MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes
Abstract
1. Introduction
2. Results
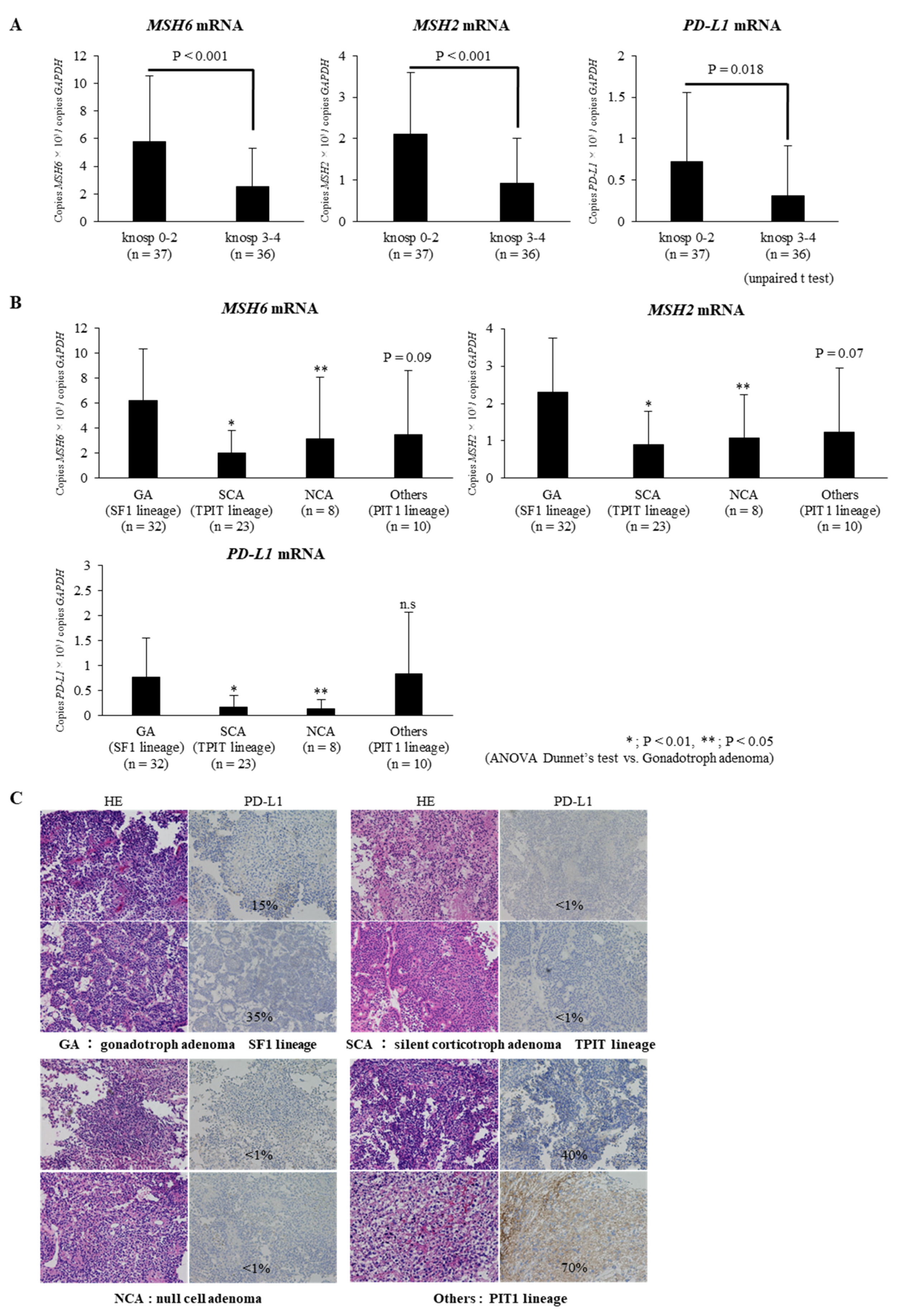
2.1. Expressions of MSH6/2 and PD-L1 mRNA Are Significantly Lower in Invasive NFPAs Than in Non-Invasive NFPAs
2.2. Expressions of MSH6/2 and PD-L1 mRNA Are Significantly Lower in Silent Corticotroph Adenomas and Null Cell Adenomas Than in Gonadotroph Adenomas
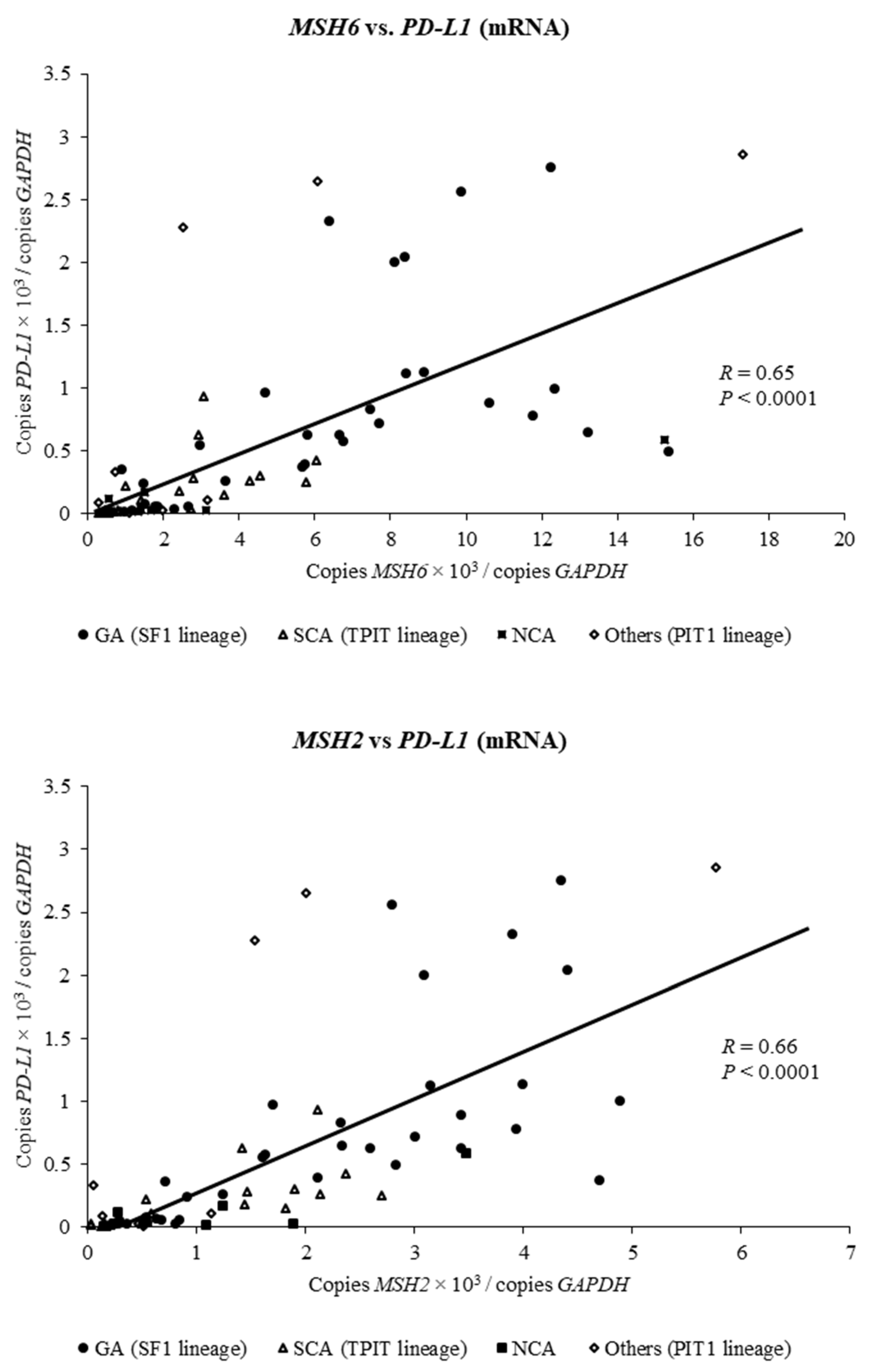
2.3. Expression of MSH6 and MSH2 mRNA Is Positively Associated with Expression of PD-L1 mRNA in Clinically Nonfunctioning Pituitary Adenomas
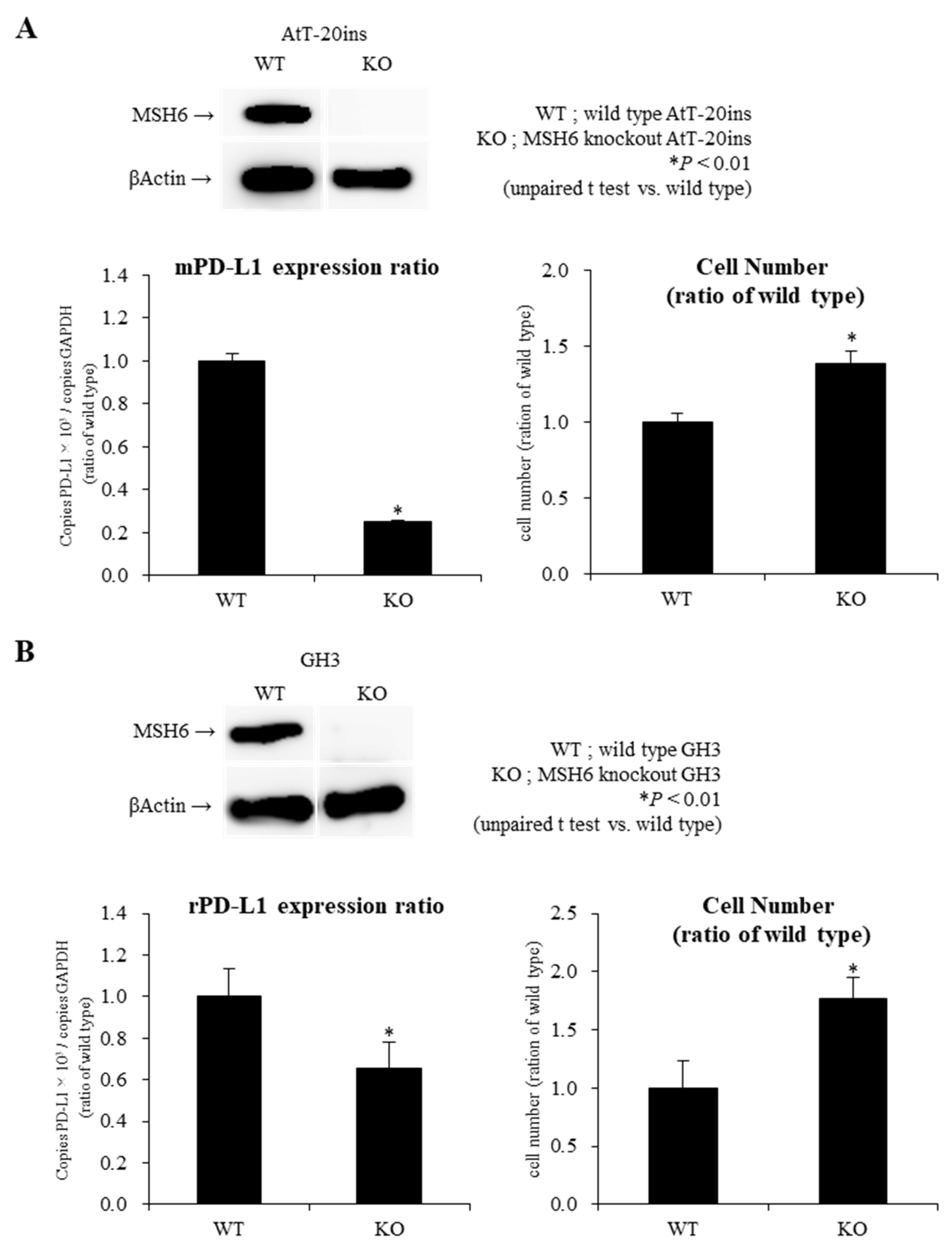
2.4. Knockout of MSH6 Expression by Cells by Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 (CRISPR-Cas9) System Reduces PD-L1 mRNA Expression with Promotion of Cell Proliferation in AtT-20ins and GH3 Cells
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Patient Samples
4.3. Immunohistochemistry (IHC)
4.4. Measurement of MSH6/2 Gene and PD-L1 Gene mRNA Expression
4.5. Generation of MSH6 Knockout AtT-20ins and GH3 Cells by Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Genome Editing Technique
4.6. Cell Culture and Treatment Schedule
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIP | aryl hydrocarbon receptor-interacting protein |
| ANOVA | analysis of variance |
| ATR-Chk1 | Ataxia telangiectasia and Rad3-related-checkpoint kinase 1 |
| GA | gonadotroph adenoma |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| ICIs | immune checkpoint inhibitors |
| LI | labeling index |
| MMR | mismatch repair |
| mRNA | messenger RNA |
| MSH2/6 | mutS homolog 2/6 |
| NCA | null cell adenoma |
| NF | nonfunctioning |
