Sulfur Deficiency Increases Phosphate Accumulation, Uptake, and Transport in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
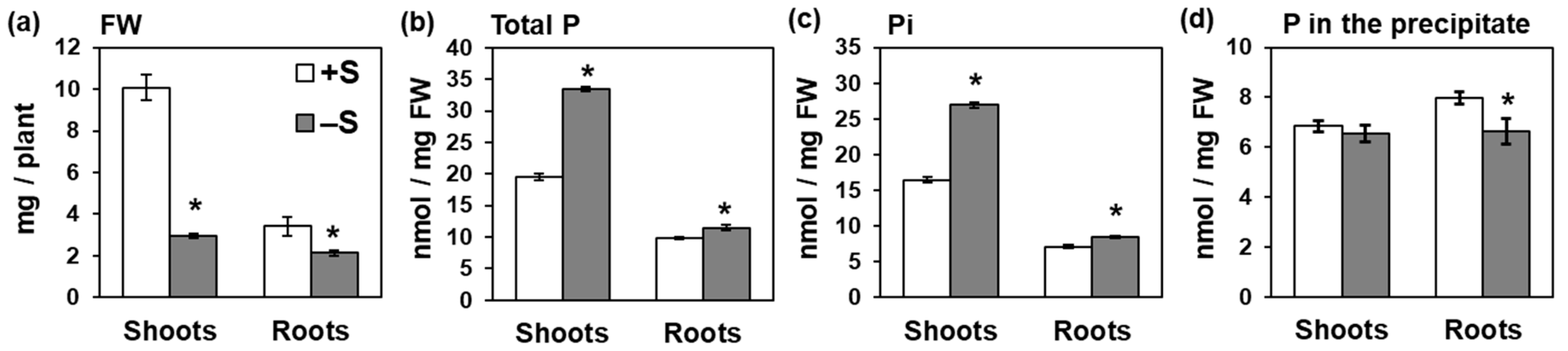
2.1. Sulfur Deficiency Increased Pi Accumulation in Shoots
2.2. Enhancement of Pi Uptake under –S
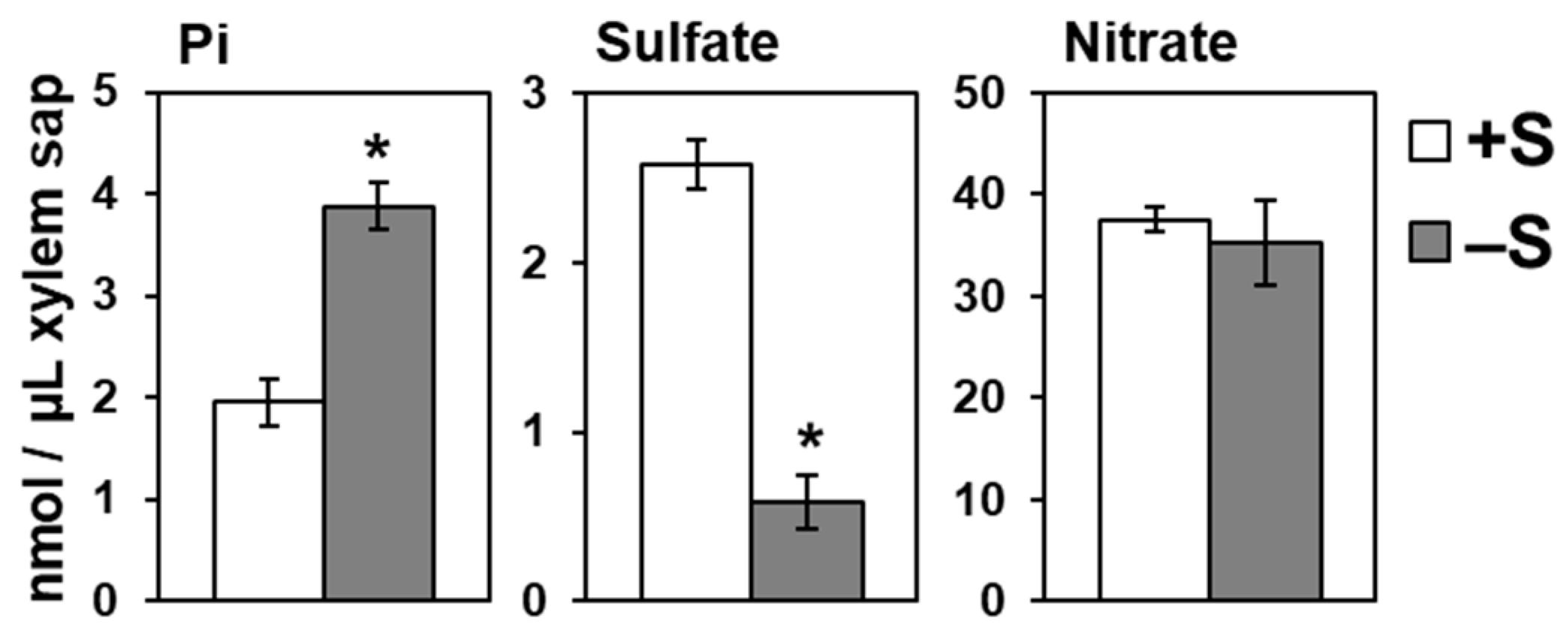
2.3. Increased Root-to-Shoot Pi Transport under –S
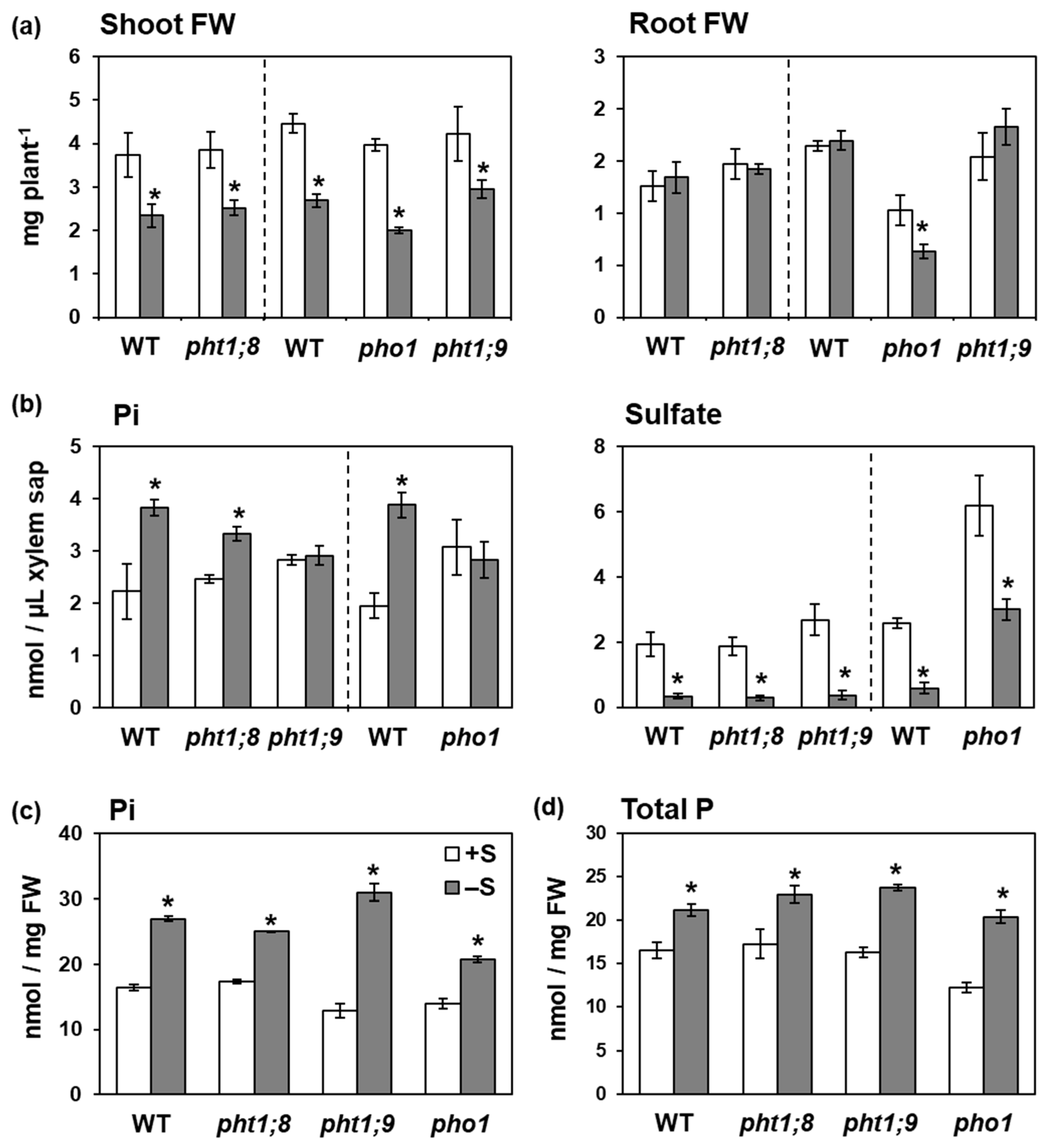
2.4. PHT1;9 and PHO1 Were Involved in the –S-Increased Pi Accumulation in Xylem Sap
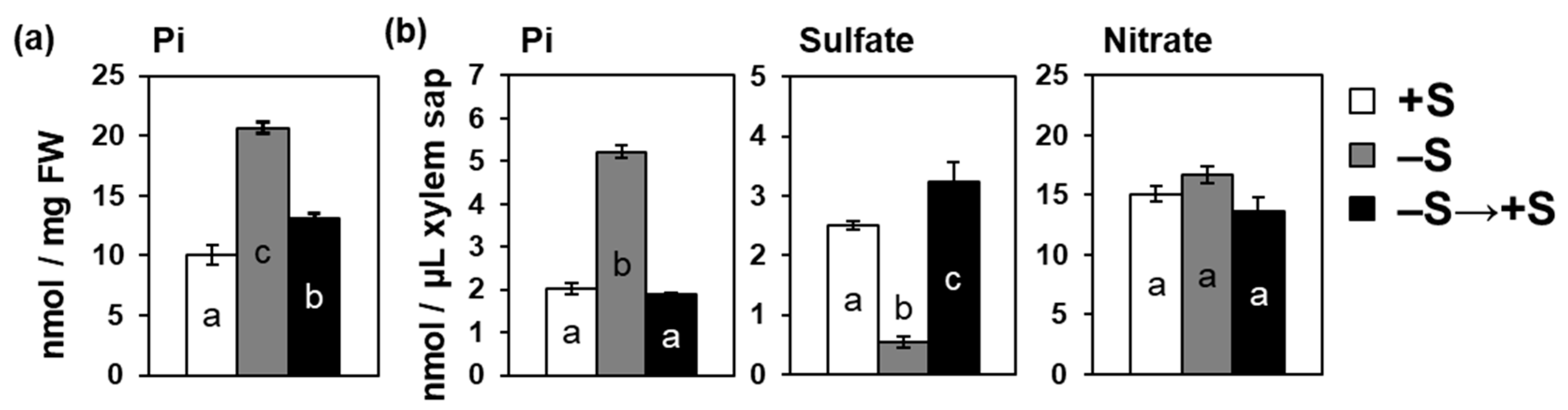
2.5. Increase in Pi Accumulation by –S Was Reversed by Sulfate Addition
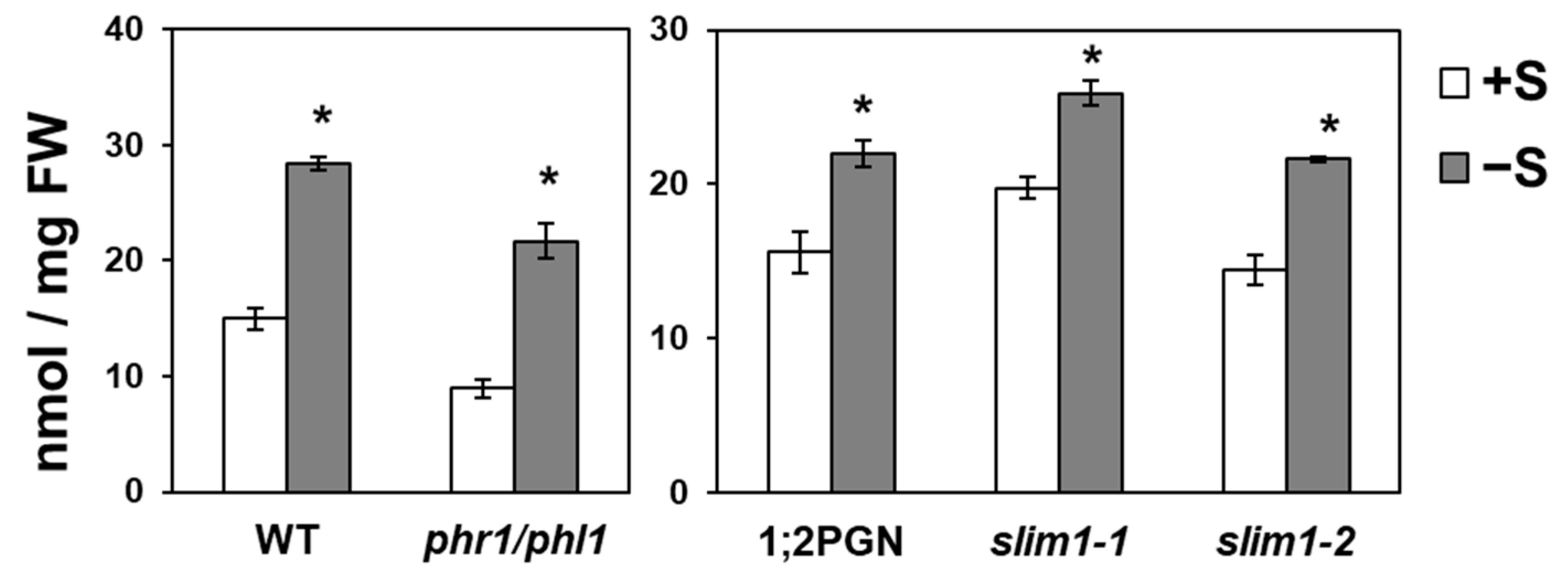
2.6. Increased Pi Accumulation under −S Was Observed in SLIM1 Mutants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of Total P, Pi, Sulfate, and Nitrate in Plant Tissues
4.3. Analysis of Transcript Level
4.4. Pi Uptake Analysis
4.5. Xylem Sap Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| P | Phosphorus |
| S | Sulfur |
| Pi | Phosphate |
| +S | Sulfur sufficiency |
| –S | Sulfur deficiency |
| –P | Phosphorus deficiency |
| PHT | Phosphate transporter |
| T-DNA | transfer DNA |
| RNA | Ribonucleic acid |
| miR | Micro ribonucleic acid |
| qPCR | Quantitative polymerase chain reaction |
| IC | Ion chromatography |
| WT | Wild-type |
| n.d. | not determined |
| SE | Standard error |
| ANOVA | Analysis of variance |
| GM | Germination media |
| µL | Microliter |
| µM | Micro molar |
| mL | Milliliter |
| mM | Millimolar |
| °C | Degrees of Celsius |
References
- Long, S.; Kahn, M.; Seefeldt, L.; Tsay, Y.; Kopriva, S. Nitrogen and sulfur. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; WILEY: Blackwell, NJ, USA, 2015; pp. 746–768. [Google Scholar]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef]
- Ticconi, C.A.; Abel, S. Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci. 2004, 9, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Bucher, M. Phosphate transport and homeostasis in Arabidopsis. Arab. Book 2002, 1, e0024. [Google Scholar] [CrossRef] [PubMed]
