A Computational Probe into the Structure and Dynamics of the Full-Length Toll-Like Receptor 3 in a Phospholipid Bilayer
Abstract
1. Introduction
2. Results
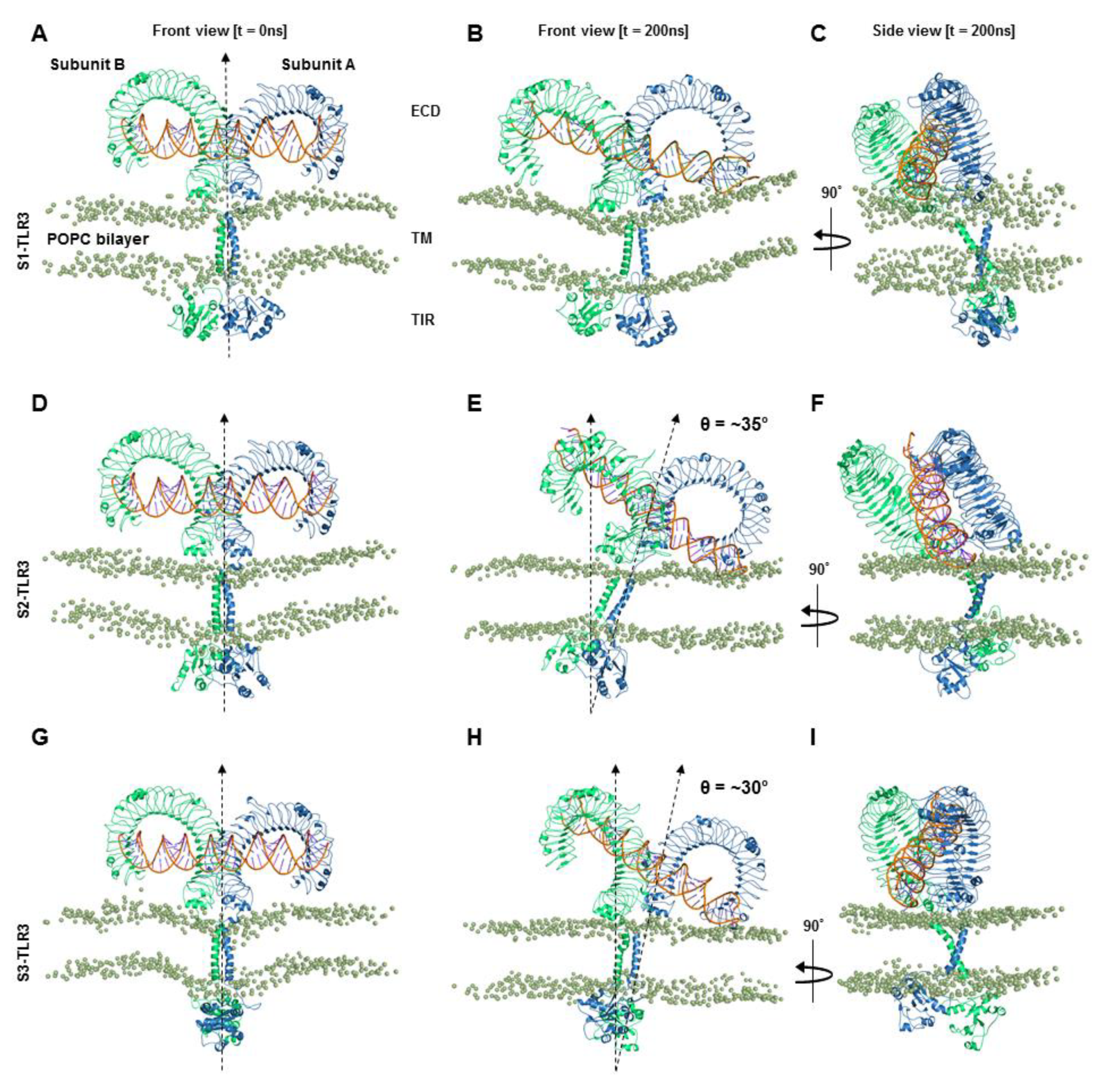
2.1. Full-length TLR3 Tilts and Wraps around the Phospholipid Bilayer
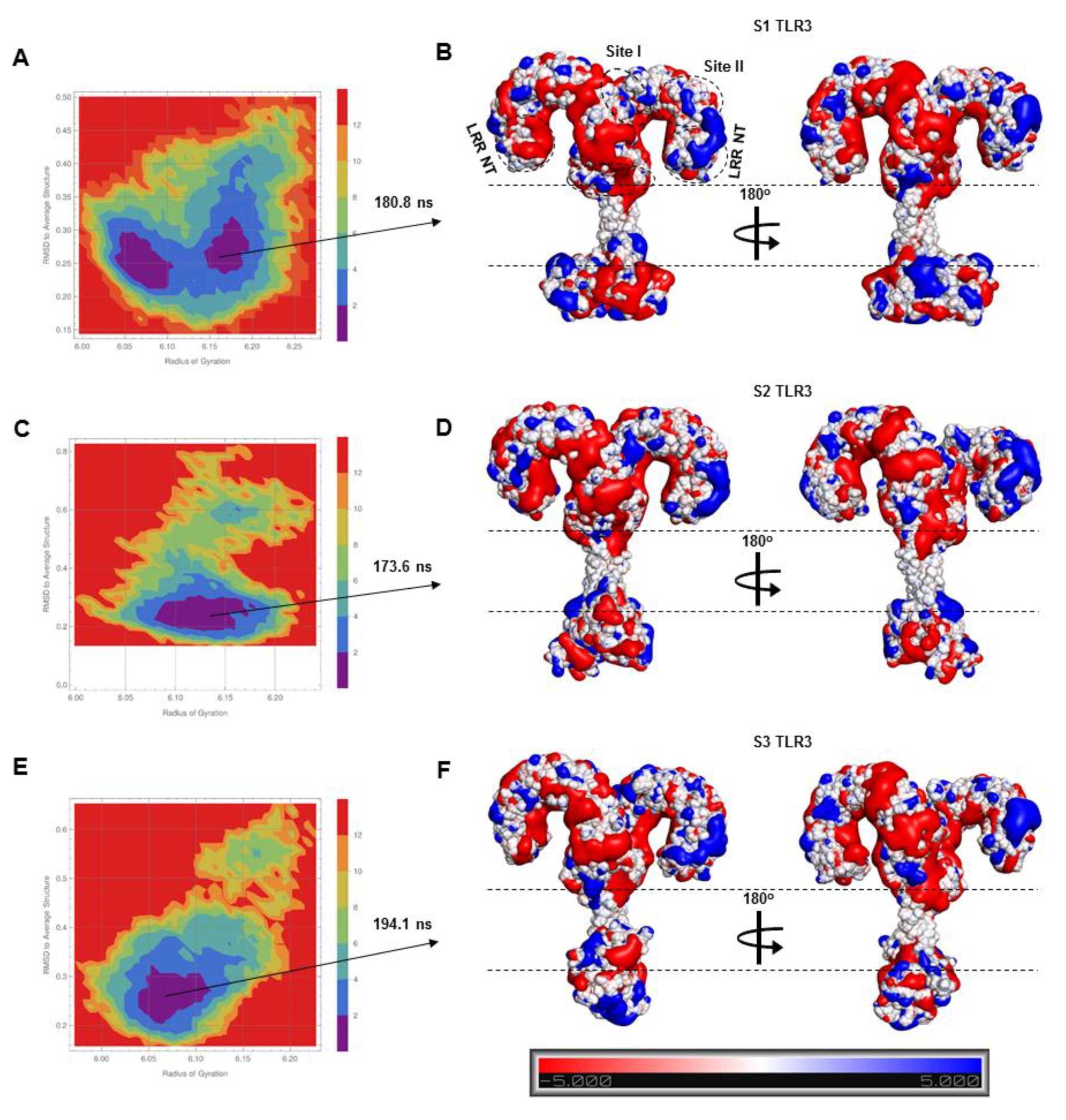
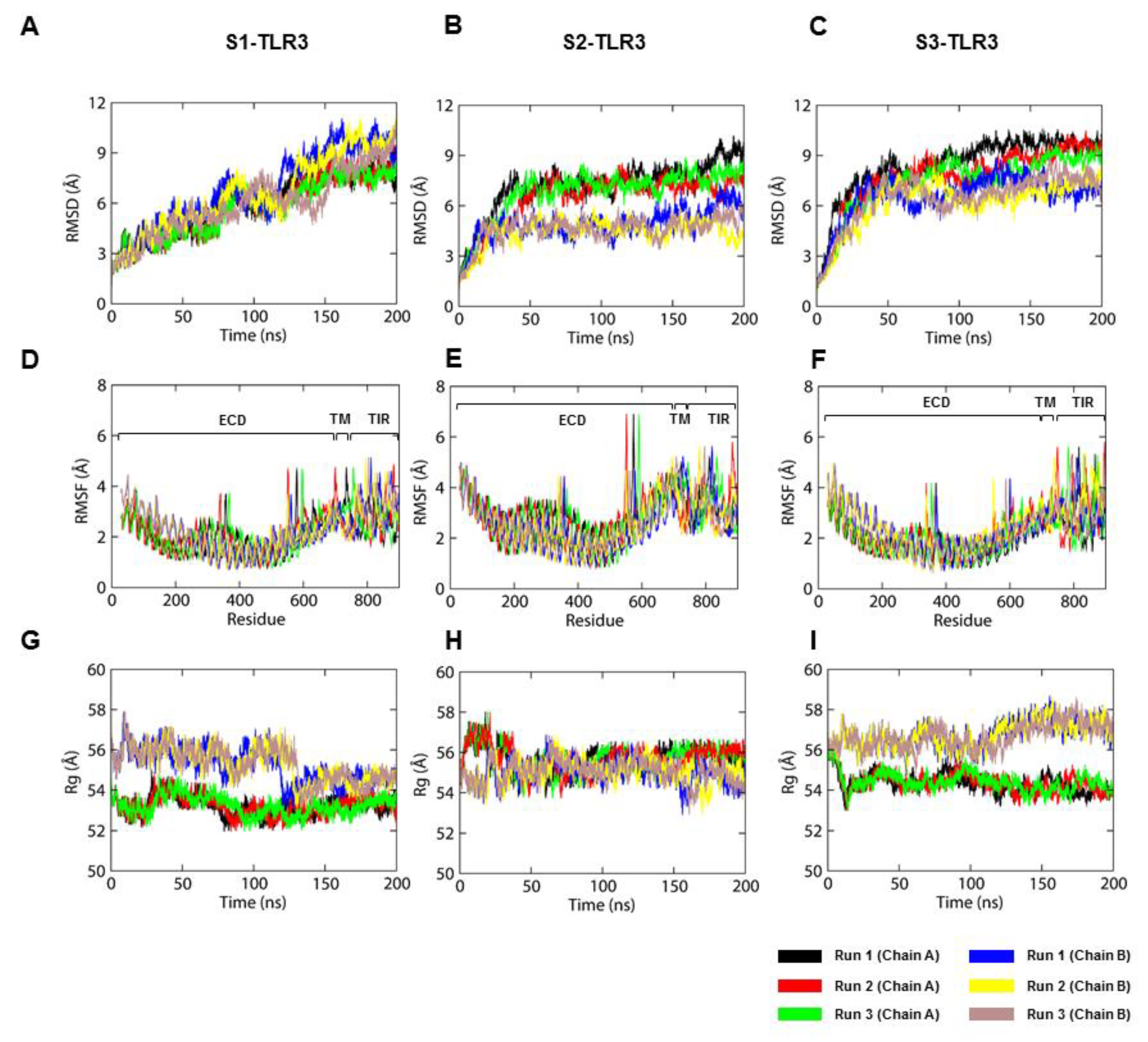
2.2. The S2-TLR3-POPC System Maintains Better Stability During MD Simulations
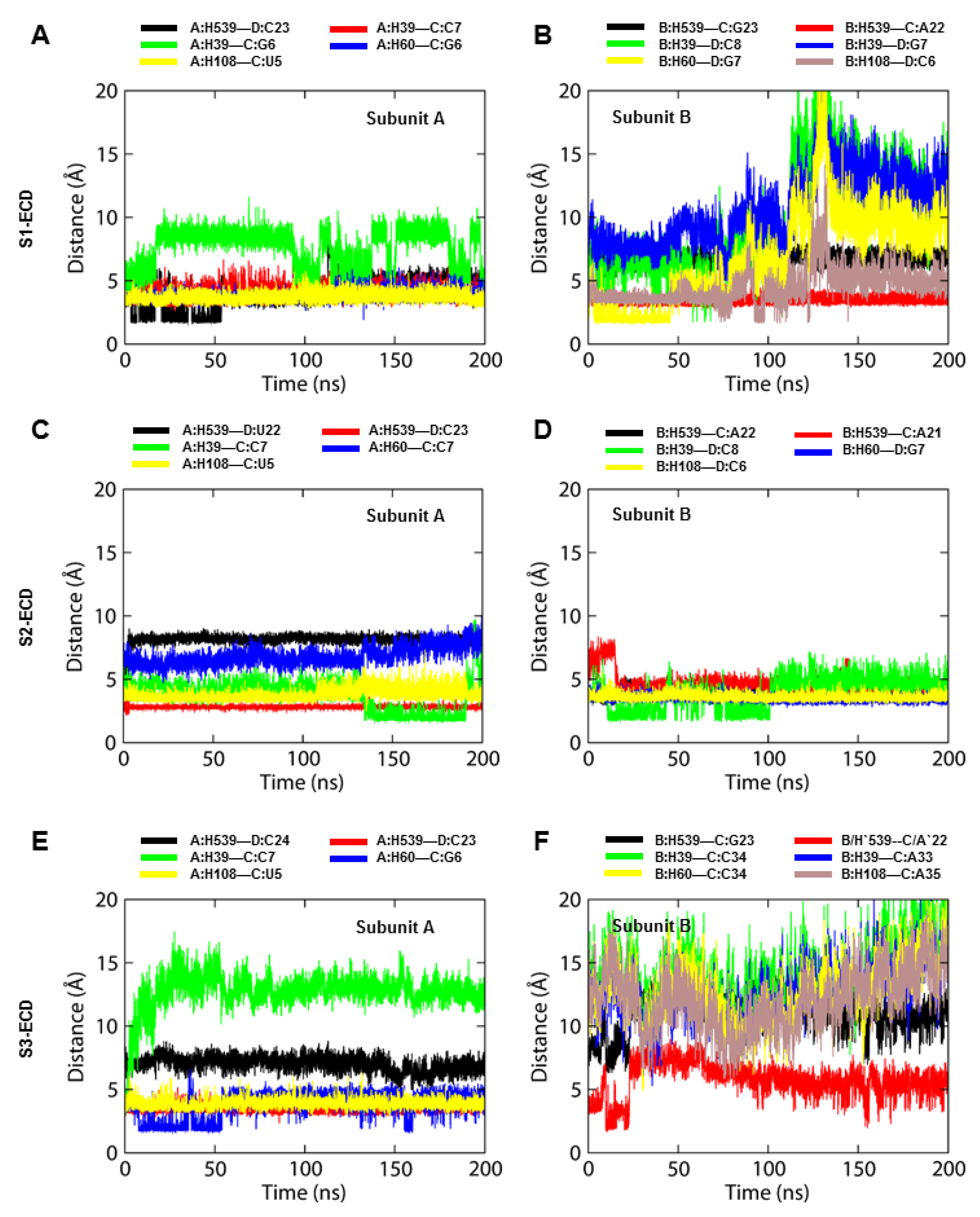
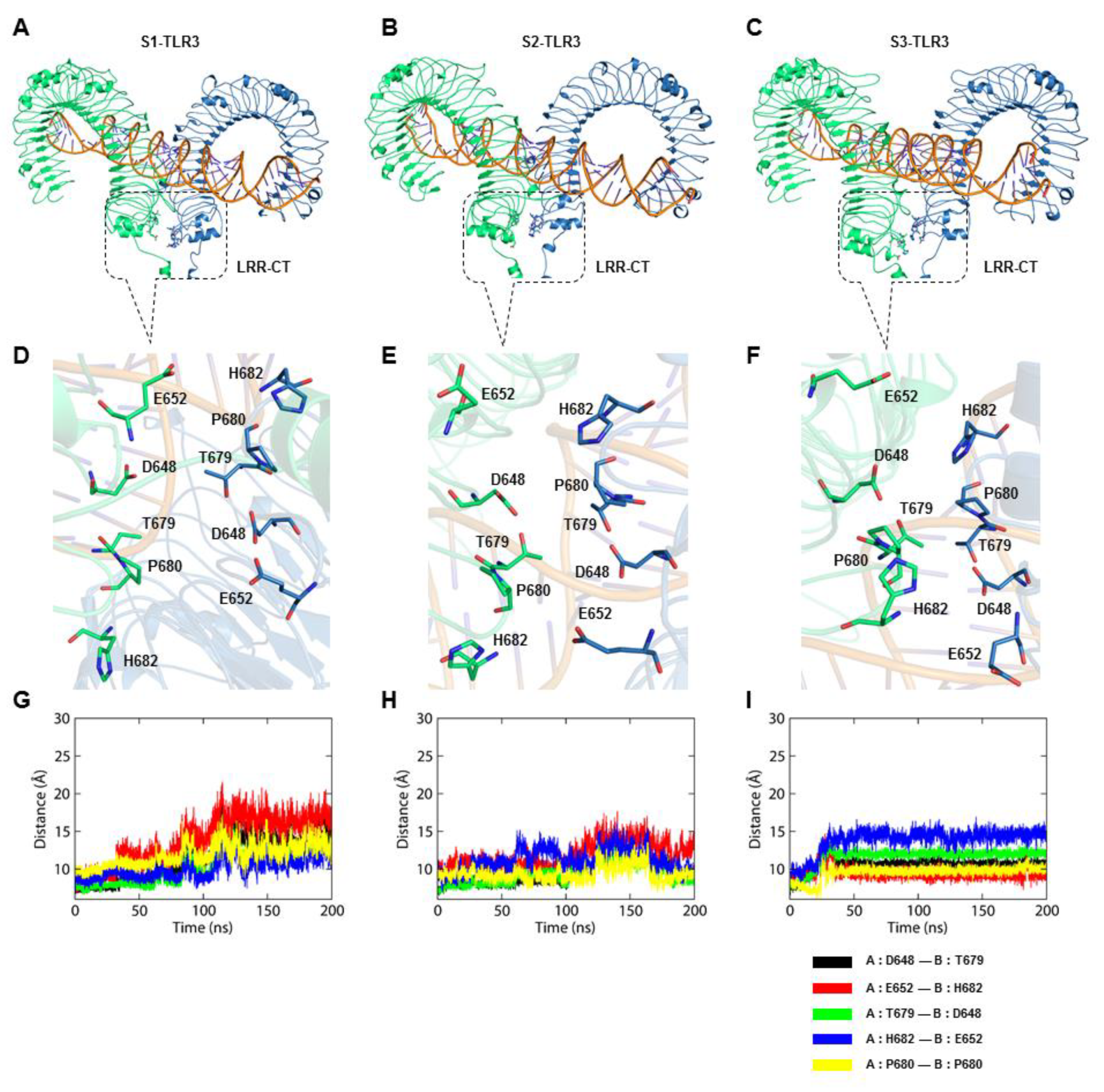
2.3. The S2-TLR3-dsRNA Complex Forms Relatively Steady Intermolecular Contacts within the Phospholipid Bilayer
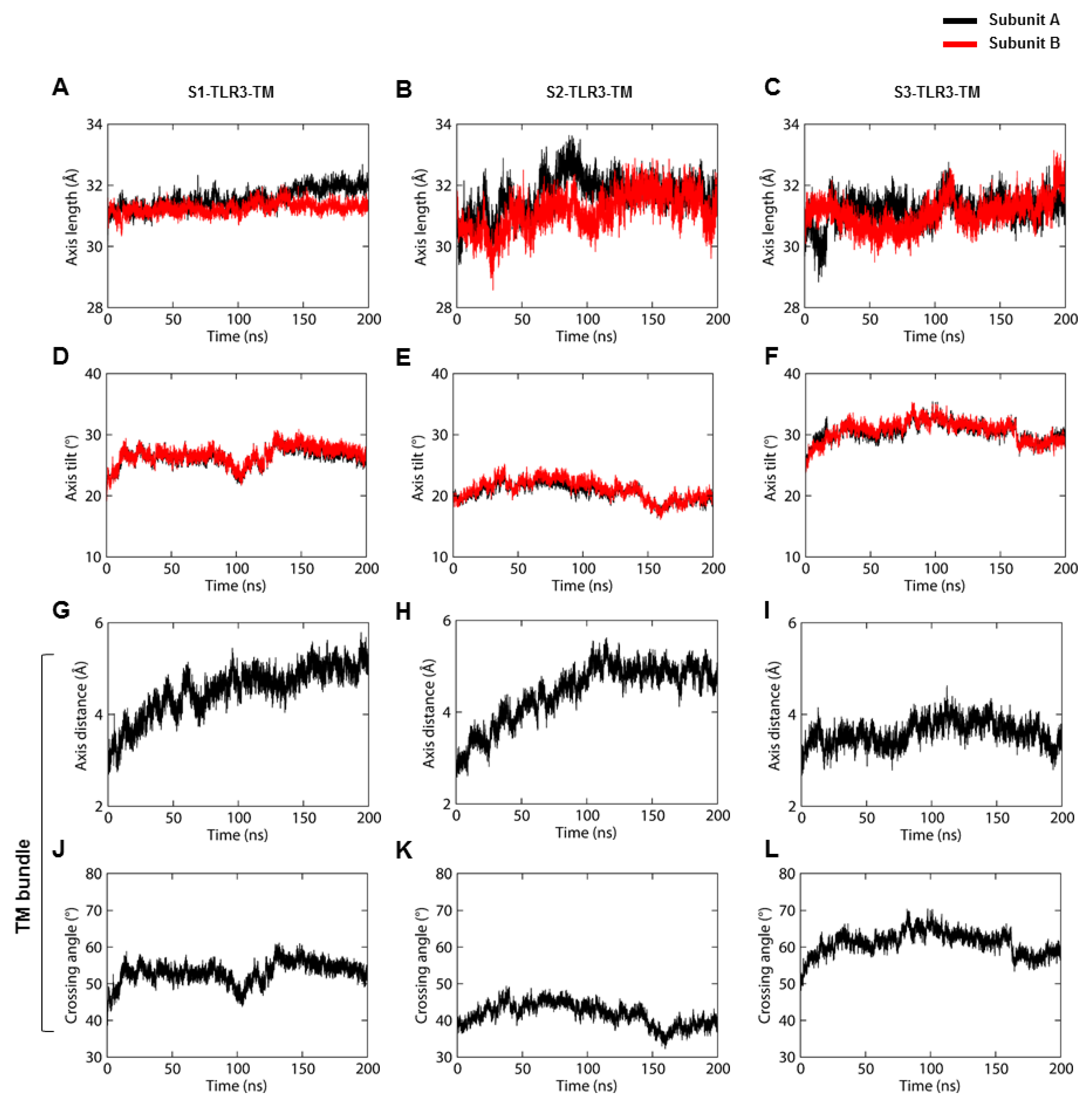
2.4. The Hydrophobic Mismatch Affects the Dynamics of TM Domains in the Membrane-Bound TLR3
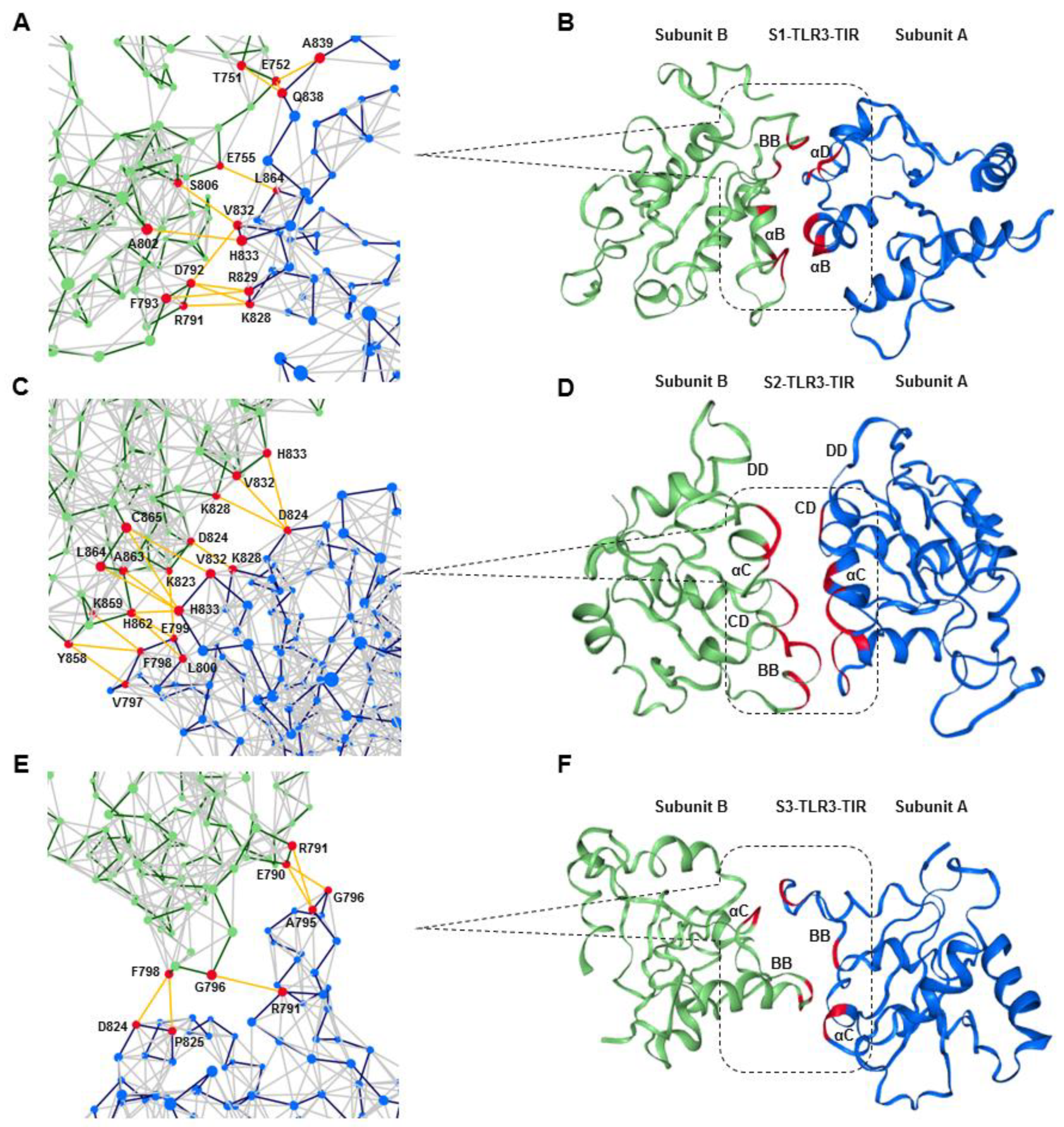
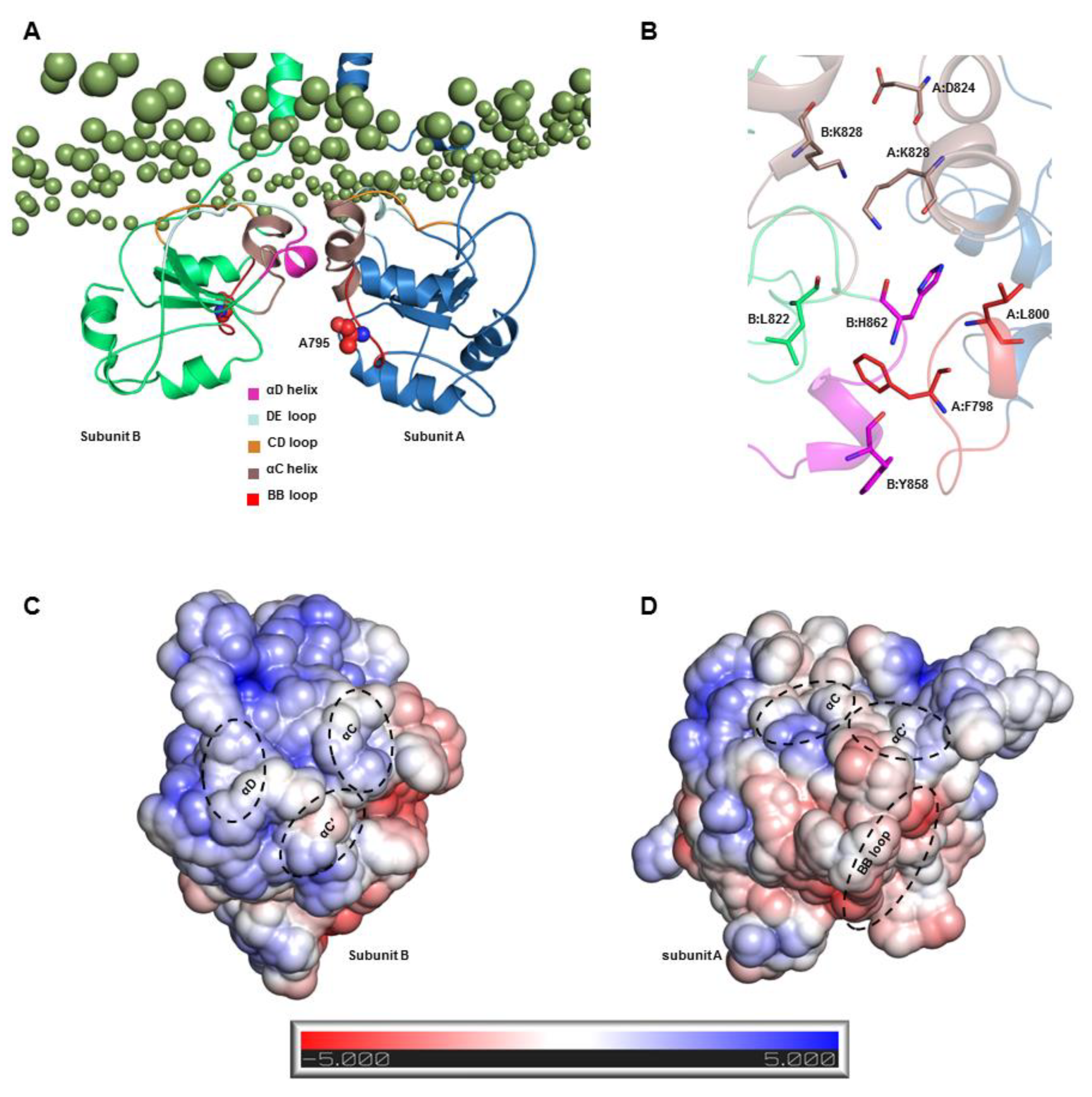
2.5. S2-TIR Domains Represent the Most Stable Dimer Interface of TLR3
3. Discussion
4. Materials and Methods
4.1. Construction of the Full-Length TLR3 Models
4.2. Construction of the Lipid Bilayers and the Packing of Lipids around the Modeled TLR3
4.3. MD Simulations of the TLR3-dsRNA Complexes
4.4. Electrostatic Potential Surface
4.5. Free Energy Landscape (FEL)
4.6. Model Validation
4.7. Binding Free Energy (BFE)
4.8. Protein Structure Network
4.9. Miscellaneous Properties
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, X.L.; Zhao, D.; Pan, L.N.; Huang, C.W.; Guo, L.J.; Lu, Q.; Wang, J. TLR3 ligand Poly IC Attenuates Reactive Astrogliosis and Improves Recovery of Rats after Focal Cerebral Ischemia. CNS Neurosci. Ther. 2015, 21, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Leonard, J.N.; Ghirlando, R.; Askins, J.; Bell, J.K.; Margulies, D.H.; Davies, D.R.; Segal, D.M. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. USA 2008, 105, 258–263. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004, 5, 987–995. [Google Scholar] [CrossRef]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003, 301, 640–643. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Botos, I.; Segal, D.M.; Davies, D.R. The structural biology of Toll-like receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef]
- Liu, L.; Botos, I.; Wang, Y.; Leonard, J.N.; Shiloach, J.; Segal, D.M.; Davies, D.R. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008, 320, 379–381. [Google Scholar] [CrossRef]