| PA | pituitary adenoma |
| PCR | polymerase chain reaction |
| PD-L1 | programmed cell death 1 ligand 1 |
| Pit-NETs | pituitary neuroendocrine tumors |
| PIT1 | pituitary transcription factor 1 |
| qRT-PCR | quantitative reverse transcriptase-polymerase chain reaction |
| SCA | silent corticotroph adenoma |
| SF1 | steroidogenic factor-1 |
| TPIT | t-box pituitary transcription factor |
| USP8 | ubiquitin specific protease 8 |
| WHO | World Health Organization |
References
- Asa, S.L.; Casar-Borota, O.; Chanson, P.; Delgrange, E.; Earls, P.; Ezzat, S.; Grossman, A.; Ikeda, H.; Inoshita, N.; Karavitaki, N.; et al. From pituitary adenoma to pituitary neuroendocrine tumor (PitNET): An International Pituitary Pathology Club proposal. Endocr. Relat. Cancer 2017, 24, C5–C8. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.F.; Rixhon, M.; Adam, C.; Dempegioti, A.; Tichomirowa, M.A.; Beckers, A. High prevalence of pituitary adenomas: A cross-sectional study in the province of Liege, Belgium. J. Clin. Endocrinol. Metab. 2006, 91, 4769–4775. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, S.; Asa, S.L. Mechanisms of disease: The pathogenesis of pituitary tumors. Nat. Clin. Pract. Endocrinol. Metab. 2006, 2, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Melmed, S. Mechanisms for pituitary tumorigenesis: The plastic pituitary. J. Clin. Investig. 2003, 112, 1603–1618. [Google Scholar] [CrossRef] [PubMed]
- Mete, O.; Gomez-Hernandez, K.; Kucharczyk, W.; Ridout, R.; Zadeh, G.; Gentili, F.; Ezzat, S.; Asa, S.L. Silent subtype 3 pituitary adenomas are not always silent and represent poorly differentiated monomorphous plurihormonal Pit-1 lineage adenomas. Mod. Pathol. 