- Saito, K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 2004, 136, 2443–2450. [Google Scholar] [CrossRef]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Sherameti, I.; Oelmüller, R. Siroheme. Plant Signal. Behav. 2010, 5, 14–20. [Google Scholar] [CrossRef]
- Leustek, T. Sulfate metabolism. Arab. Book 2002, 1, e0017. [Google Scholar] [CrossRef]
- Nikiforova, V.J.; Kopka, J.; Tolstikov, V.; Fiehn, O.; Hopkins, L.; Hawkesford, M.J.; Hesse, H.; Hoefgen, R. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 2005, 138, 304–318. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Ohkama-Ohtsu, N. Chapter 13 Sulfur assimilation and glutathione metabolism in plants. In Glutathione in Plant Growth, Development, and Stress Tolerance; Springer: New York, NY, USA, 2017; pp. 287–308. [Google Scholar]
- Stigter, K.; Plaxton, W.; Stigter, K.A.; Plaxton, W.C. Molecular Mechanisms of Phosphorus Metabolism and Transport during Leaf Senescence. Plants 2015, 4, 773–798. [Google Scholar] [CrossRef]
- Misson, J.; Raghothama, K.G.; Jain, A.; Jouhet, J.; Block, M.A.; Bligny, R.; Ortet, P.; Creff, A.; Somerville, S.; Rolland, N.; et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 2005, 102, 11934–11939. [Google Scholar] [CrossRef]
- Rouached, H.; Secco, D.; Arpat, B.; Poirier, Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Otsuki, H.; Narisawa, T.; Kobayashi, M.; Sawai, S.; Kamide, Y.; Kusano, M.; Aoki, T.; Hirai, M.Y.; Saito, K. A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 2013, 4, 1510. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Shimojima, M.; Sawada, Y.; Toyooka, K.; Narisawa, T.; Mochida, K.; Tanaka, H.; Matsuda, F.; Hirai, A.; Hirai, M.Y.; et al. A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 2009, 21, 892–909. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Ai, Q.; Yu, D. Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Sci. Rep. 2015, 5, 11813. [Google Scholar] [CrossRef]
