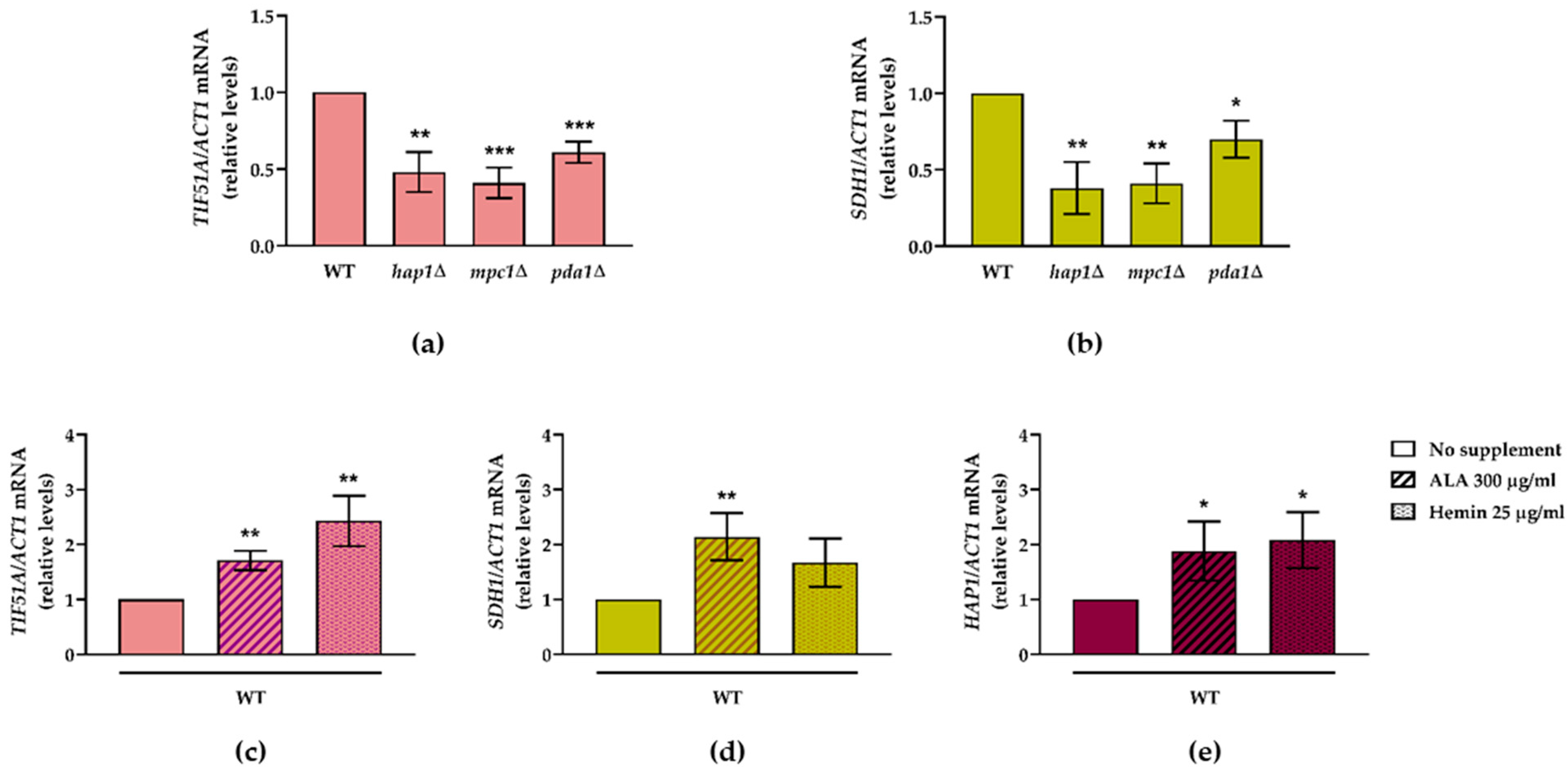
2.5. An Increase in the Metabolic Flux in the TCA Cycle and at Heme Cellular Levels Up-Regulates TIF51A in a Hap1-Dependent Manner
2.6. TIF51A Is Regulated under Iron Deficiency in a Hap1-Dependent Manner
3. Discussions
4. Materials and Methods
4.1. Yeast Strains and Growth Conditions
4.2. Western Blot Analysis
4.3. RT-qPCR Analysis
4.4. Oxygen Consumption Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| eIF2A | Eukaryotic translation initiation factor 2A |
| eIF5A | Eukaryotic translation initiation factor 5A |
| ETC | Electron transport chain |
| OXPHOS | Oxidative phosphorylation |
| PKA | Protein kinase A |
| ROS | Reactive oxygen species |
| SDH | Succinate dehydrogenase |
| TCA | Tricarboxylic acid |
| TORC1 | Target of rapamycin complex I |
References
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Alepuz, P. EIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017, 45, 7326–7338. [Google Scholar] [CrossRef] [PubMed]
- Schuller, A.P.; Wu, C.C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205.e195. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Belda-Palazón, B.; Ferrando, A.; Alepuz, P. Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae. Genetics 2014, 197, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. eif5A promotes translation of polyproline motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef]
- Muñoz-Soriano, V.; Domingo-Muelas, A.; Li, T.; Gamero, E.; Bizy, A.; Fariñas, I.; Alepuz, P.; Paricio, N. Evolutionary conserved role of eukaryotic translation factor eIF5A in the regulation of actin-nucleating formins. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation elongation and recoding in eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Schwelberger, H.G.; Hyun, A.K.; Hershey, J.W.B. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025. [Google Scholar]
- Clement, P.M.J.; Henderson, C.A.; Jenkins, Z.A.; Smit-McBride, Z.; Wolff, E.C.; Hershey, J.W.B.; Park, M.H.; Johansson, H.E. Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 2003, 270, 4254–4263. [Google Scholar] [CrossRef]
- Schnier, J.; Schwelberger, H.G.; Smit-McBride, Z.; Kang, H.A.; Hershey, J.W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 3105–3114. [Google Scholar] [CrossRef]
- Magdolen, V.; Klier, H.; Wöhl, T.; Klink, F.; Hirt, H.; Hauber, J.; Lottspeich, F. The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. MGG Mol. Gen. Genet. 1994, 244, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, Z.A.; Hååg, P.G.; Johansson, H.E. Human EIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 2001, 71, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A. Translational control of eIF5A in various diseases. Amino Acids 2012, 42, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S.; Cleveland, J.L. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 2016, 48, 2353–2362. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Wang, L.; Zhang, H.; Jiao, X.; Chen, D. Eukaryotic translation initiation factor 5A in the pathogenesis of cancers. Oncol. Lett. 2020, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Mathews, M.B.; Hershey, J.W.B. The translation factor eIF5A and human cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Zitomer, R.S.; Lowry, C.V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 1992, 56, 1–11. [Google Scholar] [CrossRef]