- Mineev, K.S.; Goncharuk, S.A.; Arseniev, A.S. Toll-like receptor 3 transmembrane domain is able to perform various homotypic interactions: An NMR structural study. FEBS Lett. 2014, 588, 3802–3807. [Google Scholar] [CrossRef]
- Roach, J.C.; Glusman, G.; Rowen, L.; Kaur, A.; Purcell, M.K.; Smith, K.D.; Hood, L.E.; Aderem, A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 9577–9582. [Google Scholar] [CrossRef] [PubMed]
- Pradere, J.P.; Dapito, D.H.; Schwabe, R.F. The Yin and Yang of Toll-like receptors in cancer. Oncogene 2014, 33, 3485–3495. [Google Scholar] [CrossRef] [PubMed]
- Hussein, W.M.; Liu, T.Y.; Skwarczynski, M.; Toth, I. Toll-like receptor agonists: A patent review (2011–2013). Expert Opin. Ther. Pat. 2014, 24, 453–470. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Lee, G.; Kim, S.G.; Choi, S. Toll-like receptor modulators: A patent review (2006-2010). Expert Opin. Ther. Pat. 2011, 21, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Perales-Linares, R.; Navas-Martin, S. Toll-like receptor 3 in viral pathogenesis: Friend or foe? Immunology 2013, 140, 153–167. [Google Scholar] [CrossRef]
- Keogh, B.; Parker, A.E. Toll-like receptors as targets for immune disorders. Trends Pharmacol. Sci. 2011, 32, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Clanchy, F.I.; Sacre, S.M. Modulation of toll-like receptor function has therapeutic potential in autoimmune disease. Expert Opin. Biol. Ther. 2010, 10, 1703–1716. [Google Scholar] [CrossRef]
- Matijevic, T.; Pavelic, J. Toll-like receptors: Cost or benefit for cancer? Curr. Pharm. Des. 2010, 16, 1081–1090. [Google Scholar] [CrossRef]
- Mullick, A.E.; Tobias, P.S.; Curtiss, L.K. Modulation of atherosclerosis in mice by Toll-like receptor 2. J. Clin. Investig. 2005, 115, 3149–3156. [Google Scholar] [CrossRef]
- Malia, T.J.; Obmolova, G.; Luo, J.; Teplyakov, A.; Sweet, R.; Gilliland, G.L. Crystallization of a challenging antigen-antibody complex: TLR3 ECD with three noncompeting Fabs. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Duffy, K.E.; Lamb, R.J.; San Mateo, L.R.; Jordan, J.L.; Canziani, G.; Brigham-Burke, M.; Korteweg, J.; Cunningham, M.; Beck, H.S.; Carton, J.; et al. Down modulation of human TLR3 function by a monoclonal antibody. Cell. Immunol. 