Abstract
KLF11 (Krüppel-like factor 11) belongs to the family of Sp1/Krüppel-like zinc finger transcription factors that play important roles in a variety of cell types and tissues. KLF11 was initially described as a transforming growth factor-beta (TGF-β) inducible immediate early gene (TIEG). KLF11 promotes the effects of TGF-β on cell growth control by influencing the TGFβ–Smads signaling pathway and regulating the transcription of genes that induce either apoptosis or cell cycle arrest. In carcinogenesis, KLF11 can show diverse effects. Its function as a tumor suppressor gene can be suppressed by phosphorylation of its binding domains via oncogenic pathways. However, KLF 11 can itself also show tumor-promoting effects and seems to have a crucial role in the epithelial–mesenchymal transition process. Here, we review the current knowledge about the function of KLF11 in cell growth regulation. We focus on its transcriptional regulatory function and its influence on the TGF-β signaling pathway. We further discuss its possible role in mediating crosstalk between various signaling pathways in normal cell growth and in carcinogenesis.
1. Introduction
The Krüppel protein regulates the body segmentation in the thorax and anterior abdomen of the Drosophila embryo in Drosophila melanogaster [1]. Krüppel-like factors (KLFs) were first discovered in 1993 and named “Krüppel-like” as mammalian homologs of the Drosophila melanogaster gene Krüppel. The first discovered KLF was named EKLF (also known as KLF1) [2]. Since then, diverse homologous genes have emerged. All of them are now called the family of KLFs [3]. Currently, there are 17 known KLFs in mammals. The functions of the family members are, in some cases, overlapping and, in others, widely distinct [4]. KLFs can be divided into three subgroups based on structural and functional features. Subgroups I and II of KLFs consist of KLF3, KLF5, KLF6, KLF7, KLF8, and KLF12 and of KLF1, KLF2, KLF4, and KLF17, respectively. The remaining KLFs (KLF9, KLF10, KLF11, KLF13, KLF14, KLF15, KLF16) belong to subgroup III [5].
Structurally, KLFs are composed of a conserved C-terminal region and widely divergent N-terminal regions. The C-terminus consists of three tandem Cys2His2 zinc finger motifs, which act as a DNA-binding domain. The zinc fingers bind GC-rich sequences (designated as Sp1 sites), including GC boxes (GGGGCGGGG), GT/CACC boxes (GGTGTGGGG), and basic transcription elements (with a preference for the 5′-CACCC-3′core motif), with varying affinities. These binding sequences are widely distributed in promoters, enhancers, and locus control regions (LCRs) of housekeeping genes as well as of tissue-specific and viral genes [6,7] that are necessary for cell proliferation, differentiation, apoptosis and morphogenesis [8,9,10,11]. The N-terminus contains the transcriptional regulatory domains that vary significantly. It consists of acidic transactivation domains, Sin3 interacting domains (SID), or C-terminal binding protein (CtBP)-binding repressor domains that can bind co-factors and contribute to the distinct functions of KLFs [5]. The functions of Sin3 and CtBP might compete because the SID and CtBP-binding domains overlap [5]. Only in several KLFs (KLF1, KLF4, KLF8, KLF11), the nuclear localization signal (NLS)—which causes the nuclear localization of KLFs—is located immediately adjacent to/within the zinc finger motifs. [12].
The transforming growth factor-beta (TGF-β) inducible immediate early gene (TIEG) encodes a protein that has the ability to inhibit cell growth in cultured osteoblastic and epithelial cell populations [13,14]. In 1998, a novel Sp1-like zinc finger encoding cDNA was identified, and the gene product was named KLF11 [15]. KLF11 shows a high level of homology with TIEG and was named TIEG2, accordingly. Since then, TIEG has been referred to as TIEG1 or KLF10 [15]. Both TIEG proteins can be assigned to subgroup III of the KLFs. TIEG2 shares 91% homology within the C-terminal zinc finger region with TIEG1 and 44% homology within the N-terminus. However, it does not share any significant homology with any other previously identified protein [15]. TIEG1 and TIEG2 share several proline-rich sequences within the N-terminal domain, a property commonly found in other transcription factors as well [2,16,17,18,19,20]. Both TIEG proteins, like all members of subgroup III, share in the N-terminus a conserved repression motif and an α-helical domain highly similar to the Sin3 interaction domain (SID) of the transcriptional repressor Mad1 [1]. This SID-like domain mediates transcriptional repression by interacting with the histone deacetylase corepressor complex mSin3A [21,22,23]. However, the repressional function can be modified: The phosphorylation of different residues in a region adjacent to the KLF11 SID-domain via the EGF–Ras–MEK1–ERK2 signaling pathway [24] and/or the EGFR/AKT–KLF11 (Thr-56 phosphorylation) signaling pathway [25] disrupts the interaction of the SID-like domain with mSin3A and results in a significant loss of the repressional function.
KLF11 plays a vital role in a variety of cell types and tissues. It potentiates TGF-β-mediated anti-proliferative signaling pathways and thereby inhibits epithelial cell growth [15,26,27]. While TIEG2/KLF11 represents the human isoform [15], Tg3/KLF11 [28] is a murine isoform. Based on the observations that KLF11 is significantly induced by TGF-β and, when artificially overexpressed, mimics TGF-β–induced effects in various epithelial cell systems in vivo and in vitro, it has been speculated that KLF11 might function as a TGF-β effector protein that participates in TGF-β signaling pathways [15,29]. Within this review, we will focus on recent advances describing the different roles and mechanisms of KLF11 between normal cell growth and cell growth disorders, which is particularly important for future analyzes regarding the role of KLF11 in carcinogenesis and which may lead to new insights and discoveries for cancer diagnosis or treatment.
2. Genomic Organization and Characteristic Structural Features of KLF11
KLF11 mRNA is ubiquitously expressed in human tissues, with the highest levels found in the pancreas and in skeletal muscle [15]. It is a nuclear protein but is excluded from the nucleolus [15]. The transcriptional regulatory function of KLF11 is mediated by potent repressor activity located within the N-terminal region of the protein [15]. Table 1 gives an overview of the genomic information of KLF11 in Homo sapiens (humans). Chromosomal localization data refers to human genes and has been obtained from the human genome database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/gene).

