Regulation of Iron Homeostasis and Use in Chloroplasts
Abstract
1. Introduction
2. Chloroplast Fe Use
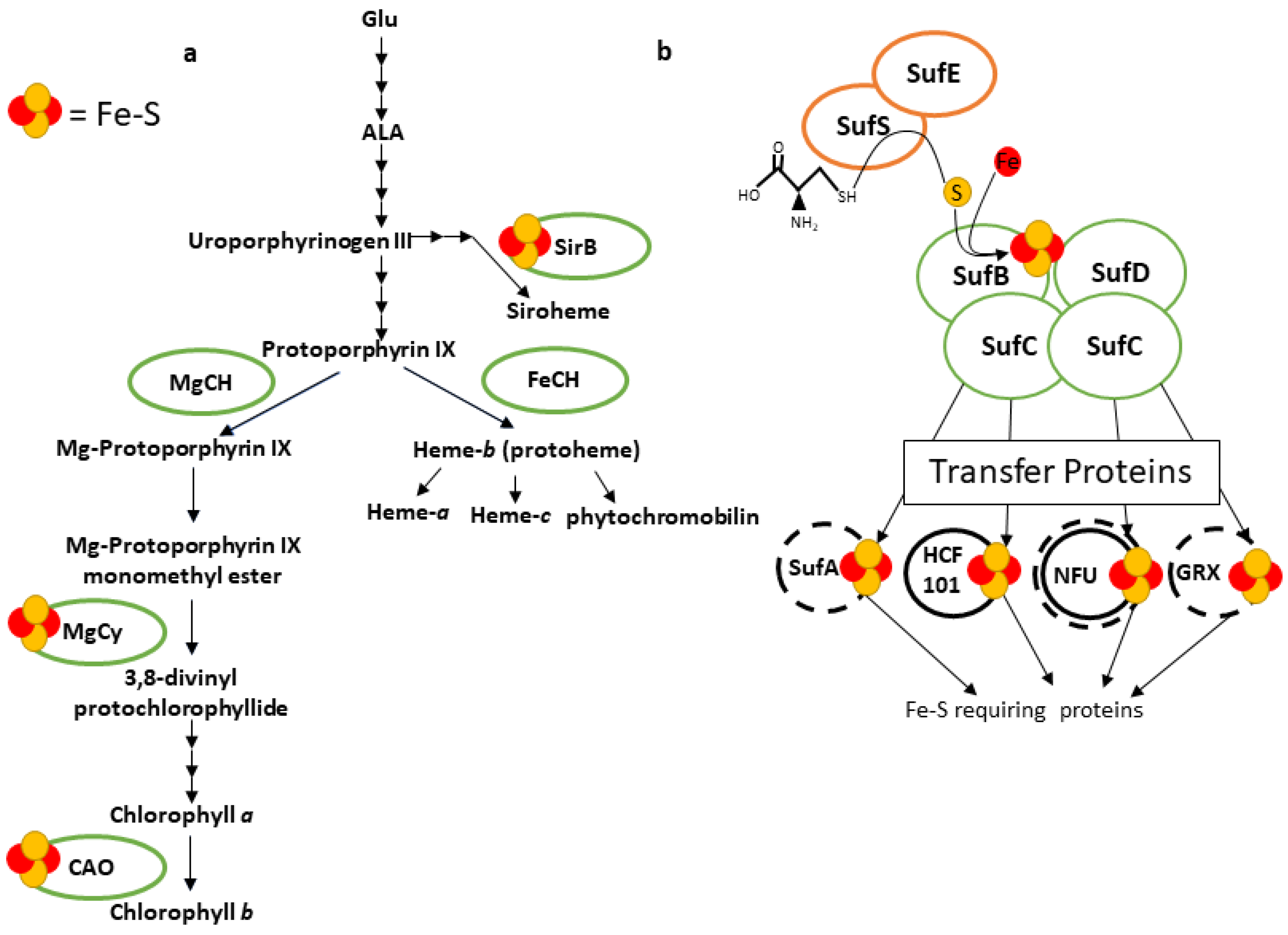
Fe Cofactor Assembly in Plastids
3. Chloroplast Fe Transport and Storage
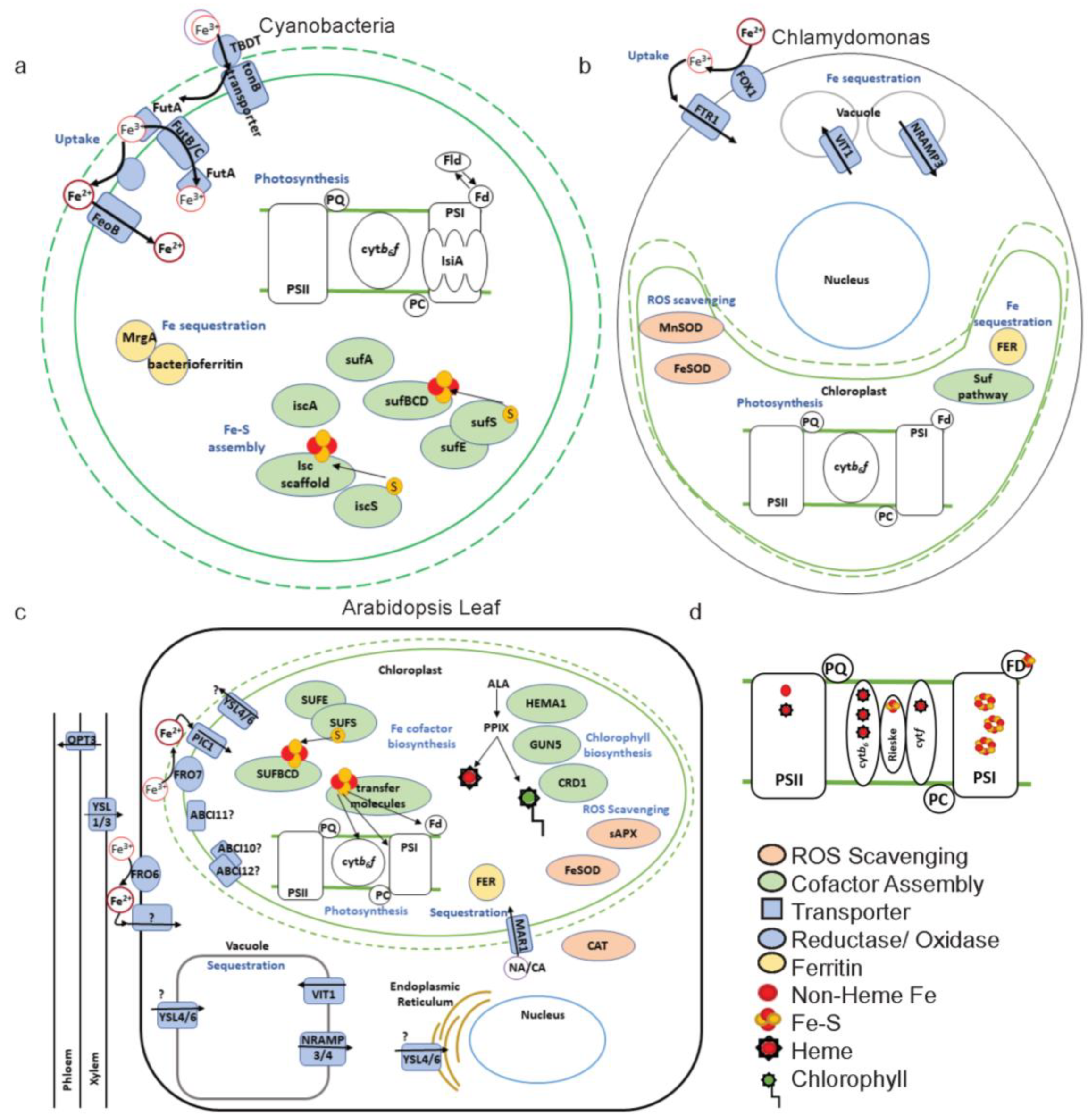
3.1. Chloroplast Fe Uptake
3.2. Chloroplast Fe Export
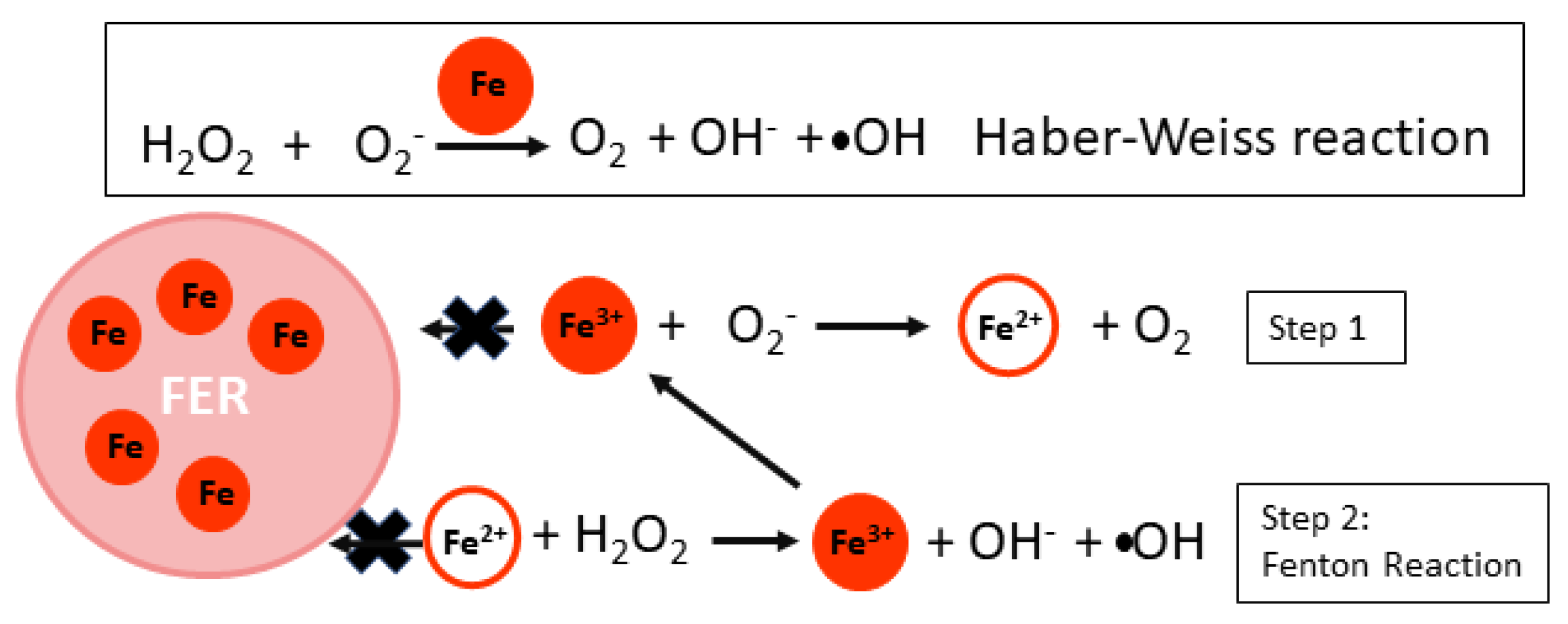
3.3. Fe Sequestration
4. Acclimation to Low Fe
4.1. Increase Fe Uptake
4.2. Metabolic Remodeling
4.3. Fe Utilization
4.4. Compensatory Responses to Fe Deficiency
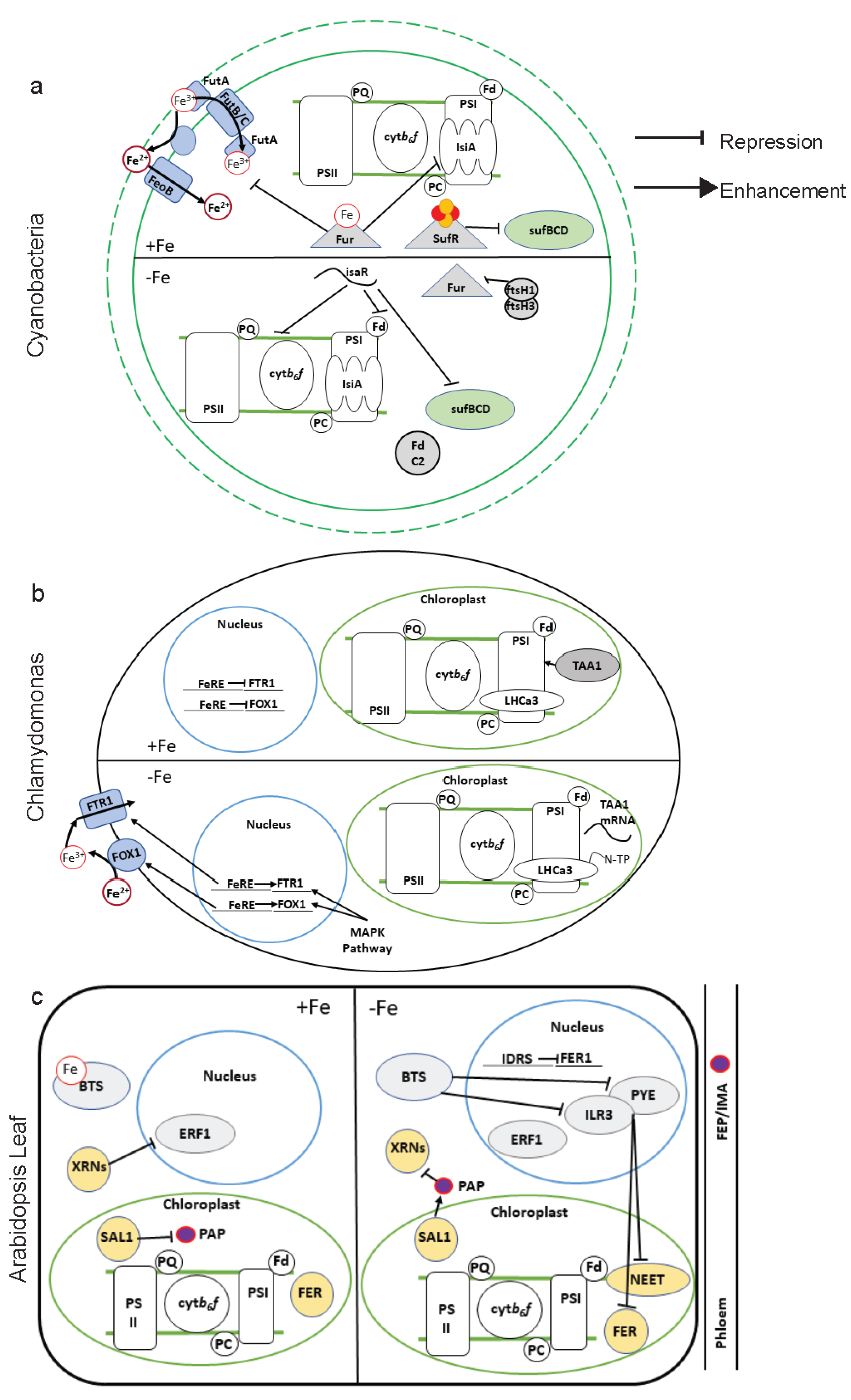
5. Regulating Acclimation to Low Fe
5.1. Root Regulation of Fe Uptake
5.2. Transcriptional Regulation
5.3. Post-Transcriptional Regulation
5.4. Post-Translational Regulation
5.5. Fe Sensing
6. Regulating Acclimation to Excess Fe
7. Conclusions
Funding
Conflicts of Interest
References
- Wacey, D.; Saunders, M.; Brasier, M.D.; Kilburn, M.R. Earliest microbially mediated pyrite oxidation in similar to 3.4 billion-year-old sediments. Earth Planet. Sci. Lett. 2011, 301, 393–402. [Google Scholar] [CrossRef]
- Fischer, W.; Hemp, J.; Johnson, J. Evolution of Oxygenic Photosynthesis. Annu. Rev. Earth Planet. Sci. 2016, 44, 647–683. [Google Scholar] [CrossRef]
- Lyons, T.W.; Reinhard, C.T.; Planavsky, N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Lenton, T.M.; Dahl, T.W.; Daines, S.J.; Mills, B.J.W.; Ozaki, K.; Saltzman, M.R.; Porada, P. Earliest land plants created modern levels of atmospheric oxygen. Proc. Natl. Acad. Sci. USA 2016, 113, 9704–9709. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Allen, J.F. Genomics and chloroplast evolution: What did Cyanobacteria do for plants? Genome Biol. 2003, 4, 209. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Lagarias, J.C.; Bhattacharya, D. Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Front. Ecol. Evol. 2014, 2, 66. [Google Scholar] [CrossRef]
- Merchant, S.; Sawaya, M.R. The Light Reactions: A Guide to Recent Acquisitions for the Picture Gallery. Plant Cell 2005, 17, 648–663. [Google Scholar] [CrossRef]
- Shikanai, T.; Muller-Moule, P.; Munekage, Y.; Niyogi, K.K.; Pilon, M. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 2003, 15, 1333–1346. [Google Scholar] [CrossRef]
- Canfield, D.E. The early history of atmospheric oxygen: Homage to Robert M. Garrels. Annu. Rev. Earth Planet. Sci. 2005, 33, 1–36. [Google Scholar] [CrossRef]
- Crichton, R.R. Iron Metabolism: From Molecular Mechanisms to Clinical Consequences, 4th ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2016. [Google Scholar]
- Thi Tuyet Le, C.; Brumbarova, T.; Bauer, P. The interplay of ROS and iron signaling. In Redox Homeostasis in Plants; Springer: Berlin, Germany, 2019; Volume 1, pp. 43–66. [Google Scholar]
- Tewari, R.K.; Hadacek, F.; Sassmann, S.; Lang, I. Iron deprivation-induced reactive oxygen species generation leads to non-autolytic PCD in Brassica napus leaves. Environ. Exp. Bot. 2013, 91, 74–83. [Google Scholar] [CrossRef]
- Yadavalli, V.; Neelam, S.; Rao, A.S.V.C.; Reddy, A.R.; Subramanyam, R. Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 2012, 169, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Terzulli, A.; Kosman, D.J. Analysis of the High-Affinity Iron Uptake System at the Chlamydomonas reinhardtii Plasma Membrane. Eukaryot. Cell 2010, 9, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; McIntyre, L.; Sherman, L. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the Cyanobacterium synechocystis sp. PCC 6803. Plant Physiol. 2003, 132, 1825–1839. [Google Scholar] [CrossRef]
- Chappell, P.D.; Webb, E.A. A molecular assessment of the iron stress response in the two phylogenetic clades of Trichodesmium. Environ. Microbiol. 2010, 12, 13–27. [Google Scholar] [CrossRef]
- Kim, S.A.; Guerinot, M.L. Mining iron: Iron uptake and transport in plants. FEBS Lett. 2007, 581, 2273–2280. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron Uptake, Translocation, and Regulation in Higher Plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Thomine, S.; Vert, G. Iron transport in plants: Better be safe than sorry. Curr. Opin. Plant Biol. 2013, 16, 322–327. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J.; Rodriguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef]
- Peers, G.; Niyogi, K.K. Pond scum genomics: The genomes of Chlamydomonas and Ostreococcus. Plant Cell 2008, 20, 502–507. [Google Scholar] [CrossRef]
- Frey, P.A.; Reed, G.H. The ubiquity of iron. ACS Chem. Biol. 2012, 7, 1477–1481. [Google Scholar] [CrossRef]
- Colombo, C.; Palumbo, G.; He, J.-Z.; Pinton, R.; Cesco, S. Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soils Sediments 2014, 14, 538–548. [Google Scholar] [CrossRef]
- Blomqvist, S.; Gunnars, A.; Elmgren, R. Why the limiting nutrient differs between temperate coastal seas and freshwater lakes: A matter of salt. Limnol. Oceanogr. 2004, 49, 2236–2241. [Google Scholar] [CrossRef]
- Gledhill, M.; Buck, K.N. The organic complexation of iron in the marine environment: A review. Front Microbiol 2012, 3, 69. [Google Scholar] [CrossRef] [PubMed]
- Sasso, S.; Stibor, H.; Mittag, M.; Grossman, A.R. The Natural History of Model Organisms: From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. eLife 2018, 7, e39233. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Baracaldo, P.; Hayes, P.K.; Blank, C.E. Morphological and habitat evolution in the Cyanobacteria using a compartmentalization approach. Geobiology 2005, 3, 145–165. [Google Scholar] [CrossRef]
- Jeong, J.; Cohu, C.; Kerkeb, L.; Pilon, M.; Connolly, E.L.; Guerinot, M.L. Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 10619–10624. [Google Scholar] [CrossRef]