2016, 29, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, H.; Inoshita, N.; Mete, O.; Asa, S.L.; Hayashi, K.; Takeshita, A.; Fukuhara, N.; Yamaguchi-Okada, M.; Takeuchi, Y.; Yamada, S. The Complementary Role of Transcription Factors in the Accurate Diagnosis of Clinically Nonfunctioning Pituitary Adenomas. Endocr. Pathol. 2015, 26, 349–355. [Google Scholar] [CrossRef]
- Nishioka, H.; Inoshita, N.; Sano, T.; Fukuhara, N.; Yamada, S. Correlation between histological subtypes and MRI findings in clinically nonfunctioning pituitary adenomas. Endocr. Pathol. 2012, 23, 151–156. [Google Scholar] [CrossRef]
- Yamada, S.; Ohyama, K.; Taguchi, M.; Takeshita, A.; Morita, K.; Takano, K.; Sano, T. A study of the correlation between morphological findings and biological activities in clinically nonfunctioning pituitary adenomas. Neurosurgery 2007, 61, 580–585. [Google Scholar] [CrossRef]
- Drummond, J.; Roncaroli, F.; Grossman, A.B.; Korbonits, M. Clinical and Pathological Aspects of Silent Pituitary Adenomas. J. Clin. Endocrinol. Metab. 2019, 104, 2473–2489. [Google Scholar] [CrossRef]
- Vandeva, S.; Daly, A.F.; Petrossians, P.; Zacharieva, S.; Beckers, A. GENETICS IN ENDOCRINOLOGY: Somatic and germline mutations in the pathogenesis of pituitary adenomas. Eur. J. Endocrinol. 2019. [Google Scholar] [CrossRef]
- Uraki, S.; Ariyasu, H.; Doi, A.; Furuta, H.; Nishi, M.; Sugano, K.; Inoshita, N.; Nakao, N.; Yamada, S.; Akamizu, T. Atypical pituitary adenoma with MEN1 somatic mutation associated with abnormalities of DNA mismatch repair genes; MLH1 germline mutation and MSH6 somatic mutation. Endocr. J. 2017, 64, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Uraki, S.; Ariyasu, H.; Doi, A.; Kawai, S.; Takeshima, K.; Morita, S.; Fukai, J.; Fujita, K.; Furuta, H.; Nishi, M.; et al. Reduced Expression of Mismatch Repair Genes MSH6/MSH2 Directly Promotes Pituitary Tumor Growth via the ATR-Chk1 Pathway. J. Clin. Endocrinol. Metab. 2018, 103, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.H.; Arvis, C.D.; Ahn, M.J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef]
- Lin, A.L.; Jonsson, P.; Tabar, V.; Yang, T.J.; Cuaron, J.; Beal, K.; Cohen, M.; Postow, M.; Rosenblum, M.; Shia, J.; et al. Marked Response of a Hypermutated ACTH-Secreting Pituitary Carcinoma to Ipilimumab and Nivolumab. J. Clin. Endocrinol. Metab. 2018, 103, 3925–3930. [Google Scholar] [CrossRef]
- Knosp, E.; Steiner, E.; Kitz, K.; Matula, C. Pituitary adenomas with invasion of the cavernous sinus space: A magnetic resonance imaging classification compared with surgical findings. Neurosurgery 1993, 33, 610–618. [Google Scholar] [CrossRef]
- Lloyd, R.; Osamura, R.; Klöppel, G.; Rosai, J. (Eds.) World Health Organization Classification of Tumours of Endocrine Organs, 4th ed.; IARC Publication: Lyon, France, 2017; Volume 10. [Google Scholar]
- Inoshita, N.; Nishioka, H. The 2017 WHO classification of pituitary adenoma: Overview and comments. Brain Tumor Pathol. 2018, 35, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, H.; Inoshita, N. New WHO classification of pituitary adenomas (4th edition): Assessment of pituitary transcription factors and the prognostic histological factors. Brain Tumor Pathol. 2018, 35, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Hewish, M.; Lord, C.J.; Martin, S.A.; Cunningham, D.; Ashworth, A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat. Rev. Clin. Oncol. 