Table 1.
Genomic information and aliases of human Krüppel-like factor 11 (KLF11).
As a member of the KLFs family, the structure of KLF11 is characterized by the highly homologous C-terminal DNA binding domain containing three Cys2His2 zinc finger motifs that bind the GC-rich sequences (the Sp1-like sites) within promoters [1]. The “Sp1 site” dependent transcription can be regulated: the activity, expression, and/or post-translation modification can be altered with cell growth. A basic tetra peptide within the second and third zinc finger of the KLF11 DNA-binding domain functions as an NLS, which is essential for it to be able to act as a site-specific transcription factor [15,30]. The N-terminal region of KLF11 consists of the three repression domains named R1, R2, and R3. They can be tethered to the DNA through a heterologous DNA binding domain (DBD) to mediate repression. The R1 domain, an alpha-helical repression motif (HRM), has been shown to be essential for the interaction with the co-repressor mSin3A, and this interaction is followed by the inhibition of the transcriptional activation of target genes [23]. Therefore, the R1 repression domain is often referred to as the mSin3A interacting domain (SID) and seems to be the most important domain for facilitating KLF11-mediated transcriptional repression [9,15,23].
3. KLF11, a Context-Dependent Transcriptional Repressor or Activator
KLFs are known to activate or repress gene expression in various organisms [31], subsequently leading to context-dependent effects that might appear contradictory. TIEG1/KLF10 has been shown to behave as a transcriptional repressor [32], and similarly, KLF11 represses promoter activity in vitro and in vivo through the Sp1-like binding sites in the three repression domains [15]. However, the zinc finger of KLF11 containing the DBD alone is able to activate the transcription [30].
KLF11 represses transcription through the recruitment of the mSin3A–histone deacetylase (HDAC) complex via the SID [23]. Sequence analysis revealed that the SID is highly conserved within the TIEG protein family, indicating that the recruitment of the HDAC complexes via binding mSin3A is a general mechanism of transcriptional repression in these proteins [30]. Nevertheless, in HeLa cells, KLF11-mediated transcriptional repression was not mSin3A-dependent, so the R2 and R3 domains might also play an important role as transcriptional repressors [9,30]. However, the inhibition of transcription by R2 and R3 is less well understood than the repression mediated by R1.
Although KLF11 was originally identified as a transcriptional repressor, recent studies have demonstrated that it is also able to activate transcription in various cell types. Binding to the insulin promotor and activating the transcription of the insulin gene, KLF11 plays a vital role in the function of the endocrine pancreas [33]. KLF11 could also interact with the co-activator p300 and activate the pancreatic-duodenal homeobox-1 gene (Pdx-1), which is an important mediator of pancreatic beta-cell activity [34].
As KLF11 can behave as a transcriptional activator or as a repressor, it has been suggested that the divergent function depends on the cell type and the promoter context [15,35]. The intramolecular interactions of the N-terminal repression domains and the C-terminal activator domain, which could be mediated by protein folding, have a counteracting activity in the context of the full-length state [30]. Thus, KLF11 might act as a context-dependent transcriptional repressor or activator by interacting with coactivators or corepressors to orchestrate the transcriptional regulation of target genes.
4. KLF11 Contributes to the Regulation of Normal Cell Growth
TIEG proteins can act as growth-inhibiting and/or pro-apoptotic proteins in different cell types [14,36,37,38,39]. Early research reported that KLF11 inhibits cell growth in pancreatic epithelial cells [14,40] in an osteoblastic cell population [13] and in prostate cells through cell cycle regulation [41]. A proposed mechanism is the interaction with the TGF-β signaling pathway. TGF-β plays an important role in the inhibition of cell growth, the regulation of extracellular matrix components, in cell differentiation and migration, as well as in the induction of apoptosis. The intracellular signaling pathway of TGF-β has been well elucidated [42,43,44]. In the TGF-β-signaling pathway, the Smad7 protein acts as a negative feedback loop that suppresses the TGF-β-induced growth inhibition. KLF11 is now able to transcriptionally silence the Smad7 gene and to disrupt the negative feedback loop. Thereby, KLF11 potentiates the TGF-β-signaling and the inhibitory effects of TGF-β on cell growth [45,46]. KLF11 can be induced by several members of the TGF-β superfamily, including all three TGF-β isoforms, by activin, by BMPs, and by the glial-cell-derived neurotrophic factor [47]. Via transcriptional regulation of further genes that induce apoptosis or cell cycle arrest, KLF11 can also inhibit proliferation [15,23,37].
The importance of KLF11 in cell growth regulation was firmly established by a study demonstrating that its overexpression blocks the proliferation of Chinese hamster ovary (CHO) cells [15]. CHO cells transfected with wild-type KLF11 showed decreased proliferation by 60% compared to cells that were transfected with SID-deleted KLF11 or to normal control CHO cells [37]. Similarly, the PANC1 exocrine pancreatic epithelial cell line transfected with KLF11 showed a significant decrease in cell proliferation and an increase in the number of apoptotic cells [37]. In vivo, transgenic expression of KLF11 in the mouse exocrine pancreas resulted in decreased cell proliferation, increased apoptosis, and reduced organ size [37]. KLF11-transgenic mice displayed downregulation of genes encoding oxidative stress scavengers, such as superoxide dismutase 2 (SOD2) and catalase1, which might contribute to an increased susceptibility to oxidative stress and increased cell death. In embryonic stem cells, cell growth inhibition and transcriptional regulation were observed when KLF11 was transiently or stably expressed [48]. In OLI-neu cells, KLF11 was described as a pro-apoptotic downstream mediator of TGF-β [28,30].