- Kastaniotis, A.J.; Mennella, T.A.; Konrad, C.; Torres, A.M.R.; Zitomer, R.S. Roles of Transcription Factor Mot3 and Chromatin in Repression of the Hypoxic Gene ANB1 in Yeast. Mol. Cell. Biol. 2000, 20, 7088–7098. [Google Scholar] [CrossRef]
- Klinkenberg, L.G.; Mennella, T.A.; Luetkenhaus, K.; Zitomer, R.S. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot. Cell 2005, 4, 649–660. [Google Scholar] [CrossRef]
- Sertil, O.; Kapoor, R.; Cohen, B.D.; Abramova, N.; Lowry, C.V. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 5831–5837. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Hach, A. Molecular mechanism of heme signaling in yeast: The transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 1999, 56, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Hickman, M.J.; Winston, F. Heme Levels Switch the Function of Hap1 of Saccharomyces cerevisiae between Transcriptional Activator and Transcriptional Repressor. Mol. Cell. Biol. 2007, 27, 7414–7424. [Google Scholar] [CrossRef] [PubMed]
- Wöhl, T.; Klier, H.; Ammer, H.; Lottspeich, F.; Magdolen, V. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: Characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. MGG Mol. Gen. Genet. 1993, 241, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhan, X.; Li, M.; Li, G.; Zhang, P.; Xiao, Z.; Shao, M.; Peng, F.; Hu, R.; Chen, Z. Mitochondrial proteomics of nasopharyngeal carcinoma metastasis. BMC Med. Genom. 2012, 5, 62. [Google Scholar] [CrossRef]
- Miyake, T.; Pradeep, S.; Wu, S.Y.; Rupaimoole, R.; Zand, B.; Wen, Y.; Gharpure, K.M.; Nagaraja, A.S.; Hu, W.; Cho, M.S.; et al. XPO1/CRM1 inhibition causes antitumor effects by mitochondrial accumulation of eIF5A. Clin. Cancer Res. 2015, 21, 3286–3297. [Google Scholar] [CrossRef]
- Pereira, K.D.; Tamborlin, L.; Meneguello, L.; de Proença, A.R.G.; Almeida, I.C.; Lourenço, R.F.; Luchessi, A.D. Alternative Start Codon Connects eIF5A to Mitochondria. J. Cell. Physiol. 2016, 231, 2682–2689. [Google Scholar] [CrossRef]
- Weir, B.A.; Yaffe, M.P. Mmd1p, a Novel, Conserved Protein Essential for Normal Mitochondrial Morphology and Distribution in the Fission Yeast Schizosaccharomyces pombe. Mol. Biol. Cell 2004, 15, 1656–1665. [Google Scholar] [CrossRef]
- Ma, D.; Zheng, B.; Liu, H.L.; Zhao, Y.B.; Liu, X.; Zhang, X.H.; Li, Q.; Shi, W.B.; Suzuki, T.; Wen, J.K. Klf5 down-regulation induces vascular senescence through eIF5a depletion and mitochondrial fission. PLoS Biol. 2020, 18, e3000808. [Google Scholar] [CrossRef]
- Tan, X.; Wang, D.B.; Lu, X.; Wei, H.; Zhu, R.; Zhu, S.S.; Jiang, H.; Yang, Z.J. Doxorubicin induces apoptosis in H9c2 cardiomyocytes: Role of overexpressed Eukaryotic translation initiation factor 5A. Biol. Pharm. Bull. 2010, 33, 1666–1672. [Google Scholar] [CrossRef]