2007, 248, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Takemura, N.; Kawasaki, T.; Kunisawa, J.; Sato, S.; Lamichhane, A.; Kobiyama, K.; Aoshi, T.; Ito, J.; Mizuguchi, K.; Karuppuchamy, T.; et al. Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nat. Commun. 2014, 5, 3492. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Wang, X.; Yin, H. Small-molecule inhibitors of the TLR3/dsRNA complex. J. Am. Chem. Soc. 2011, 133, 3764–3767. [Google Scholar] [CrossRef] [PubMed]
- Patra, M.C.; Choi, S. Recent progress in the development of Toll-like receptor (TLR) antagonists. Expert Opin. Ther. Pat. 2016, 26, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Xu, Y.; Zheng, Y.; Beg, A.A.; Tong, L. An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem. Biophys. Res. Commun. 2002, 299, 216–221. [Google Scholar] [CrossRef]
- Jang, T.H.; Park, H.H. Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR-TIR interaction for toll-like receptor signaling pathway. J. Mol. Biol. 2014, 426, 3305–3313. [Google Scholar] [CrossRef]
- Nyman, T.; Stenmark, P.; Flodin, S.; Johansson, I.; Hammarstrom, M.; Nordlund, P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J. Biol. Chem. 2008, 283, 11861–11865. [Google Scholar] [CrossRef]
- Patra, M.C.; Kwon, H.K.; Batool, M.; Choi, S. Computational Insight Into the Structural Organization of Full-Length Toll-Like Receptor 4 Dimer in a Model Phospholipid Bilayer. Front. Immunol. 2018, 9, 489. [Google Scholar] [CrossRef]
- Dave, P.C.; Tiburu, E.K.; Damodaran, K.; Lorigan, G.A. Investigating structural changes in the lipid bilayer upon insertion of the transmembrane domain of the membrane-bound protein phospholamban utilizing 31P and 2H solid-state NMR spectroscopy. Biophys. J. 2004, 86, 1564–1573. [Google Scholar] [CrossRef][Green Version]
- Ferreira, T.M.; Coreta-Gomes, F.; Ollila, O.H.; Moreno, M.J.; Vaz, W.L.; Topgaard, D. Cholesterol and POPC segmental order parameters in lipid membranes: Solid state 1H-13C NMR and MD simulation studies. Phys. Chem. Chem. Phys. PCCP 2013, 15, 1976–1989. [Google Scholar] [CrossRef]
- Jurkiewicz, P.; Cwiklik, L.; Vojtiskova, A.; Jungwirth, P.; Hof, M. Structure, dynamics, and hydration of POPC/POPS bilayers suspended in NaCl, KCl, and CsCl solutions. Biochim. Biophys. Acta 2012, 1818, 609–616. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Davies, D.R.; Segal, D.M. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J. Biol. Chem. 2010, 285, 36836–36841. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.H. Protein structure networks. Brief. Funct. Genom. 2012, 11, 469–478. [Google Scholar] [CrossRef] [PubMed]