- Waters, M.T.; Langdale, J.A. The making of a chloroplast. EMBO J. 2009, 28, 2861–2873. [Google Scholar] [CrossRef]
- Barkan, A. Expression of Plastid Genes: Organelle-Specific Elaborations on a Prokaryotic Scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef]
- Fukuyama, K.; Hase, T.; Matsumoto, S.; Tsukihara, T.; Katsube, Y.; Tanaka, N.; Kakudo, M.; Wada, K.; Matsubara, H. Structure of s-platensis 2Fe-2S ferredoxin and evolution of chloroplast-type ferredoxins. Nature 1980, 286, 522–524. [Google Scholar] [CrossRef]
- Hurt, E.; Hauska, G. A Cytochrome f/b6 Complex of Five Polypeptides with Plastoquinol-Plastocyanin-Oxidoreductase Activity from Spinach Chloroplasts. Eur. J. Biochem. 1981, 117, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Shevela, D.; Eaton-Rye, J.J.; Shen, J.R. Photosystem II and the unique role of bicarbonate: A historical perspective. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1134–1151. [Google Scholar] [CrossRef] [PubMed]
- Moulin, M.; Smith, A.G. Regulation of tetrapyrrole biosynthesis in higher plants. Biochem. Soc. Trans. 2005, 33, 737–742. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0145. [Google Scholar] [CrossRef]
- Czarnecki, O.; Hedtke, B.; Melzer, M.; Rothbart, M.; Richter, A.; Schröter, Y.; Pfannschmidt, T.; Grimm, B. An Arabidopsis GluTR Binding Protein Mediates Spatial Separation of 5-Aminolevulinic Acid Synthesis in Chloroplasts. Plant Cell 2011, 23, 4476–4491. [Google Scholar] [CrossRef]
- Kumar, A.M.; Söll, D. Antisense HEMA1 RNA Expression Inhibits Heme and Chlorophyll Biosynthesis in Arabidopsis. Plant Physiol. 2000, 122, 49–55. [Google Scholar] [CrossRef]
- Raux-Deery, E.; Leech, H.K.; Nakrieko, K.A.; McLean, K.J.; Munro, A.W.; Heathcote, P.; Rigby, S.E.J.; Smith, A.G.; Warren, M.J. Identification and characterization of the terminal enzyme of siroheme biosynthesis from Arabidopsis thaliana—A plastid-located sirohydrochlorin ferrochelatase containing a 2Fe-2S center. J. Biol. Chem. 2005, 280, 4713–4721. [Google Scholar] [CrossRef]
- Page, M.L.D.; Hamel, P.P.; Gabilly, S.T.; Zegzouti, H.; Perea, J.V.; Alonso, J.M.; Ecker, J.R.; Theg, S.M.; Christensen, S.K.; Merchant, S. A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b(6)f complex in Arabidopsis chloroplasts. J. Biol. Chem. 2004, 279, 32474–32482. [Google Scholar] [CrossRef]
- Gabilly, S.T.; Dreyfuss, B.W.; Karamoko, M.; Corvest, V.; Kropat, J.; Page, M.D.; Merchant, S.S.; Hamel, P.P. CCS5, a Thioredoxin-like Protein Involved in the Assembly of Plastid c-Type Cytochromes. J. Biol. Chem. 2010, 285, 29738–29749. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.M.; White, R.H.; Cash, V.L.; Jack, R.F.; Dean, D.R. Cysteine desulfurase activity indicates a role for nifs in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 1993, 90, 2754–2758. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.M.; Cash, V.L.; Flint, D.H.; Dean, D.R. Assembly of iron-sulfur clusters—Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 1998, 273, 13264–13272. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Tokumoto, U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 2002, 277, 28380–28383. [Google Scholar] [CrossRef]
- Outten, F.W.; Djaman, O.; Storz, G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 2004, 52, 861–872. [Google Scholar] [CrossRef]
- Frazzon, J.; Fick, J.R.; Dean, D.R. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 2002, 30, 680–685. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef]
- Lill, R.; Muhlenhoff, U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008, 77, 669–700. [Google Scholar] [CrossRef]
- Mihara, H.; Esaki, N. Bacterial cysteine desulfurases: Their function and mechanisms. Appl. Microbiol. Biotechnol. 2002, 60, 12–23. [Google Scholar]
- Balk, J.; Pilon, M. Ancient and essential: The assembly of iron–sulfur clusters in plants. Trends Plant Sci. 2011, 16, 218–226. [Google Scholar] [CrossRef]
- Kushnir, S.; Babiychuk, E.; Storozhenko, S.; Davey, M.W.; Papenbrock, J.; De Rycke, R.; Engler, G.; Stephan, U.W.; Lange, H.; Kispal, G.; et al. A Mutation of the Mitochondrial ABC Transporter Sta1 Leads to Dwarfism and Chlorosis in the Arabidopsis Mutant starik. Plant Cell 2001, 13, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Bernard, D.G.; Cheng, Y.F.; Zhao, Y.D.; Balk, J. An Allelic Mutant Series of ATM3 Reveals Its Key Role in the Biogenesis of Cytosolic Iron-Sulfur Proteins in Arabidopsis. Plant Physiol. 2009, 151, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, T.A.; Thornton, J.D.; Kruse, I.; Schwarzlander, M.; Meyer, A.J.; van Veen, H.W.; Balk, J. A Conserved Mitochondrial ATP-binding Cassette Transporter Exports Glutathione Polysulfide for Cytosolic Metal Cofactor Assembly. J. Biol. Chem. 2014, 289, 23264–23274. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Witcher, S.; Outten, F.W.; Pilon, M. The Suf System in Archea, Bacteria, and Eukaryotic Organelles. In Metalloprotein Active Site Assembly; Johnson, M.K., Scott, R.A., Eds.; Wiley: West Sussex, UK, 2017; Volume 1, pp. 37–52. [Google Scholar]
- Li, H.M.; Theg, S.M.; Bauerle, C.M.; Keegstra, K. Metal-ion-center assembly of ferredoxin and plastocyanin in isolated-chloroplasts. Proc. Natl. Acad. Sci. USA 1990, 87, 6748–6752. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M.; America, T.; van’t Hof, R.; de Kruijff, B.; Weisbeek, P. Protein translocation into chloroplasts. In Advances in Molecular and Cell Biology; Rothman, S.S., Ed.; JAI Press: Greenwich, CT, USA, 1995; pp. 229–255. [Google Scholar]
- Takahashi, Y.; Mitsui, A.; Hase, T.; Matsubara, H. Formation of the iron sulfur cluster of ferredoxin in isolated-chloroplasts. Proc. Natl. Acad. Sci. USA 1986, 83, 2434–2437. [Google Scholar] [CrossRef]