- Sun, Z.; Cheng, Z.; Taylor, C.A.; Mcconkey, B.J.; Thompson, J.E. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J. Cell. Physiol. 2010, 223, 798–809. [Google Scholar] [CrossRef]
- Melis, N.; Rubera, I.; Cougnon, M.; Giraud, S.; Mograbi, B.; Belaid, A.; Pisani, D.F.; Huber, S.M.; Lacas-Gervais, S.; Fragaki, K.; et al. Targeting eIF5A hypusination prevents anoxic cell death through mitochondrial silencing and improves kidney transplant outcome. J. Am. Soc. Nephrol. 2017, 28, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Giraud, S.; Kerforne, T.; Zely, J.; Ameteau, V.; Couturier, P.; Tauc, M.; Hauet, T. The inhibition of eIF5A hypusination by GC7, a preconditioning protocol to prevent brain death-induced renal injuries in a preclinical porcine kidney transplantation model. Am. J. Transplant. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bourourou, M.; Gouix, E.; Melis, N.; Friard, J.; Heurteaux, C.; Tauc, M.; Blondeau, N. Inhibition of eIF5A hypusination pathway as a new pharmacological target for stroke therapy. J. Cereb. Blood Flow Metab. 2020. [Google Scholar] [CrossRef] [PubMed]
- Puleston, D.J.; Buck, M.D.; Klein Geltink, R.I.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019, 30, 1–12. [Google Scholar] [CrossRef]
- Schüller, H.J. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003, 43, 139–160. [Google Scholar] [CrossRef]
- Rolland, F.; Winderickx, J.; Thevelein, J.M. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002, 2, 183–201. [Google Scholar] [CrossRef]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Turcotte, B.; Liang, X.B.; Robert, F.; Soontorngun, N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010, 10, 2–13. [Google Scholar] [CrossRef]
- Fendt, S.M.; Sauer, U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biol. 2010, 4, 1–12. [Google Scholar] [CrossRef]
- Galdieri, L.; Mehrotra, S.; Yu, S.; Vancura, A. Transcriptional regulation in yeast during diauxic shift and stationary phase. Omi. A J. Integr. Biol. 2010, 14, 629–638. [Google Scholar] [CrossRef]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 13, 2408–2420. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, H.; Whiteway, M. Increased respiration in the sch9Δ mutant is required for increasing chronological life span but not replicative life span. Eukaryot. Cell 2008, 7, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Sanz, P.; Viana, R.; Garcia-Gimeno, M.A. AMPK in Yeast: The SNF1 (Sucrose Non-fermenting 1) Protein Kinase Complex. EXS 2016, 107, 353–374. [Google Scholar] [CrossRef] [PubMed]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Perić, M.; Lovrić, A.; Šarić, A.; Musa, M.; Bou Dib, P.; Rudan, M.; Nikolić, A.; Sobočanec, S.; Mikecin, A.M.; Dennerlein, S.; et al. TORC1-mediated sensing of chaperone activity alters glucose metabolism and extends lifespan. Aging Cell 2017, 16, 994–1005. [Google Scholar] [CrossRef]
- Kunkel, J.; Luo, X.; Capaldi, A.P. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat. Commun. 2019, 10, 3558. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 1989, 5, 153–180. [Google Scholar] [CrossRef]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009, 5, 245. [Google Scholar] [CrossRef]