- del Sol, A.; O’Meara, P. Small-world network approach to identify key residues in protein-protein interaction. Proteins 2005, 58, 672–682. [Google Scholar] [CrossRef]
- Chen, J.; Sawyer, N.; Regan, L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. A Publ. Protein Soc. 2013, 22, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Verstak, B.; Arnot, C.J.; Gay, N.J. An alanine-to-proline mutation in the BB-loop of TLR3 Toll/IL-1R domain switches signalling adaptor specificity from TRIF to MyD88. J. Immunol. 2013, 191, 6101–6109. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Mirabal, H.R.; Winther, J.R. Redox characteristics of the eukaryotic cytosol. Biochim. Et Biophys. Acta 2008, 1783, 629–640. [Google Scholar] [CrossRef]
- Hughes, M.M.; Lavrencic, P.; Coll, R.C.; Ve, T.; Ryan, D.G.; Williams, N.C.; Menon, D.; Mansell, A.; Board, P.G.; Mobli, M.; et al. Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E6480–E6489. [Google Scholar] [CrossRef]
- Lin, S.C.; Lo, Y.C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef]
- Funami, K.; Matsumoto, M.; Oshiumi, H.; Akazawa, T.; Yamamoto, A.; Seya, T. The cytoplasmic ‘linker region’ in Toll-like receptor 3 controls receptor localization and signaling. Int. Immunol. 2004, 16, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Mineev, K.S.; Goncharuk, S.A.; Goncharuk, M.V.; Volynsky, P.E.; Novikova, E.V.; Aresinev, A.S. Spatial structure of TLR4 transmembrane domain in bicelles provides the insight into the receptor activation mechanism. Sci. Rep. 2017, 7, 6864. [Google Scholar] [CrossRef] [PubMed]
- Bocharov, E.V.; Mineev, K.S.; Pavlov, K.V.; Akimov, S.A.; Kuznetsov, A.S.; Efremov, R.G.; Arseniev, A.S. Helix-helix interactions in membrane domains of bitopic proteins: Specificity and role of lipid environment. Biochim. Et Biophys. Acta. Biomembr. 2017, 1859, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Nishiya, T.; Kajita, E.; Miwa, S. Ligand-independent oligomerization of TLR4 regulated by a short hydrophobic region adjacent to the transmembrane domain. Biochem. Biophys. Res. Commun. 