- Takahashi, Y.; Mitsui, A.; Matsubara, H. Formation of the Fe-S cluster of ferredoxin in lysed spinach-chloroplasts. Plant Physiol. 1991, 95, 97–103. [Google Scholar] [CrossRef]
- Takahashi, Y.; Mitsui, A.; Fujita, Y.; Matsubara, H. Roles of ATP and NADPH in formation of the fe-s cluster of spinach ferredoxin. Plant Physiol. 1991, 95, 104–110. [Google Scholar] [CrossRef][Green Version]
- Balk, J.; Schaedler, T.A. Iron Cofactor Assembly in Plants. Annu. Rev. Plant Biol. 2014, 65, 125–153. [Google Scholar] [CrossRef]
- Pilon-Smits, E.A.H.; Garifullina, G.F.; Abdel-Ghany, S.; Kato, S.-I. Characterization of a NifS-like chloroplastic protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol. 2002, 130, 1309–1318. [Google Scholar] [CrossRef]
- Leon, S.; Touraine, B.; Ribot, C.; Briat, J.F.; Lobreaux, S. Iron-sulphur cluster assembly in plants: Distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem. J. 2003, 371, 823–830. [Google Scholar] [CrossRef]
- Ye, H.; Garifullina, G.F.; Abdel-Ghany, S.E.; Zhang, L.H.; Pilon-Smits, E.A.H.; Pilon, M. The chloroplast NifS-like protein of Arabidopsis thaliana is required for iron-sulfur cluster formation in ferredoxin. Planta 2005, 220, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Van Hoewyk, D.; Abdel-Ghany, S.E.; Cohu, C.M.; Herbert, S.K.; Kugrens, P.; Pilon, M.; Pilon-Smits, E.A.H. Chloroplast iron-sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc. Natl. Acad. Sci. USA 2007, 104, 5686–5691. [Google Scholar] [CrossRef]
- Ye, H.; Abdel-Ghany, S.E.; Anderson, T.D.; Pilon-Smits, E.A.H.; Pilon, M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 2006, 281, 8958–8969. [Google Scholar] [CrossRef]
- Xu, X.M.; Moller, S.G. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 2006, 25, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Wu, H.C.; Dhalleine, T.; Pegeot, H.; Sudre, D.; Gualberto, J.M.; Jacquot, J.P.; Gaymard, F.; Vignols, F.; Rouhier, N. Monothiol GlutaredoxinBolA Interactions: Redox Control of Arabidopsis thaliana BolA2 and SufE1. Mol. Plant 2014, 7, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Murthy, N.; Ollagnier-de-Choudens, S.; Sanakis, Y.; Abdel-Ghany, S.E.; Rousset, C.; Ye, H.; Fontecave, M.; Pilon-Smits, E.A.H.; Pilon, M. Characterization of Arabidopsis thaliana SufE2 and SufE3—Functions in chloroplast iron-sulfur cluster assembly and NAD synthesis. J. Biol. Chem. 2007, 282, 18254–18264. [Google Scholar]
- Moller, S.G.; Kunkel, T.; Chua, N.H. A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 2001, 15, 90–103. [Google Scholar] [CrossRef]
- Xu, X.M.; Møller, S.G.; Cashmore, A.R. AtNAP7 Is a Plastidic SufC-Like ATP-Binding Cassette/ATPase Essential for Arabidopsis Embryogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 9143–9148. [Google Scholar] [CrossRef]
- Ahn, C.S.; Lee, J.H.; Pai, H.S. Silencing of NbNAP1 encoding a plastidic SufB-like protein affects chloroplast development in Nicotiana benthamiana. Mol. Cells 2005, 20, 112–118. [Google Scholar]
- Hu, X.Y.; Kato, Y.; Sumida, A.; Tanaka, A.; Tanaka, R. The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe-S proteins. Plant J. 2017, 90, 235–248. [Google Scholar] [CrossRef]
- Lu, Y. Assembly and Transfer of Iron-Sulfur Clusters in the Plastid. Front. Plant Sci. 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Yabe, T.; Morimoto, K.; Kikuchi, S.; Nishio, K.; Terashima, I.; Nakai, M. The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 2004, 16, 993–1007. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.E.; Ye, H.; Garifullina, G.F.; Zhang, L.H.; Pilon- Smits, E.A.H.; Pilon, M. Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 2005, 138, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Lezhneva, L.; Amann, K.; Meurer, J. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 2004, 37, 174–185. [Google Scholar] [CrossRef]
- Schwenkert, S.; Netz, D.J.A.; Frazzon, J.; Pierik, A.J.; Bill, E.; Gross, J.; Lill, R.; Meurer, J. Chloroplast HCF101 is a scaffold protein for 4Fe-4S cluster assembly. Biochem. J. 2010, 425, 207–214. [Google Scholar] [CrossRef]
- Touraine, B.; Boutin, J.P.; Marion-Poll, A.; Briat, J.F.; Peltier, G.; Lobréaux, S. Nfu2: A scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 2004, 40, 101–111. [Google Scholar] [CrossRef]
- Touraine, B.; Vignols, F.; Przybyla-Toscano, J.; Ischebeck, T.; Dhalleine, T.; Wu, H.-C.; Magno, C.; Nathalie, B.; Couturier, J.; Dubos, C.; et al. Iron-sulfur protein NFU2 is required for branched chain amino acid synthesis in Arabidopsis roots. J. Exp. Bot. 2019, 70, 1875–1889. [Google Scholar] [CrossRef]
- Yabe, T.; Nakai, M. Arabidopsis AtIscA-I is affected by deficiency of Fe–S cluster biosynthetic scaffold AtCnfU-V. Biochem. Biophys. Res. Commun. 2006, 340, 1047–1052. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Gama, F.; Molina-Navarro, M.M.; Gualberto, J.M.; Claxton, R.; Naik, S.G.; Huynh, B.H.; Herrero, E.; Jacquot, J.P.; Johnson, M.K.; et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of 2Fe-2S clusters. EMBO J. 2008, 27, 1122–1133. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Song, L.; Sengupta, S.; McInturf, S.A.; Grant, D.G.; Marjault, H.-B.; Castro-Guerrero, N.A.; Burks, D.; Azad, R.K.; Mendoza-Cozatl, D.G.; et al. Expression of a dominant-negative AtNEET-H89C protein disrupts iron-sulfur metabolism and iron homeostasis in Arabidopsis. Plant J. Cell Mol. Biol. 2019, 101, 1152–1169. [Google Scholar] [CrossRef]