Members of the Bcl-2 family are important regulators of apoptosis, deciding whether cells live or die. The ratio between anti-apoptotic (e.g., Bcl-XL, Bcl-2) and pro-apoptotic (e.g., Bax, Bak) proteins determines the susceptibility of cells to apoptosis [49]. The promoter region of Bcl-XL contains several Sp1 sites serving as binding elements of KLFs [50]. KLF11 bridges the TGF-β-signaling to the repression of anti-apoptotic Bcl-XL expression and thereby increases apoptosis. Overexpression of KLF11 in OLI-neu cells resulted in apoptotic cell death accompanied by decreased levels of the anti-apoptotic Bcl-XL [39]. On the other hand, KLF11 increased leiomyoma cell proliferation and abolished the anti-proliferative effect of the progesterone antagonist RU486 via integrating with progesterone receptor (PR) signaling [51].
However, one other in vivo study showed contrary effects with less importance of KLF11: The KLF11 gene was inactivated in mice by gene knockout technology, and KLF11-/- mice were normal in growth, bred according to the normal Mendelian genetics, and lived as long as KLF11+/+ mice. Hematological analyzes also revealed no abnormalities in KLF11-/- mice [48].
In summary, KLF11 is highly induced by TGF-β and, when overexpressed, mimics the TGF-β induced cell cycle arrest in epithelial cells. However, in vivo, the effects of KLF11 are less consistent and show either inhibition of proliferation or normal cell growth (Table 2).

Table 2.
KLF11 in normal cells/tissues.
5. The Relation of KLF11 to Cancers
Given the described role of KLF11 in growth regulation, it is not surprising that KLF11 has also been implicated in the development of tumors. In carcinogenesis, the TGF-β signaling pathway plays a dual role characterized by tumor suppression at early tumor stages and enhanced tumor progression at the late stages of the disease [43,59,60]. TGF-β mediates tumor suppression via Smad-dependent signaling, which then regulates the transcription of cell-cycle-associated genes like p15 and p21 [44,61]. But Smad7 also exerts a negative feedback loop by binding to the activated TβR-I, blocking it and preventing phosphorylation [62,63]. Disturbances in the Smad-dependent signaling have recently been shown in human cancers (including pancreatic cancer), and are associated with the ability of tumor cells to escape from the TGF-β–induced growth inhibition [64,65]. TGF-β may also signal through Smad-independent signaling cascades (e.g., Rho-like guanosine triphosphatases, p38, mitogen-activated protein kinase [MAPK], phosphatidylinositol-3-kinase or c-Jun-N-terminal kinase) and induce an epithelial-to-mesenchymal transition (EMT)—which is a key process in the formation of cancer metastasis—of tumor cells, leading to enhanced tumor cell migration and invasion [66,67,68,69,70].
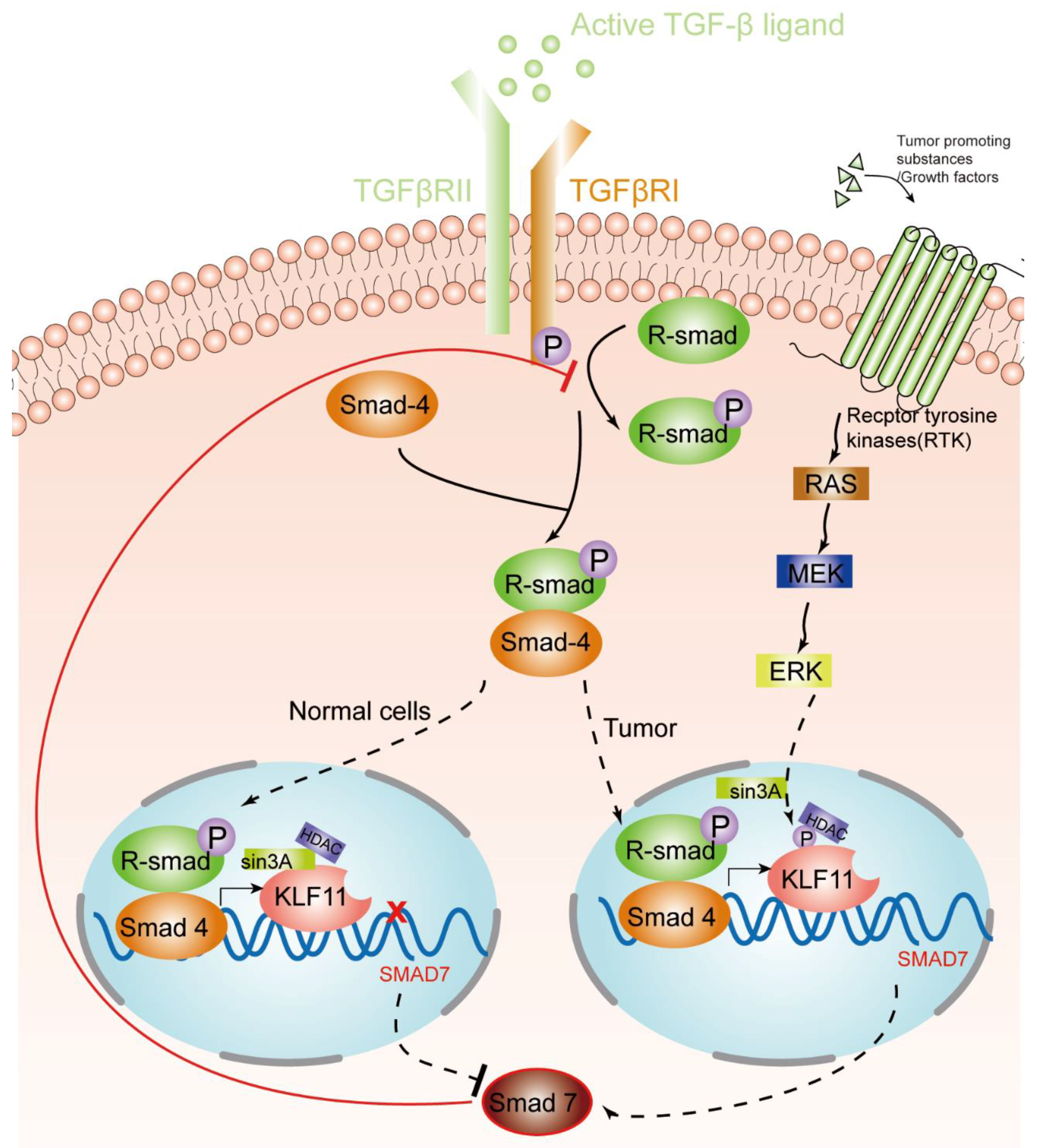
KLF11, as discussed above, is an early response transcription factor that potentiates the TGF-β induced growth inhibition in normal epithelial cells by terminating the inhibitory Smad7 loop. In pancreatic cancer cells with oncogenic Ras mutations, this function of KLF11 is inhibited by the oncogenic Erk/MAPK: Erk/MAPK phosphorylates KLF11, which leads to the disruption of the KLF11–mSin3a interaction [46]. This inhibits the binding of the KLF11–Smads complex to the TIE element and leads to a reduced TGF-β-induced c-myc repression and to a reduction of the anti-proliferative effects of TGF-β [40]. Another study showsd that KLF11 is inhibited in CHO cells by the epidermal growth factor (EGF)–Ras–MEK1–ERK2 signaling pathway. Like in the Erk/MAPK pathway, phosphorylation of four serine/threonine sites adjacent to the SID leads to the disruption of the SID–mSin3A interaction [24]. In KRAS oncogenic mutant cancer cells, KLF11 inhibits BrdU incorporation, increases apoptosis, and inhibits the KRAS-mediated foci and agar colony formation. In vivo, KLF11 partly inhibits the growth of pancreatic tumor cells of KRAS mutant xenografts, by inducing cell cycle arrest at the S phase via downregulation of cyclin A2 [11]. Therefore, KLF11 might participate in the functional switch of TGF-β from a tumor suppressor to a tumor promoter. Figure 1 gives an overview of the different roles of the KLF11-mediated TGF-β–TGF-receptor–Smad signaling pathway in normal cells and tumors.