2006, 341, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Pohar, J.; Pirher, N.; Bencina, M.; Mancek-Keber, M.; Jerala, R. The ectodomain of TLR3 receptor is required for its plasma membrane translocation. PLOS ONE 2014, 9, e92391. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Obmolova, G.; Malia, T.J.; Wu, S.J.; Duffy, K.E.; Marion, J.D.; Bell, J.K.; Ge, P.; Zhou, Z.H.; Teplyakov, A.; et al. Lateral clustering of TLR3:dsRNA signaling units revealed by TLR3ecd:3Fabs quaternary structure. J. Mol. Biol. 2012, 421, 112–124. [Google Scholar] [CrossRef]
- Gosu, V.; Son, S.; Shin, D.; Song, K.D. Insights into the dynamic nature of the dsRNA-bound TLR3 complex. Sci. Rep. 2019, 9, 3652. [Google Scholar] [CrossRef]
- Botos, I.; Liu, L.; Wang, Y.; Segal, D.M.; Davies, D.R. The toll-like receptor 3:dsRNA signaling complex. Biochim. Et Biophys. Acta 2009, 1789, 667–674. [Google Scholar] [CrossRef]
- Koberlin, M.S.; Heinz, L.X.; Superti-Furga, G. Functional crosstalk between membrane lipids and TLR biology. Curr. Opin. Cell Biol. 2016, 39, 28–36. [Google Scholar] [CrossRef]
- Mobarak, E.; Haversen, L.; Manna, M.; Rutberg, M.; Levin, M.; Perkins, R.; Rog, T.; Vattulainen, I.; Boren, J. Glucosylceramide modifies the LPS-induced inflammatory response in macrophages and the orientation of the LPS/TLR4 complex in silico. Sci. Rep. 2018, 8, 13600. [Google Scholar] [CrossRef]
- Bugge, K.; Lindorff-Larsen, K.; Kragelund, B.B. Understanding single-pass transmembrane receptor signaling from a structural viewpoint-what are we missing? FEBS J. 2016, 283, 4424–4451. [Google Scholar] [CrossRef] [PubMed]
- Ruysschaert, J.M.; Lonez, C. Role of lipid microdomains in TLR-mediated signalling. Biochim. Biophys. Acta 2015, 1848, 1860–1867. [Google Scholar] [CrossRef] [PubMed]
- Sal-Man, N.; Gerber, D.; Bloch, I.; Shai, Y. Specificity in transmembrane helix-helix interactions mediated by aromatic residues. J. Biol. Chem. 2007, 282, 19753–19761. [Google Scholar] [CrossRef] [PubMed]
- Vyncke, L.; Bovijn, C.; Pauwels, E.; Van Acker, T.; Ruyssinck, E.; Burg, E.; Tavernier, J.; Peelman, F. Reconstructing the TIR Side of the Myddosome: A Paradigm for TIR-TIR Interactions. Structure 2016, 24, 437–447. [Google Scholar] [CrossRef]