- Nechushtai, R.; Conlan, A.; Harir, Y.; Song, L.; Yogev, O.; Eisenberg-Domovich, Y.; Livnah, O.; Michaeli, D.; Rosen, R.; Ma, V.; et al. Characterization of Arabidopsis NEET Reveals an Ancient Role for NEET Proteins in Iron Metabolism. Plant Cell 2012, 24, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- González-Vallejo, E.B.; Morales, F.; Cistué, L.; Abadía, A.; Abadía, J. Iron Deficiency Decreases the Fe(III)-Chelate Reducing Activity of Leaf Protoplasts. Plant Physiol. 2000, 122, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; An, F.; Zhang, S.; Ji, Z.; Ling, H.-Q.; Zuo, J. Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant Physiol. 2006, 140, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Bughio, N.; Takahashi, M.; Yoshimura, E.; Nishizawa, N.K.; Mori, S. Characteristics of light-regulated iron transport system in barley chloroplasts. Soil Sci. Plant Nutr. 1997, 43, 959–963. [Google Scholar] [CrossRef]
- Shingles, R.; North, M.; McCarty, R.E. Ferrous ion transport across chloroplast inner envelope membranes. Plant Physiol. 2002, 128, 1022–1030. [Google Scholar] [CrossRef]
- Salome, P.A.; Oliva, M.; Weigel, D.; Kramer, U. Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO J. 2013, 32, 511–523. [Google Scholar] [CrossRef]
- Duc, C.; Cellier, F.; Lobréaux, S.; Briat, J.-F.; Gaymard, F. Regulation of iron homeostasis in Arabidopsis thaliana by the clock regulator time for coffee. J. Biol. Chem. 2009, 284, 36271–36281. [Google Scholar] [CrossRef]
- Lopez-Millan, A.F.; Duy, D.; Philippar, K. Chloroplast Iron Transport Proteins—Function and Impact on Plant Physiology. Front. Plant Sci. 2016, 7, 178. [Google Scholar] [CrossRef]
- Vigani, G.; Solti, Á.; Thomine, S.; Philippar, K. Essential and Detrimental—An Update on Intracellular Iron Trafficking and Homeostasis. Plant Cell Physiol. 2019, 60, 1420–1439. [Google Scholar] [CrossRef]
- Hopkinson, B.; Morel, F. The role of siderophores in iron acquisition by photosynthetic marine microorganisms. BioMetals 2009, 22, 659–669. [Google Scholar] [CrossRef]
- Kranzler, C.; Lis, H.; Shaked, Y.; Keren, N. The role of reduction in iron uptake processes in a unicellular, planktonic Cyanobacterium. Environ. Microbiol. 2011, 13, 2990–2999. [Google Scholar] [CrossRef] [PubMed]
- Kranzler, C.; Lis, H.; Finkel, O.M.; Schmetterer, G.; Shaked, Y.; Keren, N. Coordinated transporter activity shapes high-affinity iron acquisition in Cyanobacteria. ISME J. 2013, 8, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Mirus, O.; Strauss, S.; Nicolaisen, K.; von Haeseler, A.; Schleiff, E. TonB-dependent transporters and their occurrence in Cyanobacteria. BMC Biol. 2009, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Lammers, P.J.; Sanders-Loehr, J. Active transport of ferric schizokinen in Anabaena sp. J. Bacteriol. 1982, 151, 288–294. [Google Scholar] [CrossRef]
- Nicolaisen, K.; Moslavac, S.; Samborski, A.; Valdebenito, M.; Hantke, K.; Maldener, I.; Muro-Pastor, A.M.; Flores, E.; Schleiff, E. Alr0397 is an outer membrane transporter for the siderophore schizokinen in Anabaena sp. strain PCC 7120. J. Bacteriol. 2008, 190, 7500–7507. [Google Scholar] [CrossRef]
- Qiu, G.-W.; Lou, W.-J.; Sun, C.-Y.; Yang, N.; Li, Z.-K.; Li, D.-L.; Zang, S.-S.; Fu, F.-X.; Hutchins, D.A.; Jiang, H.-B.; et al. Outer Membrane Iron Uptake Pathways in the Model Cyanobacterium Synechocystis sp. Strain PCC 6803. Appl. Environ. Microbiol. 2018, 84, e01512–e01518. [Google Scholar] [CrossRef]
- Breuers, F.K.H.; Brautigam, A.; Weber, A.P.M. The plastid outer envelope—A highly dynamic interface between plastid and cytoplasm. Front. Plant Sci. 2011, 14, 97. [Google Scholar] [CrossRef]
- Solti, Á.; Müller, B.; Czech, V.; Sárvári, É.; Fodor, F. Functional characterization of the chloroplast ferric chelate oxidoreductase enzyme. New Phytol. 2014, 202, 920–928. [Google Scholar] [CrossRef]
- Duy, D.; Wanner, G.; Meda, A.R.; von Wiren, N.; Soll, J.; Philippar, K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 2007, 19, 986–1006. [Google Scholar] [CrossRef]
- Teng, Y.S.; Su, Y.S.; Chen, L.J.; Lee, Y.J.; Hwang, I.; Li, H.M. Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 2006, 18, 2247–2257. [Google Scholar] [CrossRef]
- Duy, D.; Stube, R.; Wanner, G.; Philippar, K. The Chloroplast Permease PIC1 Regulates Plant Growth and Development by Directing Homeostasis and Transport of Iron. Plant Physiol. 2011, 155, 1709–1722. [Google Scholar] [CrossRef] [PubMed]
- Shimoni-Shor, E.; Hassidim, M.; Yuval-Naeh, N.; Keren, N. Disruption of Nap14, a plastid-localized non-intrinsic ABC protein in Arabidopsis thaliana results in the over-accumulation of transition metals and in aberrant chloroplast structures. Plant Cell Environ. 2010, 33, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Voith von Voithenberg, L.; Park, J.; Stube, R.; Lux, C.; Lee, Y.; Philippar, K. A Novel Prokaryote-Type ECF/ABC Transporter Module in Chloroplast Metal Homeostasis. Front. Plant Sci. 2019, 10, 1264. [Google Scholar] [CrossRef] [PubMed]
- Divol, F.; Couch, D.; Conejero, G.; Roschzttardtz, H.; Mari, S.; Curie, C. The Arabidopsis YELLOW STRIPE LIKE4 and 6 Transporters Control Iron Release from the Chloroplast. Plant Cell 2013, 25, 1040–1055. [Google Scholar] [CrossRef] [PubMed]
- Conte, S.S.; Echu, H.-H.; Rodriguez, D.C.; Punshon, T.; Vasques, K.A.; Salt, D.E.; Walker, E.L. Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis. Front. Plant Sci. 2013, 4, 283. [Google Scholar] [CrossRef]
- Müller, B.; Kovács, K.; Pham, H.-D.; Kavak, Y.; Pechoušek, J.; Machala, L.; Zbořil, R.; Szenthe, K.; Abadía, J.; Fodor, F.; et al. Chloroplasts preferentially take up ferric-citrate over iron-nicotianamine complexes in Brassica napus. Planta 2019, 249, 751–763. [Google Scholar] [CrossRef]
- Solti, A.; Kovacs, K.; Basa, B.; Vertes, A.; Sarvari, E.; Fodor, F. Uptake and incorporation of iron in sugar beet chloroplasts. Plant Physiol. Biochem. 2012, 52, 91–97. [Google Scholar] [CrossRef]