Figure 1.
KLF11-mediated modulation of the TGF-β signaling pathway in normal cells and tumors. An activated TGF-β ligand binds to the type 2 domain of the TGF-β receptor, which then recruits and phosphorylates a type 1 receptor. The type 1 receptor then recruits and phosphorylates a receptor-regulated Smad (R-smad). The R-smad then binds to the common smad, Smad 4, and forms a heterodimeric complex. The Smad complex translocates to the nucleus to induce the expression of KLF11. In normal cells, the KLF11-Sin3A-HDAC complex binds to the promoters of Smad7 and represses its expression, which acts as a negative feedback loop of the TGF-β–Smads signaling pathway. However, in some tumors, RAS was activated by tumor-promoting substances/growth factors. Ras phosphates KLF11 by the RAS–MEK–ERK pathway, which leads to the disruption of the KLF11–mSin3a interaction, resulting in the termination of the inhibitory KLF11-mediated Smad7 loop.
In addition to the inhibition of KLF11 by SID phosphorylation via Erk/MAPK, Buttar et al. [25] suggested an additional model of KLF11-mediated tumor suppression and its antagonism by an oncogenic pathway. KLF11 binds to the GC-rich consensus sequences in the promoter region of cPLA2α, the key rate-limiting enzyme of the oncogenic PGE2 cascade. Following binding, KLF11 represses the cPLA2α promoter by recruiting the chromatin-remodeling complex Sin3a-HDAC to the promotor. In this way, KLF11 behaves as a tumor suppressor gene by repression of the cPLA2α–PGE2 pathway. This mechanism was shown in Barrett’s epithelial cells. EGFR-AKT signaling, which is upregulated in a subset of patients during carcinogenesis in Barrett’s esophagus cancer, leads to the phosphorylation of threonine at position 56 in the R1 domain (SID) of KLF11. This phosphorylation inhibits the KLF11 binding, and the repression of the cPLA2α promoter and the tumor-suppressing effects of KLF11 are inhibited. Probably EGFR can use two different intracellular pathways (ERK2 versus AKT) to inactivate KLF11 via phosphorylation. These phosphorylation events expand our biochemical knowledge about KLF11 to the post-translational effects.
On the contrary, direct tumor-promoting effects of KLF11 have also been described. In hepatocellular carcinoma, KLF11 had a significant influence on proliferation and apoptosis. It also promoted local invasion and distant migration by suppressing the Smad7 transcription via binding to the Smad7 promoter or by directly upregulating the Smad2/3 expression [71,72]. Ji Q et al. demonstrated that the relative Twist1 promoter region activity increased gradually with increasing KLF11 levels in the plasma [73]. Therefore, they speculated that KLF11 might regulate gastric cancer migration and invasion by increasing the Twist1 expression, which is essential for EMT [73].
KLF11-methylation-dependent inactivation and downregulation occurs in several malignancies, including leukemia, myeloproliferative disorders, esophageal adenocarcinoma, pancreatic cancer, germ cell tumors, ovarian cancer, and head and neck cancer, supporting its candidacy as an actual tumor suppressor gene in humans [25,37,74,75,76,77,78]. KLF11 is involved in the progression of a wide variety of cancers, such as ovarian cancer and pancreatic cancer [40,78]. In breast cancer, the KLF11 promotor is also hypermethylated, and the hypermethylation is associated with low expression of KLF11. KLF11 hypermethylation might be associated with higher rates of metastases [79]. Similar results were found in uterine fibroids and in myelodysplastic syndrome [74,80], suggesting that DNA methylation to regulate KLF11 expression might be a key event that directly contributes to tumorigenesis.
It was recently reported that the microRNA miR-30d increased the survival of BT474 and MDA-MB-231 breast cancer cells. It was shown that it inhibited apoptosis and increased Bcl-2 expression, while it reduced the Bax protein levels. This influence of miR-30d on breast cancer cell growth, metastasis, and EMT is dependent on a low level of KLF11 and on a high level of pSTAT3. KLF11 is a direct target of miR-30d, and KLF11 and pSTAT3 expression are regulated by miR-30d [81].