- Mahita, J.; Sowdhamini, R. Probing subtle conformational changes induced by phosphorylation and point mutations in the TIR domains of TLR2 and TLR3. Proteins 2018, 86, 524–535. [Google Scholar] [CrossRef]
- Mahita, J.; Sowdhamini, R. Investigating the effect of key mutations on the conformational dynamics of toll-like receptor dimers through molecular dynamics simulations and protein structure networks. Proteins 2018, 86, 475–490. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Sarkar, S.N.; Elco, C.P.; Peters, K.L.; Chattopadhyay, S.; Sen, G.C. Two tyrosine residues of Toll-like receptor 3 trigger different steps of NF-kappa B activation. J. Biol. Chem. 2007, 282, 3423–3427. [Google Scholar] [CrossRef]
- Sarkar, S.N.; Peters, K.L.; Elco, C.P.; Sakamoto, S.; Pal, S.; Sen, G.C. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 2004, 11, 1060–1067. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Schmidt, T.H.; Kandt, C. LAMBADA and InflateGRO2: Efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012, 52, 2657–2669. [Google Scholar] [CrossRef] [PubMed]
- Cordomi, A.; Caltabiano, G.; Pardo, L. Membrane Protein Simulations Using AMBER Force Field and Berger Lipid Parameters. J. Chem. Theory Comput. 2012, 8, 948–958. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. A Publ. Protein Soc. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins 2007, 66, 778–795. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Open Source Drug Discovery, C.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, B.; Parekh, N. NAPS: Network Analysis of Protein Structures. Nucleic Acids Res. 2016, 44, W375–W382. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008, 3, 1213–1227. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]

| 1 ΔvdW | 2 Δelec | 3 Δps | 4 ΔSASA | 5 ΔGTotal | |
|---|---|---|---|---|---|
| Subunit A | |||||
| S1-TLR3-dsRNA | −62.13 ± 17.88 | −5754.51 ± 128.30 | 1267.30 ± 120.76 | −10.79 ± 1.91 | −4560.13 ± 65.34 |
| S2-TLR3-dsRNA | −65.09 ± 11.99 | −5744.62 ± 124.09 | 1170.13 ± 110.45 | −11.68 ± 4.13 | −4651.26 ± 68.97 |
| S3-TLR3-dsRNA | −55.04 ± 13.05 | −5236.47 ± 125.15 | 1256.26 ± 129.08 | −18.23 ± 2.03 | −4053.48 ± 66.08 |
| Subunit B | |||||
| S1-TLR3-dsRNA | −97.99 ± 14.01 | −5474.26 ± 104.82 | 1811.79 ± 37.17 | −17.72 ± 16.45 | −3778.18 ± 62.93 |
| S2-TLR3-dsRNA | −92.51 ± 18.44 | −5394.85 ± 111.91 | 1863.41 ± 47.51 | −14.43 ± 16.75 | −3638.38 ± 65.44 |