- Conte, S.; Stevenson, D.; Furner, I.; Lloyd, A. Multiple Antibiotic Resistance in Arabidopsis Is Conferred by Mutations in a Chloroplast-Localized Transport Protein. Plant Physiol. 2009, 151, 559–573. [Google Scholar] [CrossRef]
- Ravet, K.; Touraine, B.; Boucherez, J.; Briat, J.F.; Gaymard, F.; Cellier, F. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 2009, 57, 400–412. [Google Scholar] [CrossRef]
- Petit, J.M.; Briat, J.F.; Lobreaux, S. Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 2001, 359, 575–582. [Google Scholar] [CrossRef]
- Proudhon, D.; Wei, J.; Briat, J.; Theil, E. Ferritin gene organization: Differences between plants and animals suggest possible kingdom-specific selective constraints. J. Mol. Evol. 1996, 42, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.F.; Duc, C.; Ravet, K.; Gaymard, F. Ferritins and iron storage in plants. Biochem. Biophys. Acta 2010, 1800, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B. Soybean ferritin: Implications for iron status of vegetarians. Am. J. Clin. Nutr. 2009, 89, 1680S–1685S. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shcolnick, S.; Summerfield, T.; Reytman, L.; Sherman, L.; Keren, N. The Mechanism of Iron Homeostasis in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803 and Its Relationship to Oxidative Stress. Plant Physiol. 2009, 150, 2045–2056. [Google Scholar] [CrossRef]
- Adams, D.G.; Duggan, P.S. Tansley Review No. 107. Heterocyst and akinete differentiation in Cyanobacteria. New Phytol. 1999, 144, 3–33. [Google Scholar] [CrossRef]
- Glaesener, A.G.; Merchant, S.S.; Blaby-Haas, C.E. Iron economy in Chlamydomonas reinhardtii. Front. Plant Sci. 2013, 4, 337. [Google Scholar] [CrossRef]
- Lopez-Millan, A.; Grusak, M.A.; Abadia, A.; Abadia, J. Iron deficiency in plants: An insight from proteomic approaches. Front. Plant Sci. 2013, 4, 254. [Google Scholar] [CrossRef]
- Kobayashi, T. Understanding the Complexity of Iron Sensing and Signaling Cascades in Plants. Plant Cell Physiol. 2019, 60, 1440–1446. [Google Scholar] [CrossRef]
- Rodriguez-Celma, J.; Pan, I.; Li, W.; Lan, P.; Buckhout, T.; Schmidt, W. The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci. 2013, 4, 246. [Google Scholar] [CrossRef]
- Larbi, A.; Abadía, A.; Abadía, J.; Morales, F. Down co-regulation of light absorption, photochemistry, and carboxylation in Fe-deficient plants growing in different environments. Photosynth. Res. 2006, 89, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Hantzis, L.J.; Kroh, G.E.; Jahn, C.E.; Cantrell, M.; Peers, G.; Pilon, M.; Ravet, K. A Program for Iron Economy during Deficiency Targets Specific Fe Proteins. Plant Physiol. 2018, 176, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Gomez, S.M.; Whitelegge, J.P.; Nishio, J.N. Hydroponics on a chip: Analysis of the Fe deficient Arabidopsis thylakoid membrane proteome. J. Proteom. 2009, 72, 397–415. [Google Scholar] [CrossRef] [PubMed]
- Godman, J.; Balk, J. Genome analysis of Chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics 2008, 179, 59–68. [Google Scholar] [CrossRef]
- Keren, N.; Aurora, R.; Pakrasi, H.B. critical roles of bcterioferritins in iron storage and proliferation of Cyanobacteria. Plant Physiol. 2004, 135, 1666–1673. [Google Scholar] [CrossRef]
- Shcolnick, S.; Shaked, Y.; Keren, N. A Role for mrgA, a DPS Family Protein, in the Internal Transport of Fe in the Cyanobacterium Synechocystis Sp. PCC6803. Biochim. Biophys. Acta 2007, 1767, 814–819. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dedaldechamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef]
- Chen, J.-C.; Hsieh, S.I.; Kropat, J.; Merchant, S.S. A Ferroxidase Encoded by FOX1 Contributes to Iron Assimilation under Conditions of Poor Iron Nutrition in Chlamydomonas. Eukaryot. Cell 2008, 7, 541–545. [Google Scholar] [CrossRef]
- Jeong, J.; Merkovich, A.; Clyne, M.; Connolly, E.L. Directing iron transport in dicots: Regulation of iron acquisition and translocation. Curr. Opin. Plant Biol. 2017, 39, 106–113. [Google Scholar] [CrossRef]
- Thimm, O.; Essigmann, B.; Altmann, T.; Buckhout, T.J. Response of Arabidopsis to Iron Deficiency Stress as Revealed by Microarray Analysis. Plant Physiol. 2001, 127, 1030–1043. [Google Scholar] [CrossRef]
- Borlotti, A.; Vigani, G.; Zocchi, G. Iron deficiency affects nitrogen metabolism in cucumber (Cucumis sativus L.) plants. BMC Plant Biol. 2012, 12, 189. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Urzica, E.I.; Adler, L.N.; Page, M.D.; Linster, C.L.; Arbing, M.A.; Casero, D.; Pellegrini, M.; Merchant, S.S.; Clarke, S.G. Impact of oxidative stress on ascorbate biosynthesis in Chlamydomonas via regulation of the VTC2 gene encoding a GDP-L-galactose phosphorylase. J. Biol. Chem. 2012, 287, 14234–14245. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, A.M.; Peers, G.; Kobayashi, M.; Niyogi, K.K.; Merchant, S.S. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 2010, 105, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Adams, S.; Chuam, N.H.; Moller, S.G. AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J. Biol. Chem. 2005, 280, 6648–6654. [Google Scholar] [CrossRef]
- Georg, J.; Kostova, G.; Vuorijoki, L.; Schön, V.; Kadowaki, T.; Huokko, T.; Baumgartner, D.; Müller, M.; Klähn, S.; Allahverdiyeva, Y.; et al. Acclimation of Oxygenic Photosynthesis to Iron Starvation Is Controlled by the sRNA IsaR1. Curr. Biol. 2017, 27, 1425–1436. [Google Scholar] [CrossRef]
- Andaluz, S.; Lopez-Millan, A.-F.; De las Rivas, J.; Abadia, J.; Abadia, A. Proteomic profiles of thylakoid membranes and changes in response to iron deficiency. Photosynth. Res. 2006, 89, 141–155. [Google Scholar] [CrossRef]