MiR-30 also reduced the profibrogenic TGF-β signaling in hepatic stellate cells by suppressing the KLF11 expression and thus enhancing the negative feedback loop of TGF-β signaling imposed by Smad7 [82]. LincRNA-p21 reduced the availability of miR-30 [83]. Besides miR-30d, overexpression of miR-10b in hepatocellular carcinoma (HCC) promoted HCC cell migration and invasion. MiR-10b downregulated KLF4, which is the inhibitory transcriptional factor of KLF11. In this way, KLF11 was upregulated, which promoted HCC EMT [72].
All these results suggest that KLF11 plays a crucial role during tumorigenesis and development. (Table 3.).

Table 3.
KLF11 in cancers.
6. Conclusions
In the last decade, the transcription factor KLF11 has come into focus in cancer research. However, the characterization of the functions of KLF11 is still in its infancy. Recent data indicates that KLF11 can affect cell growth and carcinogenesis through multiple mechanisms. KLF11 is not only a basal promoter element involved in constitutive gene transcription but also plays multiple roles in the modulation of transcription. It may respond to a particular signal in one cell type but not in another, respond differently to the same signal, or respond to different signals differently in different cells. It might act as a context-dependent transcription regulator and is linked to growth-related biological processes.
Moreover, KLF11 might play an important role in the development of tumors. However, the role of KLF11 in normal cell growth regulation and cancer is diverse, and other KLFs might induce relevant transcription responses as well. Therefore, a full understanding of the role of KLF11 in growth control and tumor progression will not only requires an analysis of the individual factor KLF11 but also the identification of all KLFs expressed in the cells of interest and the characterization of the transcriptional context.
Author Contributions
Conceptualization, U.J. and A.H.; methodology, S.M.; software, S.M.; validation, S.M., L.L.; writing—original draft preparation, A.H.; writing—review and editing, U.J.; supervision, U.J.; project administration, L.L.; funding acquisition. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We thank Michael Semmlinger for English proofreading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AKT | protein kinase B (PKB) |
| AhR | aryl hydrocarbon receptor |
| BC | Breast Cancer |
| cDNA | complementary Deoxyribonucleic Acid |
| CHO cells | Chinese hamster ovary cells |
| COX1 | cyclooxygenase-1 |
| COX2 | cyclooxygenase-2 |
| cPLA2α | cytosolic phospholipase A2α |
| CPTAC | Clinical Proteomics Tumor Analysis Consortium |
| CtBP | C-Terminal Binding Protein |
| DNA | Deoxyribonucleic Acid |
| DNMT | DNA methyl-transferase |
| DNMTis | DNA methyltranferase inhibitors |
| EGFR | Epidermal Growth Factor Receptor |
| EMT | Epithelial–mesenchymal transition |
| EP3 | prostaglandin E2 receptor 3 |
| ER | Estrogen receptor |
| ERK | extracellular signal–regulated kinases |
| FKLF | fetal-beta like globin activating krüppel-like factor |
| HDACis | histone deacetylase inhibitors |
| KLF11 | Krüppel like factors 11 |
| LCRs | locus control regions |
| LMU | Ludwig Maximilians University |
| MAPK | Mitogen-activated protein kinase |
| MEK | Mitogen-activated protein kinase kinase |
| miRNA | micro Ribonucleic Acid |
| MODY7 | maturity-onset diabetes of the young type 7 |
| NAC | neo-adjuvant chemotherapy |
| PGE2 | Prostaglandin E2 |
| PI3K | phosphatidylinositol 3 kinase |
| PPAR | peroxisome proliferator-activated receptor |
| SIN3A | SIN3 transcription regulator homologue A |
| SID | mSin3 interaction domain |
| SMAD | Mothers against decapentaplegic homolog |
| TFs | transcriptional factors |
| TGF-β | Transforming growth factor-beta |
| TIEG | TGF-β inducible early gene |
| TLA | Three letter acronym |
| LD | linear dichroism |
References
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Bieker, J.J. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell Biol. 1993, 13, 2776–2786. [Google Scholar] [CrossRef] [PubMed]
- McConnell, B.B.; Yang, V.W. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef] [PubMed]
- Tetreault, M.P.; Yang, Y.; Katz, J.P. Kruppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, G.; Feng, L.; Lu, H.; Wang, X. Kruppel-like factors in breast cancer: Function, regulation and clinical relevance. Biomed. Pharmacother. 2019, 123, 109778. [Google Scholar] [CrossRef]
- Black, A.R.; Black, J.D.; Azizkhan-Clifford, J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 2001, 188, 143–160. [Google Scholar] [CrossRef]
- Kadonaga, J.T.; Tjian, R. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 1986, 83, 5889–5893. [Google Scholar] [CrossRef]
- Liu, C.; Calogero, A.; Ragona, G.; Adamson, E.; Mercola, D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit. Rev. Oncog. 1996, 7, 101–125. [Google Scholar]
- Cook, T.; Gebelein, B.; Urrutia, R. Sp1 and its likes: Biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 1999, 880, 94–102. [Google Scholar] [CrossRef]
- Bureau, C.; Calogero, A.; Ragona, G.; Adamson, E.; Mercola, D. Expression and Function of Kruppel Like-Factors (KLF) in Carcinogenesis. Curr. Genom. 2009, 10, 353–360. [Google Scholar] [CrossRef]
- Fernandez-Zapico, M.E.; Lomberk, G.A.; Tsuji, S.; Demars, C.J.; Bardsley, M.R.; Lin, Y.; Almada, L.L.; Han, J.; Mukhopadhyay, D.; Ordog, T.; et al. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem. J. 2011, 435, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Swamynathan, S.K. Kruppel-like factors: Three fingers in control. Hum. Genom. 2010, 4, 263–270. [Google Scholar] [CrossRef]