- Hsieh, L.-C.; Lin, S.-I.; Shih, A.C.-C.; Chen, J.-W.; Lin, W.-Y.; Tseng, C.-Y.; Li, W.-H.; Chiou, T.-J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef]
- Hirsch, J.; Marin, E.; Floriani, M.; Chiarenza, S.; Richaud, P.; Nussaume, L.; Thibaud, M.C. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 2006, 88, 1767–1771. [Google Scholar] [CrossRef]
- Nocito, F.F.; Pirovano, L.; Cocucci, M.; Sacchi, G.A. Cadmium-induced sulfate uptake in maize roots. Plant Physiol. 2002, 129, 1872–1879. [Google Scholar] [CrossRef]
- Kandlbinder, A.; Finkemeier, I.; Wormuth, D.; Hanitzsch, M.; Dietz, K.-J. The antioxidant status of photosynthesizing leaves under nutrient deficiency: Redox regulation, gene expression and antioxidant activity in Arabidopsis thaliana. Physiol. Plant. 2004, 120, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008, 177, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Nussaume, L.; Kanno, S.; Javot, H.; Marin, E.; Pochon, N.; Ayadi, A.; Nakanishi, T.M.; Thibaud, M.C. Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2011, 2, 83. [Google Scholar] [CrossRef]
- Kisko, M.; Shukla, V.; Kaur, M.; Bouain, N.; Chaiwong, N.; Lacombe, B.; Pandey, A.; Rouached, H.; Kisko, M.; Shukla, V.; et al. Phosphorus transport in Arabidopsis and Wheat: Emerging strategies to improve P pool in seeds. Agriculture 2018, 8, 27. [Google Scholar] [CrossRef]
- Misson, J.; Thibaud, M.-C.; Bechtold, N.; Raghothama, K.; Nussaume, L. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 2004, 55, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Shin, H.-S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef]
- Lapis-Gaza, H.R.; Jost, R.; Finnegan, P.M. Arabidopsis Phosphate Transportar1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 2014, 14, 334. [Google Scholar] [CrossRef]