| S3-TLR3-dsRNA | −94.09 ± 19.72 | −5264.13 ± 119.56 | 1972.11 ± 75.21 | −17.17 ± 16.73 | −3403.28 ± 69.40 |
| TLR3 | dsRNA | TLR3 | dsRNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subunit | Residue | Subunit | Residue | Subunit | Residue | Subunit | Residue | |||
| * Dist. (Å) | Dist. (Å) | |||||||||
| S1-TLR3-ECD | ||||||||||
| A | H539 | 3.93 ± 1.17 | D | U22 | B | H539 | 6.69 ± 0.48 | C | G23 | |
| A | H539 | 4.02 ± 0.53 | D | C23 | B | H539 | 3.5 ± 0.27 | C | A22 | |
| A | H39 | 7.55 ± 1.55 | C | C7 | B | H39 | 10.0 ± 4.59 | D | C8 | |
| A | H60 | 3.71 ± 0.34 | C | C7 | B | H39 | 10.7 ± 2.93 | D | G7 | |
| A | H108 | 3.74 ± 0.35 | C | U5 | B | H60 | 6.9 ± 3.76 | D | G7 | |
| B | H108 | 4.46 ± 1.35 | D | C6 | ||||||
| S2-TLR3-ECD | ||||||||||
| A | H539 | 8.13 ± 0.42 | D | C23 | B | H539 | 4.12 ± 0.33 | C | A22 | |
| A | H39 | 2.79 ± 0.1 | C | C7 | B | H539 | 4.64 ± 0.78 | C | A21 | |
| A | H39 | 3.66 ± 1.2 | C | G6 | B | H39 | 3.83 ± 1.19 | D | C8 | |
| A | H60 | 6.83 ± 0.73 | C | G6 | B | H60 | 3.48 ± 0.24 | D | G7 | |
| A | H108 | 3.8 ± 0.47 | C | U5 | B | H108 | 3.6 ± 0.25 | D | C6 | |
| S3-TLR3-ECD | ||||||||||
| A | H539 | 7.03 ± 0.57 | D | C24 | B | H539 | 11.2 ± 1.44 | C | G23 | |
| A | H539 | 3.47 ± 0.26 | D | C23 | B | H539 | 5.75 ± 1.36 | C | A22 | |
| A | H39 | 12.73 ± 1.45 | C | C7 | B | H39 | 14.14 ± 2.36 | C | C34 | |
| A | H60 | 3.78 ± 1.1 | C | G6 | B | H39 | 12.68 ± 2.19 | C | A33 | |
| A | H108 | 3.89 ± 0.36 | C | U5 | B | H60 | 12.71 ± 2.27 | C | C34 | |
| B | H108 | 12.03 ± 2.31 | C | A35 |
| Subunit | Residue | * Dist. (Å) | Subunit | Residue | |
|---|---|---|---|---|---|
| S1-TLR3-TM | |||||
| A | F718 | 3.96 ± 0.41 | B | I715 | |
| A | I715 | 3.74 ± 0.31 | B | F718 | |
| A | F706 | 7.4 ± 1.19 | B | F706 | |
| A | M707 | 7.57 ± 2.71 | B | F706 | |
| A | F706 | 4.30 ± 0.91 | B | M707 | |
| A | L722 | 3.77 ± 0.60 | B | L722 | |
| S2-TLR3-TM | |||||
| A | F718 | 5.21 ± 1.03 | B | F718 | |
| A | L714 | 7.61 ± 1.07 | B | I715 | |
| A | L714 | 4.39 ± 1.15 | B | L714 | |
| A | F706 | 7.80 ± 1.79 | B | F706 | |
| A | M707 | 3.33 ± 0.56 | B | F706 | |
| A | F706 | 5.15 ± 1.93 | B | M707 | |
| A | L722 | 3.22 ± 0.32 | B | L722 | |
| S3-TLR3-TM | |||||
| A | F718 | 4.03 ± 0.48 | B | I715 | |
| A | I715 | 7.09 ± 0.59 | B | L714 | |
| A | F706 | 5.02 ± 1.68 | B | F706 | |