- Tau, K.R.; Hefferan, T.E.; Waters, K.M.; Robinson, J.A.; Subramaniam, M.; Riggs, B.L.; Spelsberg, T.C. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology 1998, 139, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, I.; Imoto, M.; Adjei, P.N.; Gores, G.J.; Subramaniam, M.; Spelsberg, T.C.; Urrutia, R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Investig. 1997, 99, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.; Gebelein, B.; Mesa, K.; Mladek, A.; Urrutia, R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 1998, 273, 25929–25936. [Google Scholar] [CrossRef]
- Sogawa, K.; Kikuchi, Y.; Imataka, H.; Fujii-Kuriyama, Y. Comparison of DNA-binding properties between BTEB and Sp1. J. Biochem. 1993, 114, 605–609. [Google Scholar] [CrossRef]
- Anderson, K.P.; Kern, C.B.; Crable, S.C.; Lingrel, J.B. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: Identification of a new multigene family. Mol. Cell. Biol. 1995, 15, 5957–5965. [Google Scholar] [CrossRef]
- Gerber, H.P.; Seipel, K.; Georgiev, O.; Hofferer, M.; Hug, M.; Rusconi, S.; Schaffner, W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 1994, 263, 808–811. [Google Scholar] [CrossRef]
- Alevizopoulos, A.; Mermod, N. Antagonistic regulation of a proline-rich transcription factor by transforming growth factor beta and tumor necrosis factor alpha. J. Biol. Chem. 1996, 271, 29672–29681. [Google Scholar] [CrossRef]
- Kojima, S.; Kobayashi, A.; Gotoh, O.; Ohkuma, Y.; Fujii-Kuriyama, Y.; Sogawa, K. Transcriptional activation domain of human BTEB2, a GC box-binding factor. J. Biochem. 1997, 121, 389–396. [Google Scholar] [CrossRef]
- Kaczynski, J.; Zhang, J.-S.; Ellenrieder, V.; Conley, A.; Duenes, T.; Kester, H.; Van Der Burg, B.; Urrutia, R. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J. Biol. Chem. 2001, 276, 36749–36756. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.A.; Conley, A.A.; Zapico, M.F.; Delgado, S.M.; Zhang, J.-S.; Urrutia, R. Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem. J. 2002, 366 Pt 3, 873–882. [Google Scholar] [CrossRef]
- Zhang, J.S.; Moncrieffe, M.C.; Kaczynski, J.; Ellenrieder, V.; Prendergast, F.G.; Urrutia, R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 2001, 21, 5041–5049. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Zhang, J.; Kaczynski, J.; Urrutia, R. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J. 2002, 21, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Buttar, N.S.; Demars, C.J.; Lomberk, G.; Rizvi, S.; Bonilla-Velez, J.; Achra, S.; Rashtak, S.; Wang, K.K.; Fernandez-Zapico, M.E.; Urrutia, R. Distinct role of Kruppel-like factor 11 in the regulation of prostaglandin E2 biosynthesis. J. Biol. Chem. 2010, 285, 11433–11444. [Google Scholar] [CrossRef] [PubMed]
- Chiarugi, V.; Magnelli, L.; Cinelli, M. Complex interplay among apoptosis factors: RB, p53, E2F, TGF-beta, cell cycle inhibitors and the bcl2 gene family. Pharmacol. Res. 1997, 35, 257–261. [Google Scholar] [CrossRef]
- Alevizopoulos, A.; Mermod, N. Transforming growth factor-beta: The breaking open of a black box. Bioessays 1997, 19, 581–591. [Google Scholar] [CrossRef]
- Wang, Z.; Peters, B.; Klussmann, S.; Bender, H.; Herb, A.; Krieglstein, K. Gene structure and evolution of Tieg3, a new member of the Tieg family of proteins. Gene 2004, 325, 25–34. [Google Scholar] [CrossRef]
- Cook, T.; Urrutia, R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am. J Physiol. Gastrointest. Liver Physiol. 2000, 278, G513–G521. [Google Scholar] [CrossRef]
- Spittau, B.; Wang, Y.; Boinska, D.; Krieglstein, K. Functional domains of the TGF-beta-inducible transcription factor Tieg3 and detection of two putative nuclear localization signals within the zinc finger DNA-binding domain. J. Cell Biochem. 2007, 101, 712–722. [Google Scholar] [CrossRef]
- Yin, K.J.; Hamblin, M.; Fan, Y.; Zhang, J.; Chen, Y.E. Krupple-like factors in the central nervous system: Novel mediators in stroke. Metab. Brain Dis. 2015, 30, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Yajima, S.; Lammers, C.-H.; Lee, S.-H.; Hara, Y.; Mizuno, K.; Mouradian, M.M. Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF). J. Neurosci. 1997, 17, 8657–8666. [Google Scholar] [CrossRef] [PubMed]
- Noti, J.D.; Johnson, A.K.; Dillon, J.D. The zinc finger transcription factor transforming growth factor beta-inducible early gene-1 confers myeloid-specific activation of the leukocyte integrin CD11d promoter. J. Biol. Chem. 2004, 279, 26948–26958. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Zapico, M.E.; van Velkinburgh, J. C.; Gutierrez-Aguilar, R.; Neve, B.; Froguel, P.; Urrutia, R.; Stein, R. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. J. Biol. Chem. 2009, 284, 36482–36490. [Google Scholar] [CrossRef]
- Asano, H.; Li, X.S.; Stamatoyannopoulos, G. FKLF, a novel Kruppel-like factor that activates human embryonic and fetal beta-like globin genes. Mol. Cell Biol. 1999, 19, 3571–3579. [Google Scholar] [CrossRef]
- Chalaux, E.; López-Rovira, T.; Rosa, J.L.; Pons, G.; Boxer, L.M.; Bartrons, R.; Ventura, F. A zinc-finger transcription factor induced by TGF-beta promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS Lett. 1999, 457, 478–482. [Google Scholar] [CrossRef]
- Fernandez-Zapico, M.E.; Mladek, A.; Ellenrieder, V.; Folch-Puy, E.; Miller, L.; Urrutia, R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003, 22, 4748–4758. [Google Scholar] [CrossRef]