- Poirier, Y.; Thoma, S.; Somerville, C.; Schiefelbein, J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991, 97, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, D.; Rezzonico, E.; MacDonald-Comber Petétot, J.; Somerville, C.; Poirier, Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ribot, C.; Rezzonico, E.; Poirier, Y. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 2004, 135, 400–411. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Kimura, Y.; Ushiwatari, T.; Suyama, A.; Tominaga-Wada, R.; Wada, T.; Maruyama-Nakashita, A. Contribution of root hair development to sulfate uptake in Arabidopsis. Plants 2019, 8, 106. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A. Metabolic changes sustain the plant life in low-sulfur environments. Curr. Opin. Plant Biol. 2017, 39, 144–151. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Inoue, E.; Watanabe-Takahashi, A.; Yamaya, T.; Takahashi, H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003, 132, 597–605. [Google Scholar] [CrossRef]
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004, 136, 4198–4204. [Google Scholar]
- Hirai, M.Y.; Klein, M.; Fujikawa, Y.; Yano, M.; Goodenowe, D.B.; Yamazaki, Y.; Kanaya, S.; Nakamura, Y.; Kitayama, M.; Suzuki, H.; et al. Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 2005, 280, 25590–25595. [Google Scholar] [CrossRef]
- Aarabi, F.; Kusajima, M.; Tohge, T.; Konishi, T.; Gigolashvili, T.; Takamune, M.; Sasazaki, Y.; Watanabe, M.; Nakashita, H.; Fernie, A.R.; et al. Sulfur deficiency–induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci. Adv. 2016, 2, e1601087. [Google Scholar] [CrossRef]
- Zhang, L.; Kawaguchi, R.; Morikawa-Ichinose, T.; Allahham, A.; Kim, S.-J.; Maruyama-Nakashita, A. Sulfur deficiency-induced glucosinolate catabolism attributed to two β-Glucosidases, BGLU28 and BGLU30, is required for plant growth maintenance under sulfur deficiency. Plant Cell Physiol. 2020, 61, 803–813. [Google Scholar] [CrossRef]