- Timperio, A.M.; D’Amici, G.M.; Barta, C.; Loreto, F.; Zolla, L. Proteomics, pigment composition, and organization of thylakoid membranes in iron-deficient spinach leaves. J. Exp. Bot. 2007, 58, 3695–3710. [Google Scholar] [CrossRef]
- Ivanov, A.; Krol, M.; Sveshnikov, D.; Selstam, E. Iron Deficiency in Cyanobacteria Causes Monomerization of Photosystem I Trimers and Reduces the Capacity for State Transitions and the Effective Absorption Cross Section of Photosystem I in Vivo1. Plant Physiol. 2006, 141, 1436–1445. [Google Scholar] [CrossRef]
- Merchant, S.S.; Allen, M.D.; Kropat, J.; Moseley, J.L.; Long, J.C.; Tottey, S.; Terauchi, A.M. Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 2006, 1763, 578–594. [Google Scholar] [CrossRef]
- Liang, X.J.; Qin, L.; Liu, P.W.; Wang, M.H.; Ye, H. Genes for iron-sulphur cluster assembly are targets of abiotic stress in rice, Oryza sativa. Plant Cell Environ. 2014, 37, 780–794. [Google Scholar] [CrossRef]
- Pan, I.C.; Tsai, H.-H.; Cheng, Y.-T.; Wen, T.-N.; Buckhout, T.J.; Schmidt, W. Post-Transcriptional Coordination of the Arabidopsis Iron Deficiency Response is Partially Dependent on the E3 Ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2). Mol. Cell. Proteom. MCP 2015, 14, 2733–2752. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Shen, G.; Bryant, D.A.; Golbeck, J.H. Regulatory Roles for IscA and SufA in Iron Homeostasis and Redox Stress Responses in the Cyanobacterium synechococcus sp. Strain PCC 7002. J. Bacteriol. 2006, 188, 3182–3191. [Google Scholar] [CrossRef] [PubMed]
- Buckhout, T.J.; Yang, T.J.W.; Schmidt, W. Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genom. 2009, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; McInturf, S.A.; Stein, R.J. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5903–5918. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Sommer, F.; Allen, M.; Lu, S.-F.; Merchant, S. FER1 and FER2 Encoding Two Ferritin Complexes in Chlamydomonas reinhardtii Chloroplasts Are Regulated by Iron. Genetics 2008, 179, 137–147. [Google Scholar] [CrossRef]
- Busch, A.; Rimbauld, B.; Naumann, B.; Rensch, S.; Hippler, M. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 2008, 55, 201–211. [Google Scholar] [CrossRef]
- Urzica, E.I.; Casero, D.; Yamasaki, H.; Hsieh, S.I.; Adler, L.N.; Karpowicz, S.J.; Blaby-Haas, C.E.; Clarke, S.G.; Loo, J.A.; Pellegrini, M.; et al. Systems and Trans-System Level Analysis Identifies Conserved Iron Deficiency Responses in the Plant Lineage. Plant Cell 2012, 24, 3921–3948. [Google Scholar] [CrossRef]
- Zheng, L.; Huang, F.; Narsai, R.; Wu, J.; Giraud, E.; He, F.; Cheng, L.; Wang, F.; Wu, P.; Whelan, J.; et al. Physiological and Transcriptome Analysis of Iron and Phosphorus Interaction in Rice Seedlings. Plant Physiol. 2009, 151, 262–274. [Google Scholar] [CrossRef]
- Stein, R.J.; Waters, B.M. Use of natural variation reveals core genes in the transcriptome of iron-deficient roots. J. Exp. Bot. 2012, 63, 1039–1055. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein–Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Page, M.; Allen, M.; Kropat, J.; Urzica, E.; Karpowicz, S.; Hsieh, S.; Loo, J.; Merchant, S. Fe Sparing and Fe Recycling Contribute to Increased Superoxide Dismutase Capacity in Iron-Starved Chlamydomonas reinhardtii. Plant Cell 2012, 24, 2649–2665. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lodeyro, A.F.; Ceccoli, R.D.; Pierella Karlusich, J.J.; Carrillo, N. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett. 2012, 586, 2917–2924. [Google Scholar] [CrossRef]
- Pierella Karlusich, J.J.; Lodeyro, A.F.; Carrillo, N. The long goodbye: The rise and fall of flavodoxin during plant evolution. J. Exp. Bot. 2014, 65, 5161–5178. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Zurbriggen, M.D.; Morandi, E.N.; Fillat, M.F.; Valle, E.M.; Hajirezaei, M.-R.; Carrillo, N. Enhanced plant tolerance to iron starvation by functional substitution of chloroplast ferredoxin with a bacterial flavodoxin. Proc. Natl. Acad. Sci. USA 2007, 104, 11495–11500. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, X.; Zhu, X.; Li, T.; Zhan, J.; Chen, H.; He, C.; Wang, Q. Dynamic Changes of IsiA-Containing Complexes during Long-Term Iron Deficiency in Synechocystis sp. PCC 6803. Mol. Plant 2017, 10, 143–154. [Google Scholar] [CrossRef]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef]
- Gao, F.; Robe, K.; Gaymard, F.; Izquierdo, E.; Dubos, C. The Transcriptional Control of Iron Homeostasis in Plants: A Tale of bHLH Transcription Factors? Front. Plant Sci. 2019, 10, 6. [Google Scholar] [CrossRef]
- Moran Lauter, A.; Peiffer, G.; Yin, T.; Whitham, S.; Cook, D.; Shoemaker, R.; Graham, M. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max ) roots and leaves. BMC Genom. 2014, 15, 702. [Google Scholar] [CrossRef]
- Kastoori Ramamurthy, R.; Xiang, Q.; Hsieh, E.-J.; Liu, K.; Zhang, C.; Waters, B.M. New aspects of ironcopper crosstalk uncovered by transcriptomic characterization of Col-0 and the copper uptake mutant spl7 in Arabidopsis thaliana. Metallomics 2018, 10, 1824–1840. [Google Scholar] [CrossRef] [PubMed]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH Transcription Factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 Reveals Molecular Linkage between the Regulation of Iron Acquisition and Ethylene Signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Balparda, M.; Armas, A.M.; Estavillo, G.M.; Roschzttardtz, H.; Pagani, M.A.; Gomez-Casati, D.F. The PAP/SAL1 Retrograde Signaling Pathway Is Involved in Iron Homeostasis. Plant Mol. Biol. 2020, 102, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Fillat, M.F.; Bes, M.; Peleato, M.; Sevilla, E. The challenge of iron stress in Cyanobacteria. In Cyanobacteria; Tiwari, A., Ed.; IntechOpen: London, UK, 2018; pp. 109–138. [Google Scholar]