- Bender, H.; Wang, Z.; Schuster, N.; Krieglstein, K. TIEG1 facilitates transforming growth factor-beta-mediated apoptosis in the oligodendroglial cell line OLI-neu. J. Neurosci. Res. 2004, 75, 344–352. [Google Scholar] [CrossRef]
- Wang, Z.; Spittau, B.; Behrendt, M.; Peters, B.; Krieglstein, K. Human TIEG2/KLF11 induces oligodendroglial cell death by downregulation of Bcl-XL expression. J. Neural. Transm. 2007, 114, 867–875. [Google Scholar] [CrossRef]
- Buck, A.; Buchholz, M.; Wagner, M.; Adler, G.; Gress, T.; Ellenrieder, V. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol. Cancer Res. 2006, 4, 861–872. [Google Scholar] [CrossRef]
- Blok, L.J.; Grossmann, M.E.; Perry, J.E.; Tindall, D.J. Characterization of an early growth response gene, which encodes a zinc finger transcription factor, potentially involved in cell cycle regulation. Mol. Endocrinol. 1995, 9, 1610–1620. [Google Scholar] [PubMed]
- Massague, J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Wotton, D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000, 19, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Spittau, B.; Krieglstein, K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012, 347, 65–72. [Google Scholar] [CrossRef]
- Johnsen, S.A.; Subramaniam, M.; Janknecht, R.; Spelsberg, T.C. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 2002, 21, 5783–5790. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Buck, A.; Harth, A.; Jungert, K.; Buchholz, M.; Adler, G.; Urrutia, R.; Gress, T.M. KLF11 mediates a critical mechanism in TGF-beta signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology 2004, 127, 607–620. [Google Scholar] [CrossRef]
- Song, C.Z.; Gavriilidis, G.; Asano, H.; Stamatoyannopoulos, G. Functional study of transcription factor KLF11 by targeted gene inactivation. Blood Cells Mol. Dis. 2005, 34, 53–59. [Google Scholar] [CrossRef]
- Oltvai, Z.N.; Milliman, C.L.; Korsmeyer, S.J. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993, 74, 609–619. [Google Scholar] [CrossRef]
- Grillot, D.A.; González-García, M.; Ekhterae, D.; Duan, L.; Inohara, N.; Ohta, S.; Seldin, M.F.; Nuñez, G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J. Immunol. 1997, 158, 4750–4757. [Google Scholar]
- Yin, P.; Lin, Z.; Reierstad, S.; Wu, J.; Ishikawa, H.; Marsh, E.E.; Innes, J.; Cheng, Y.; Pearson, K.; Coon, J.S.; et al. Transcription factor KLF11 integrates progesterone receptor signaling and proliferation in uterine leiomyoma cells. Cancer Res. 2010, 70, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, X.; Ma, F.; Fan, Y.; Sun, P.; Zhu, T.; Zhang, J.; Hamblin, M.H.; Chen, Y.E.; Yin, K.-J. Endothelium-targeted overexpression of kruppel-like factor 11 protects blood-brain barrier function after ischemic brain injury. Brain Pathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Tan, Z.; Gan, M.; Li, Q.; Chen, L.; Niu, L.; Jiang, D.; Jiang, J.; Wang, J.; Li, X.; et al. tRNA-Derived Small Non-Coding RNAs as Novel Epigenetic Molecules Regulating Adipogenesis. Biomolecules 2019, 9, 274. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Li, J.; Zhang, M.; Yu, B. Knockdown of KLF11 attenuates hypoxia/reoxygenation injury via JAK2/STAT3 signaling in H9c2. Apoptosis 2017, 22, 510–518. [Google Scholar] [CrossRef]
- Zheng, Y.; Khan, Z.; Zanfagnin, V.; Correa, L.F.; Delaney, A.A.; Daftary, G.S. Epigenetic Modulation of Collagen 1A1: Therapeutic Implications in Fibrosis and Endometriosis. Biol. Reprod. 2016, 94, 87. [Google Scholar] [CrossRef] [PubMed]
- Loft, A.; Forss, I.; Siersbaek, M.S.; Schmidt, S.F.; Larsen, A.S.; Madsen, J.G.; Pisani, D.F.; Nielsen, R.; Aagaard, M.M.; Matgison, A.; et al. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes Dev. 2015, 29, 7–22. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Q.; Jiao, T.; Cui, A.; Sun, X.; Fang, W.; Xie, L.; Liu, Y.; Fang, F.; Chang, Y. Involvement of KLF11 in hepatic glucose metabolism in mice via suppressing of PEPCK-C expression. PLoS ONE 2014, 9, e89552. [Google Scholar] [CrossRef]
- Mathison, A.; Grzenda, A.; Lomberk, G.; Velez, G.; Buttar, N.; Tietz, P.; Hendrickson, H.; Liebl, A.; Xiong, Y.Y.; Gores, G.; et al. Role for Kruppel-like transcription factor 11 in mesenchymal cell function and fibrosis. PLoS ONE 2013, 8, e75311. [Google Scholar] [CrossRef]
- Fan, Y.; Konermann, C.; Aulmann, S.; Bermejo, J.L.; Brugger, M.; Diederichs, S.; Rom, J.; Weichenhan, D.; Claus, R.; Rehli, M.; et al. Kruppel-like factor-11, a transcription factor involved in diabetes mellitus, suppresses endothelial cell activation via the nuclear factor-kappaB signaling pathway. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2981–2988. [Google Scholar] [CrossRef]
- Grunewald, M.; Johnson, S.; Lu, D.; Wang, Z.; Lomberk, G.; Albert, P.R.; Stockmeier, C.A.; Meyer, J.H.; Urrutia, R.; Miczek, K.A.; et al. Mechanistic role for a novel glucocorticoid-KLF11 (TIEG2) protein pathway in stress-induced monoamine oxidase A expression. J. Biol. Chem. 2012, 287, 24195–24206. [Google Scholar] [CrossRef]
- Yamamoto, K.; Sakaguchi, M.; Medina, R.J.; Niida, A.; Sakaguchi, Y.; Miyazaki, M.; Kataoka, K.; Huh, N.-H. Transcriptional regulation of a brown adipocyte-specific gene, UCP1, by KLF11 and KLF15. Biochem. Biophys. Res. Commun. 2010, 400, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Perakakis, N.; Laubner, K.; Limbert, C.; Stahl, T.; Brendel, M.D.; Bretzel, R.G.; Seufert, J.; Päth, G. Human Kruppel-like factor 11 inhibits human proinsulin promoter activity in pancreatic beta cells. Diabetologia 2007, 50, 1433–1441. [Google Scholar] [CrossRef]