- Maruyama-Nakashita, A.; Nakamura, Y.; Tohge, T.; Saito, K.; Takahashi, H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 2006, 18, 3235–3251. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kanno, S.; Mabuchi, A.; Monda, K.; Iba, K.; Yanagisawa, S. A phytochrome-B-mediated regulatory mechanism of phosphorus acquisition. Nat. Plants 2018, 4, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.; Cuyas, L.; Javot, H.; Bligny, R.; Gout, E.; Dartevelle, T.; Hanchi, M.; Nakanishi, T.M.; Thibaud, M.-C.; Nussaume, L. Performance and limitations of phosphate wuantification: Guidelines for plant biologists. Plant Cell Physiol. 2016, 57, 690–706. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed]
- Maruyama-Nakashita, A.; Nakamura, Y.; Yamaya, T.; Takahashi, H. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: Implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 2004, 38, 779–789. [Google Scholar] [CrossRef]
- Lin, W.Y.; Huang, T.K.; Chiou, T.J. Nitrogen Limitation Adaptation, a target of MicroRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Wang, F.; Yang, J.; Gao, M.; Li, C.; Liu, Y.; Liu, Y.; Yamaji, N.; Ma, J.F.; et al. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 2015, 27, 711–723. [Google Scholar] [CrossRef]
- Yue, W.; Ying, Y.; Wang, C.; Zhao, Y.; Dong, C.; Whelan, J.; Shou, H. OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 2017, 90, 1040–1051. [Google Scholar] [CrossRef]
- Bari, R.P.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, microRNA399 and PHR1 define a phosphate signalling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-Y.; Huang, T.-K.; Tseng, C.-Y.; Lai, Y.-S.; Lin, S.-I.; Lin, W.-Y.; Chen, J.-W.; Chiou, T.-J. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 2012, 24, 2168–2183. [Google Scholar] [CrossRef]
- Huang, T.-K.; Han, C.-L.; Lin, S.-I.; Chen, Y.-J.; Tsai, Y.-C.; Chen, Y.-R.; Chen, J.-W.; Lin, W.-Y.; Chen, P.-M.; Liu, T.-Y.; et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Gu, M.; Xia, Y.W.; Dai, X.L.; Chang, R.D.; Zhang, J.; Wang, S.C.; Qu, H.Y.; Yamaji, N.; Ma, J.F.; et al. OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol. 2019, 179, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.P.; Kumar, H.; Waight, A.B.; Risenmay, A.J.; Roe-Zurz, Z.; Chau, B.H.; Schlessinger, A.; Bonomi, M.; Harries, W.; Sali, A.; et