- Deng, X.; Eriksson, M. Two Iron-Responsive Promoter Elements Control Expression of FOX1 in Chlamydomonas reinhardtii. Eukaryot. Cell 2007, 6, 2163–2167. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Eriksson, M.; Yang, J.; Deng, X. An Fe Deficiency Responsive Element with a Core Sequence of TGGCA Regulates the Expression of FEA1 in Chlamydomonas reinharditii. J. Biochem. 2009, 146, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Eriksson, M.; Li, Y.; Deng, X. A Novel Negative Fe-Deficiency-Responsive Element and a TGGCA-Type-Like FeRE Control the Expression of FTR1 in Chlamydomonas reinhardtii. J. Biomed. Biotechnol. 2010, 2010, 790247. [Google Scholar] [CrossRef]
- Fei, X.; Yu, J.; Li, Y.; Deng, X. CrMAPK3 regulates the expression of iron-deficiency-responsive genes in Chlamydomonas reinhardtii. BMC Biochem. 2017, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sivitz, A.B.; Hermand, V.; Curie, C.; Vert, G. Arabidopsis bHLH100 and bHLH101 Control Iron Homeostasis via a FIT-Independent Pathway. PLoS ONE 2012, 7, e44843. [Google Scholar] [CrossRef]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH Transcription Factor POPEYE Regulates Response to Iron Deficiency in Arabidopsis Roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef]
- Khan, M.A.; Castro-Guerrero, N.A.; McInturf, S.A.; Nguyen, N.T.; Dame, A.N.; Wang, J.J.; Bindbeutel, R.K.; Joshi, T.; Jurisson, S.S.; Nusinow, D.A.; et al. Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ. 2018, 41, 2263–2276. [Google Scholar] [CrossRef]
- Kim, S.A.; Lacroix, I.S.; Gerber, S.A.; Guerinot, M.L. The iron deficiency response in requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. USA 2019, 116, 24933–24942. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, J.; Jin, H.; Feng, D.; Wang, J.; Wang, H.-B.; Liu, B. The Iron Deficiency Response Regulators IAA-LEUCINE RESISTANT3 and bHLH104 Possess Different Targets and Have Distinct Effects on Photosynthesis in Arabidopsis. J. Plant Biol. 2019, 62, 109–119. [Google Scholar] [CrossRef]
- Tissot, N.; Robe, K.; Gao, F.; Grant-Grant, S.; Boucherez, J.; Bellegarde, F.; Maghiaoui, A.; Marcelin, R.; Izquierdo, E.; Benhamed, M.; et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 2019, 223, 1052–1055. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, M.; Feng, D.; Jin, H.; Wang, P.; Liu, J.; Xiong, F.; Wang, J.; Wang, H.B. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 2015, 27, 787–805. [Google Scholar] [CrossRef] [PubMed]
- Estavillo, G.; Crisp, P.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef]
- Lefebvre-Legendre, L.; Choquet, Y.; Kuras, R.; Loubery, S.; Douchi, D.; Goldschmidt-Clermont, M. A nucleus-encoded helical-repeat protein which is regulated by iron availability controls chloroplast psaA mRNA expression in Chlamydomonas. Plant Physiol. 2015, 114, 253906. [Google Scholar]
- Naumann, B.; Stauber, E.J.; Busch, A.; Sommer, F.; Hippler, M. N-terminal processing of Lhca3 Is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 2005, 280, 20431–20441. [Google Scholar] [CrossRef]
- Selote, D.; Samira, R.; Matthiadis, A.; Gillikin, J.W.; Long, T.A. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 2015, 167, 273–286. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Connorton, J.M.; Kruse, I.; Green, R.T.; Franceschetti, M.; Chen, Y.-T.; Cui, Y.; Ling, H.-Q.; Yeh, K.-C.; Balk, J. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. USA 2019, 116, 17584–17591. [Google Scholar] [CrossRef]
- Hindt, M.N.; Akmakjian, G.Z.; Pivarski, K.L.; Punshon, T.; Baxter, I.; Salt, D.E.; Guerinot, M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 2017, 9, 876–890. [Google Scholar] [CrossRef]
- Yamaguchi-Iwai, Y.; Stearman, R.; Dancis, A.; Klausner, R.D. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996, 15, 3377–3384. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, J.C.; Jaron, S.; Ray, E.; Brown, P.O.; Winge, D.R. A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. USA 2001, 98, 14322–14327. [Google Scholar] [CrossRef] [PubMed]
- Blaiseau, P.L.; Lesuisse, E.; Camadro, J.M. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 2001, 276, 34221–34226. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Mapolelo, D.T.; Dingra, N.N.; Naik, S.G.; Lees, N.S.; Hoffman, B.M.; Riggs-Gelasco, P.J.; Huynh, B.H.; Johnson, M.K.; Outten, C.E. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 2009, 48, 9569–9581. [Google Scholar] [CrossRef]
- Kumanovics, A.; Chen, O.S.; Li, L.T.; Bagley, D.; Adkins, E.M.; Lin, H.L.; Dingra, N.N.; Outten, C.E.; Keller, G.; Winge, D.; et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 2008, 283, 10276–10286. [Google Scholar] [CrossRef]
- Poor, C.B.; Wegner, S.V.; Li, H.; Dlouhy, A.C.; Schuermann, J.P.; Sanishvili, R.; Hinshaw, J.R.; Riggs-Gelasco, P.J.; Outten, C.E.; He, C. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc. Natl. Acad. Sci. USA 2014, 111, 4043–4048. [Google Scholar] [CrossRef]
- Schorsch, M.; Kramer, M.; Goss, T.; Eisenhut, M.; Robinson, N.; Osman, D.; Wilde, A.; Sadaf, S.; Brückler, H.; Walder, L.; et al. A unique ferredoxin acts as a player in the low-iron response of photosynthetic organisms. Proc. Natl. Acad. Sci. USA 2018, 115, E12111. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef]
- Larkin, R.M. Tetrapyrrole Signaling in Plants. Front. Plant Sci. 2016, 7, 1586. [Google Scholar] [CrossRef]
- Susek, R.E.; Ausubel, F.M.; Chory, J. Signal-transduction mutants of arabidopsis uncouple nuclear cab and rbcs gene-expression from chloroplast development. Cell 1993, 74, 787–799. [Google Scholar] [CrossRef]