- DeRisi, J.L.; Iyer, V.R.; Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997, 278, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Bu, P.; Zeng, J.; Vancura, A. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J. Biol. Chem. 2017, 292, 16942–16954. [Google Scholar] [CrossRef] [PubMed]
- Valentini, S.R.; Casolari, J.M.; Oliveira, C.C.; Silver, P.A.; McBride, A.E. Genetic interactions of yeast eukaryotic translation initiation factor 5a (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 2002, 160, 393–405. [Google Scholar] [PubMed]
- Park, J.H.; Aravind, L.; Wolff, E.C.; Kaevel, J.; Kim, Y.S.; Park, M.H. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: A HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 51–56. [Google Scholar] [CrossRef]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998, 17, 3556–3564. [Google Scholar] [CrossRef]
- Boy-Marcotte, E.; Perrot, M.; Bussereau, F.; Boucherie, H.; Jacquet, M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 1998, 180, 1044–1052. [Google Scholar] [CrossRef]
- Pfanzagl, V.; Görner, W.; Radolf, M.; Parich, A.; Schuhmacher, R.; Strauss, J.; Reiter, W.; Schüller, C. A constitutive active allele of the transcription factor Msn2 mimicking low PKA activity dictates metabolic remodeling in yeast. Mol. Biol. Cell 2018, 29, 2848–2862. [Google Scholar] [CrossRef]
- Schneider, J.C.; Guarente, L. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 1991, 11, 4934–4942. [Google Scholar] [CrossRef][Green Version]
- Cardenas, M.E.; Cutler, N.S.; Lorenz, M.C.; Di Como, C.J.; Heitman, J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999, 13, 3271–3279. [Google Scholar] [CrossRef]
- Hardwick, J.S.; Kuruvilla, F.G.; Tong, J.K.; Shamji, A.F.; Schreiber, S.L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 14866–14870. [Google Scholar] [CrossRef]
- Proud, C.G. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 2002, 268, 5338–5349. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Alonso, L.; Romero, A.M.; Martínez-Pastor, M.T.; Puig, S. Iron Regulatory Mechanisms in Saccharomyces cerevisiae. Front. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ihrig, J.; Hausmann, A.; Hain, A.; Richter, N.; Hamza, I.; Lill, R.; Mühlenhoff, U. Iron regulation through the back door: Iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in saccharomyces cerevisiae. Eukaryot. Cell 2010, 9, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Kroemer, G.; Pouyssegur, J. Tumor Cell Metabolism: Cancer’s Achilles’ Heel. Cancer Cell 2008, 13, 472–482. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Hukelmann, J.L.; Anderson, K.E.; Sinclair, L.V.; Grzes, K.M.; Murillo, A.B.; Hawkins, P.T.; Stephens, L.R.; Lamond, A.I.; Cantrell, D.A. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 2016, 17, 104–112. [Google Scholar] [CrossRef]
- Zanelli, C.F.; Valentini, S.R. Is there a role for eIF5A in translation? Amino Acids 2007, 33, 351–358. [Google Scholar] [CrossRef]