al. Crystal structure of a eukaryotic phosphate transporter. Nature 2013, 496, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Liu, H.; Harrison, M.J. The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 2001, 25, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, E.B.; DiTusa, S.F.; Kato, N.; Olivier, D.M.; Dale, R.; Lin, W.Y.; Chiou, T.J.; Macnaughtan, M.A.; Smith, A.P. Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site-directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant Cell Environ. 2015, 38, 2012–2022. [Google Scholar] [CrossRef]
- Gilroy, S.; Jones, D.L. Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 2000, 5, 56–60. [Google Scholar] [CrossRef]
- Ma, Z.; Bielenberg, D.G.; Brown, K.M.; Lynch, J.P. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 2001, 24, 459–467. [Google Scholar] [CrossRef]
- Wege, S.; Khan, G.A.; Jung, J.Y.; Vogiatzaki, E.; Pradervand, S.; Aller, I.; Meyer, A.J.; Poirier, Y. The EXS Domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016, 170, 385–400. [Google Scholar] [CrossRef]
- Vogiatzaki, E.; Baroux, C.; Jung, J.Y.; Poirier, Y. PHO1 exports phosphate from the chalazal seed coat to the embryo in developing Arabidopsis seeds. Curr. Biol. 2017, 27, 2893–2900. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H.; Arpat, A.B.; Poirier, Y. Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 2010, 3, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Młodzińska, E.; Zboińska, M. Phosphate uptake and allocation—A closer look at arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 2016, 7, 1198. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Lessard, P.A.; Beachy, R.N. Seed-specific repression of GUS activity in tobacco plants by antisense RNA. Plant Mol. Biol. 1992, 20, 1059–1069. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Fujiwara, T.; Chino, M.; Naito, S. Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1995, 36, 1331–1339. [Google Scholar] [PubMed]