- Zhang, Z.-W.; Yuan, S.; Feng, H.; Xu, F.; Cheng, J.; Shang, J.; Zhang, D.-W.; Lin, H.-H. Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs—New evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 2011, 168, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Vigani, G.; Zocchi, G.; Bashir, K.; Philippar, K.; Briat, J.F. Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci. 2013, 18, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Balasubramanian, R.; Wang, T.; Wu, Y.; Hoffart, L.M.; Krebs, C.; Bryant, D.A.; Golbeck, J.H. SufR coordinates two 4Fe-4S (2+,1+) clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J. Biol. Chem. 2007, 282, 31909–31919. [Google Scholar] [CrossRef] [PubMed]
- Vuorijoki, L.; Tiwari, A.; Kallio, P.; Aro, E.M. Inactivation of iron-sulfur cluster biogenesis regulator SufR in Synechocystis sp PCC 6803 induces unique iron-dependent protein-level responses. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Lin, W.-D.; Schmidt, W. Transcriptional Profiling of the Arabidopsis Iron Deficiency Response Reveals Conserved Transition Metal Homeostasis Networks. Plant Physiol. 2010, 152, 2130–2141. [Google Scholar] [CrossRef]
- Grusak, M.A.; Pezeshgi, S. Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 1996, 110, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.A.; Briat, J.F.; Curie, C. Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 2003, 132, 796–804. [Google Scholar] [CrossRef]
- Zhai, Z.Y.; Gayomba, S.R.; Jung, H.I.; Vimalakumari, N.K.; Pineros, M.; Craft, E.; Rutzke, M.A.; Danku, J.; Lahner, B.; Punshon, T.; et al. OPT3 Is a Phloem-Specific Iron Transporter That Is Essential for Systemic Iron Signaling and Redistribution of Iron and Cadmium in Arabidopsis. Plant Cell 2014, 26, 2249–2264. [Google Scholar] [CrossRef]
- Kumar, R.K.; Chu, H.H.; Abundis, C.; Vasques, K.; Rodriguez, D.C.; Chia, J.C.; Huang, R.; Vatamaniuk, O.K.; Walker, E.L. Iron-Nicotianamine Transporters Are Required for Proper Long Distance Iron Signaling. Plant Physiol. 2017, 175, 1254–1268. [Google Scholar] [CrossRef]
- Grillet, L.; Lan, P.; Li, W.; Mokkapati, G.; Schmidt, W. IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 2018, 4, 953–963. [Google Scholar] [CrossRef]
- Hirayama, T.; Lei, G.J.; Yamaji, N.; Nakagawa, N.; Ma, J.F. The Putative Peptide Gene FEP1 Regulates Iron Deficiency Response in Arabidopsis. Plant Cell Physiol. 2018, 59, 1739–1752. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Reyt, G.; Arnaud, N.; Krouk, G.; Djouani, E.B.; Boucherez, J.; Briat, J.F.; Gaymard, F. Iron and ROS control of the DownSTream mRNA decay pathway is essential for plant fitness. EMBO J. 2012, 31, 175–186. [Google Scholar] [CrossRef] [PubMed]

| Function | Arabidopsis Gene Name | A. thaliana | C. reinhardtii | Synechocyst Is PCC 6803 |
|---|---|---|---|---|
| Transport | FRO7 | AT5G49740 | Cre04.g227400.t1.2 | |
| PIC1 | AT2G15290 | Cre10.g454734.t2.1 | sll1656 [103] | |
| ATP-Binding Cassette I11 (ABCI11)/NAP14 | AT5G14100 | PNW71978/Cre16.g687550.t1.1 [107] | slr0354 [107] | |
| ABCI10 | AT4G33460 | XP_001703542/Cre03.g164150.t1.1 [107] | sll1623 [107] | |
| ABCI12 | AT3G21580 | PNW75614/Cre12.g533950.t1.1 [107] | slr1978 [107] | |
| Multiple Antibiotic Resistance 1 (MAR1) | AT5G26820 | Cre03.g175200.t1.2 | lap75 | |
| Yellow Stripe Like 4/6 (YSL4/6) | (4) AT5G41000, (6) AT3G27020 | |||
| ROS homeostasis | Fe SuperOxide Dismutase (FSD) | AT4G25100 AT5G51100 AT5G23310 | Cre10.g436050.t1.2 | WP_010872652 |
| Stromal Ascorbate Peroxidase (sAPX) | AT4G08390 | Cre02.g087700.t1.2 | ||
| Conserved in Green Linage and Diatoms 27 (CGLD27) | AT5G67370 | Cre05.g237050.t1.1 | WP_010873853 | |
| Suf Fe–S assembly | SUFB | AT4G04770 | Cre15.g643600.t1.2 [128] | slr0074 [128] |
| SUFC | AT3G10670 | Cre07.g339700.t1.2 [128] | slr0075 [128] | |
| SUFD | AT5G44316 | Cre12.g513950.t1.2 * [128] | slr0076 [128] | |
| SUFS | AT1G08490 | Cre12.g525650.t1.2 [128] | slr0077 [128] | |
| SUFE | AT4G26500 | Cre06.g309717.t1.1 [128] | slr1419 [128] | |
| SUFA | AT1G10500 | Cre07.g349600.t1.2 [128] | slr1417 [128] | |
| Nitrogen Fixation U-Like (NFU) | (1) AT4G01940 (2) AT5G49940 (3) AT4G25910 | (1) Cre18.g748447.t1.1 (2) Cre12.g504150.t2.1 (3) Cre17.g710800.t2.1 [128] | ssl2667 [128] | |
| High Chlorophyll Fluorescence 101 (HCF101) | At3g24430 | Cre01.g045902.t1.1 [128] | slr0067 [128] | |
| Monothiol Glutaredoxins (GRXS) | (14) AT3G54900 (16) AT2G38270 | (14) Cre07.g325743.t1.1 (16) Cre01.g047800.t1.1 [128] | WP_010871706 | |
| NEET | AT5G51720 | Cre01.g050550.t1.2 | ||
| Heme biosynthesis | Ferrochelatase (FC) | AT5G26030 AT2G30390 | Cre07.g339750.t2.1 | WP_010873751 |
| Sirohydrochlorin Ferrochelatase B (SirB) | Cre04.g214100.t1.2 | WP_010874018 | ||
| Sequestration | Ferritin (FER) | (1) AT5G01600 (3) AT3G56090 (4) AT2G40300 | Cre09.g387800.t1.2, Cre13.g574500.t1.1 | sll1341 ** [129] slr1890 ** slr1894 ** [130] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroh, G.E.; Pilon, M. Regulation of Iron Homeostasis and Use in Chloroplasts. Int. J. Mol. Sci. 2020, 21, 3395. https://doi.org/10.3390/ijms21093395
Kroh GE, Pilon M. Regulation of Iron Homeostasis and Use in Chloroplasts. International Journal of Molecular Sciences. 2020; 21(9):3395. https://doi.org/10.3390/ijms21093395
Chicago/Turabian StyleKroh, Gretchen E., and Marinus Pilon. 2020. "Regulation of Iron Homeostasis and Use in Chloroplasts" International Journal of Molecular Sciences 21, no. 9: 3395. https://doi.org/10.3390/ijms21093395
APA StyleKroh, G. E., & Pilon, M. (2020). Regulation of Iron Homeostasis and Use in Chloroplasts. International Journal of Molecular Sciences, 21(9), 3395. https://doi.org/10.3390/ijms21093395