- Neve, B.; Fernandez-Zapico, M.E.; Ashkenazi-Katalan, V.; Dina, C.; Hamid, Y.H.; Joly, E.; Vaillant, E.; Benmezroua, Y.; Durand, E.; Bakaher, N.; et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc. Natl. Acad. Sci. USA 2005, 102, 4807–4812. [Google Scholar] [CrossRef] [PubMed]
- Akhurst, R.J.; Derynck, R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001, 11, S44–S51. [Google Scholar] [PubMed]
- Jakowlew, S.B. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 2006, 25, 435–457. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H.; Miyazono, K.; Dijke, P.T. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Abdollah, S.; Qiu, Y.; Cai, J.; Xu, Y.Y.; Grinnell, B.W.; A Richardson, M.; Topper, J.N.; A Gimbrone, M.; Wrana, J.L.; et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 1997, 89, 1165–1173. [Google Scholar] [CrossRef]
- Nakao, A.; Afrakhte, M.; Morn, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Ltoh, S.; Kawabata, M.; Heldin, N.-E.; Heldin, C.-H.; et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997, 389, 631–635. [Google Scholar] [CrossRef]
- de Caestecker, M.P.; Piek, E.; Roberts, A.B. Role of transforming growth factor-beta signaling in cancer. J. Natl. Cancer Inst. 2000, 92, 1388–1402. [Google Scholar] [CrossRef]
- Dumont, N.; Arteaga, C.L. Targeting the TGF beta signaling network in human neoplasia. Cancer Cell 2003, 3, 531–536. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Ghiassi, M.; Bakin, A.; Aakre, M.; Lundquist, C.A.; Engel, M.E.; Arteaga, C.L.; Moses, H.L. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 2001, 12, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bakin, A.V.; Tomlinson, A.K.; Bhowmick, N.A.; Moses, H.L.; Arteaga, C.L. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000, 275, 36803–36810. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Hebert, M.C.; Zhang, Y.E. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. EMBO J. 2002, 21, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Fowlis, D.J.; Bryson, S.; Duffie, E.; Ireland, H.; Balmain, A.; Akhurst, R.J. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 1996, 86, 531–542. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Hendler, S.F.; Boeck, W.; Seufferlein, T.; Menke, A.; Ruhland, C.; Adler, G.; Gress, T.M. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001, 61, 4222–4228. [Google Scholar] [PubMed]
- Jung, C.J.; Liengar, S.; lyengar, S.; Blahbik, K.R.; Jiang, J.X.; Tahimic, C.; Torok, N.J.; de vere White, R. W.; Farnham, P.J.; Zern, M. Human ESC self-renewal promoting microRNAs induce epithelial-mesenchymal transition in hepatocytes by controlling the PTEN and TGFbeta tumor suppressor signaling pathways. Mol. Cancer Res. 2012, 10, 979–991. [Google Scholar] [CrossRef]
- Hujie, G.; Zhou, S.H.; Zhang, H.; Qu, J.; Xiong, X.W.; Hujie, O.; Liao, C.G.; Yang, S.E. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/KLF11/Smads in hepatocellular carcinoma. Cancer Cell Int. 2018, 18, 10. [Google Scholar] [CrossRef]
- Ji, Q.; Li, Y.; Zhao, Q.; Fan, L.Q.; Tan, B.B.; Zhang, Z.D.; Zhao, X.F.; Liu, Y.; Wang, D.; Jia, N. KLF11 promotes gastric cancer invasion and migration by increasing Twist1 expression. Neoplasma 2019, 66, 92–100. [Google Scholar] [CrossRef]
- Potapova, A.; Hasemeier, B.; Römermann, D.; Metzig, K.; Göhring, G.; Schlegelberger, B.; Länger, F.; Kreipe, H.; Lehmann, U. Epigenetic inactivation of tumour suppressor gene KLF11 in myelodysplastic syndromes*. Eur. J. Haematol. 2010, 84, 298–303. [Google Scholar] [CrossRef]
- Wermann, H.; Stoop, H.; Gillis, A.J.; Honecker, F.; Van Gurp, R.J.; Ammerpohl, O.; Richter, J.; Oosterhuis, J.W.; Bokemeyer, C.; Looijenga, L.H.J. Global DNA methylation in fetal human germ cells and germ cell tumours: Association with differentiation and cisplatin resistance. J. Pathol. 2010, 221, 433–442. [Google Scholar] [CrossRef]
- Gebhard, C.; Schwarzfischer, L.; Pham, T.-H.; Schilling, E.; Klug, M.; Andreesen, R.; Rehli, M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006, 66, 6118–6128. [Google Scholar] [CrossRef] [PubMed]
- Belbin, T.J.; Singh, B.; Barber, l.; Socci, N.; Wenig, B.; Smith, R.; Prystowsky, M.B.; Childs, G. Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res. 2002, 62, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Tian, W.; Wang, Y.; Wu, D.; Sun, Z.; Zhao, E. Promoter DNA methylation is associated with KLF11 expression in epithelial ovarian cancer. Genes Chromosomes Cancer 2015, 54, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Faryna, M.; Konermann, C.; Aulmann, S.; Bermejo, J.L.; Brugger, M.; Diederichs, S.; Rom, J.; Weichenhan, D.; Claus, R.; Rehli, M.; et al. Genome-wide methylation screen in low-grade breast cancer identifies novel epigenetically altered genes as potential biomarkers for tumor diagnosis. FASEB J. 2012, 26, 4937–4950. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Yin, P.; Monsivais, D.; Lin, S.M.; Du, P.; Wei, J.-J.; E Bulun, S. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS ONE 2012, 7, e33284. [Google Scholar] [CrossRef]
- Han, M.; Wang, Y.; Guo, G.; Li, L.; Dou, D.; Ge, X.; Lv, P.; Wang, F.; Gu, Y. microRNA-30d mediated breast cancer invasion, migration, and EMT by targeting KLF11 and activating STAT3 pathway. J. Cell Biochem. 2018, 119, 8138–8145. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).