| A | M707 | 7.11 ± 1.56 | B | F706 | |
| A | F706 | 11.81 ± 1.25 | B | M707 | |
| A | L722 | 8.68 ± 1.02 | B | L722 |
| System | 1 ΔvdW | 2 Δelec | 3 Δps | 4 ΔSASA | 5 ΔGTotal |
|---|---|---|---|---|---|
| S1-TM | −115.49 ± 17.31 | 9.24 ± 2.85 | 46.47 ± 10.1 | −15.55 ± 2.23 | −75.33 ± 12.85 |
| S2-TM | −183.38 ± 21.29 | 11.74 ± 4.34 | 75.14 ± 12.3 | −22.2 ± 1.8 | −118.7 ± 22.32 |
| S3-TM | −132.91 ± 15.77 | 7.31 ± 2.5 | 55.29 ± 16.25 | −17.62 ± 1.72 | −87.93 ± 18.4 |
| 6 S4-TM | −147.03 ± 15.38 | 9.29 ± 4.11 | 59.81 ± 15.56 | −21.87 ± 2.3 | −99.8 ± 15.02 |
| Domain | BSA (Ų) | Energy (kcal mol−1) |
|---|---|---|
| S1-TLR3-TIR | 722.22 ± 15.23 | −3.48 ± 0.29 |
| S2- TLR3-TIR | 885.56 ± 14.2 | −5.77 ± 0.25 |
| S3- TLR3-TIR | 480.45 ± 16.12 | −2.64 ± 0.63 |
| System | 1 ΔvdW | 2 Δelec | 3 Δps | 4 ΔSASA | 5 ΔGTotal |
|---|---|---|---|---|---|
| S1-TLR3-TIR | −289.06 ± 12.06 | −459.99 ± 19.11 | 958.13 ± 12.14 | −38.94 ± 3.72 | 170.44 ± 14.24 |
| S2-TLR3-TIR | −223.41 ± 19.12 | −509.07 ± 19.38 | 643.17 ± 18.59 | −31.00 ± 5.29 | −120.31 ± 17.44 |
| S3-TLR3-TIR | −155.66 ± 12.29 | −123.57 ± 19.45 | 478.11 ± 21.03 | −20.56 ± 4.34 | 178.32 ± 17.35 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patra, M.C.; Batool, M.; Haseeb, M.; Choi, S. A Computational Probe into the Structure and Dynamics of the Full-Length Toll-Like Receptor 3 in a Phospholipid Bilayer. Int. J. Mol. Sci. 2020, 21, 2857. https://doi.org/10.3390/ijms21082857
Patra MC, Batool M, Haseeb M, Choi S. A Computational Probe into the Structure and Dynamics of the Full-Length Toll-Like Receptor 3 in a Phospholipid Bilayer. International Journal of Molecular Sciences. 2020; 21(8):2857. https://doi.org/10.3390/ijms21082857
Chicago/Turabian StylePatra, Mahesh Chandra, Maria Batool, Muhammad Haseeb, and Sangdun Choi. 2020. "A Computational Probe into the Structure and Dynamics of the Full-Length Toll-Like Receptor 3 in a Phospholipid Bilayer" International Journal of Molecular Sciences 21, no. 8: 2857. https://doi.org/10.3390/ijms21082857
APA StylePatra, M. C., Batool, M., Haseeb, M., & Choi, S. (2020). A Computational Probe into the Structure and Dynamics of the Full-Length Toll-Like Receptor 3 in a Phospholipid Bilayer. International Journal of Molecular Sciences, 21(8), 2857. https://doi.org/10.3390/ijms21082857