- Williams, C.C.; Jan, C.H.; Weissman, J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014, 346, 748–751. [Google Scholar] [CrossRef]
- Zhang, L.; Guarente, L. The yeast activator HAP1-A GAL4 family member-binds DNA in a directly repeated orientation. Genes Dev. 1994, 8, 2110–2119. [Google Scholar] [CrossRef]
- Cao, T.T.; Lin, S.H.; Fu, L.; Tang, Z.; Che, C.M.; Zhang, L.Y.; Ming, X.Y.; Liu, T.F.; Tang, X.M.; Tan, B.B.; et al. Eukaryotic translation initiation factor 5A2 promotes metabolic reprogramming in hepatocellular carcinoma cells. Carcinogenesis 2017, 38, 94–104. [Google Scholar] [CrossRef] [PubMed]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Peltz, S.W. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 2001, 7, 1191–1200. [Google Scholar] [CrossRef]
- Puig, S.; Askeland, E.; Thiele, D.J. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 2005, 120, 99–110. [Google Scholar] [CrossRef]
- Puig, S.; Vergara, S.V.; Thiele, D.J. Cooperation of Two mRNA-Binding Proteins Drives Metabolic Adaptation to Iron Deficiency. Cell Metab. 2008, 7, 555–564. [Google Scholar] [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Soler, M.À.; Perea-García, A.; Alepuz, P.; Puig, S.; Martínez-Pastor, M.T. Yeast Cth2 protein represses the translation of ARE-containing mRNAs in response to iron deficiency. PLoS Genet. 2018, 14, e1007476. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A.; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Gietz, D.; Jean, A.S.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Zuzuarregui, A.; Li, T.; Friedmann, C.; Ammerer, G.; Alepuz, P. Msb2 is a Ste11 membrane concentrator required for full activation of the HOG pathway. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 722–730. [Google Scholar] [CrossRef]
- Garre, E.; Romero-Santacreu, L.; Barneo-Muñoz, M.; Miguel, A.; Pérez-Ortín, J.E.; Alepuz, P. Nonsense-Mediated mRNA Decay Controls the Changes in Yeast Ribosomal Protein Pre-mRNAs Levels upon Osmotic Stress. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barba-Aliaga, M.; Villarroel-Vicente, C.; Stanciu, A.; Corman, A.; Martínez-Pastor, M.T.; Alepuz, P. Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways. Int. J. Mol. Sci. 2021, 22, 219. https://doi.org/10.3390/ijms22010219
Barba-Aliaga M, Villarroel-Vicente C, Stanciu A, Corman A, Martínez-Pastor MT, Alepuz P. Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways. International Journal of Molecular Sciences. 2021; 22(1):219. https://doi.org/10.3390/ijms22010219
Chicago/Turabian StyleBarba-Aliaga, Marina, Carlos Villarroel-Vicente, Alice Stanciu, Alba Corman, María Teresa Martínez-Pastor, and Paula Alepuz. 2021. "Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways" International Journal of Molecular Sciences 22, no. 1: 219. https://doi.org/10.3390/ijms22010219
APA StyleBarba-Aliaga, M., Villarroel-Vicente, C., Stanciu, A., Corman, A., Martínez-Pastor, M. T., & Alepuz, P. (2021). Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways. International Journal of Molecular Sciences, 22(1), 219. https://doi.org/10.3390/ijms22010219