- Yamaguchi, C.; Takimoto, Y.; Ohkama-Ohtsu, N.; Hokura, A.; Shinano, T.; Nakamura, T.; Suyama, A.; Maruyama-Nakashita, A. Effects of cadmium treatment on the uptake and translocation of sulfate in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 2353–2366. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; De Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodríguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef]
- Stefanovic, A.; Ribot, C.; Rouached, H.; Wang, Y.; Chong, J.; Belbahri, L.; Delessert, S.; Poirier, Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007, 50, 982–994. [Google Scholar] [CrossRef]
- Valvekens, D.; Van Montagu, M.; Van Lijsebettens, M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 1988, 85, 5536–5540. [Google Scholar] [CrossRef]
- Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.-Y.; Leung, H.-Y.; Hattori, K.; Konomi, M.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allahham, A.; Kanno, S.; Zhang, L.; Maruyama-Nakashita, A. Sulfur Deficiency Increases Phosphate Accumulation, Uptake, and Transport in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2971. https://doi.org/10.3390/ijms21082971
Allahham A, Kanno S, Zhang L, Maruyama-Nakashita A. Sulfur Deficiency Increases Phosphate Accumulation, Uptake, and Transport in Arabidopsis thaliana. International Journal of Molecular Sciences. 2020; 21(8):2971. https://doi.org/10.3390/ijms21082971
Chicago/Turabian StyleAllahham, Alaa, Satomi Kanno, Liu Zhang, and Akiko Maruyama-Nakashita. 2020. "Sulfur Deficiency Increases Phosphate Accumulation, Uptake, and Transport in Arabidopsis thaliana" International Journal of Molecular Sciences 21, no. 8: 2971. https://doi.org/10.3390/ijms21082971
APA StyleAllahham, A., Kanno, S., Zhang, L., & Maruyama-Nakashita, A. (2020). Sulfur Deficiency Increases Phosphate Accumulation, Uptake, and Transport in Arabidopsis thaliana. International Journal of Molecular Sciences, 21(8), 2971. https://doi.org/10.3390/ijms21082971