2010, 7, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Kober, P.; Zbijewska, J.; Kunicki, J.; Bonicki, W. Microsatellite instability analysis in pituitary adenomas. Neuroendocrinol. Lett. 2015, 36, 511–514. [Google Scholar]
- García-Martínez, A.; Sottile, J.; Sánchez-Tejada, L.; Fajardo, C.; Cámara, R.; Lamas, C.; Barberá, V.M.; Picó, A. DNA Methylation of Tumor Suppressor Genes in Pituitary Neuroendocrine Tumors. J. Clin. Endocrinol. Metab. 2019, 104, 1272–1282. [Google Scholar] [CrossRef]
- Bengtsson, D.; Joost, P.; Aravidis, C.; Askmalm Stenmark, M.; Backman, A.S.; Melin, B.; von Salomé, J.; Zagoras, T.; Gebre-Medhin, S.; Burman, P. Corticotroph Pituitary Carcinoma in a Patient With Lynch Syndrome (LS) and Pituitary Tumors in a Nationwide LS Cohort. J. Clin. Endocrinol. Metab. 2017, 102, 3928–3932. [Google Scholar] [CrossRef]
- Serioli, S.; Doglietto, F.; Fiorindi, A.; Biroli, A.; Mattavelli, D.; Buffoli, B.; Ferrari, M.; Cornali, C.; Rodella, L.; Maroldi, R.; et al. Pituitary Adenomas and Invasiveness from Anatomo-Surgical, Radiological, and Histological Perspectives: A Systematic Literature Review. Cancers 2019, 4, 1936. [Google Scholar] [CrossRef]
- Rosenbaum, M.W.; Bledsoe, J.R.; Morales-Oyarvide, V.; Huynh, T.G.; Mino-Kenudson, M. PD-L1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod. Pathol. 2016, 29, 1104–1112. [Google Scholar] [CrossRef]
- Sloan, E.A.; Ring, K.L.; Willis, B.C.; Modesitt, S.C.; Mills, A.M. PD-L1 Expression in Mismatch Repair-deficient Endometrial Carcinomas, Including Lynch Syndrome-associated and MLH1 Promoter Hypermethylated Tumors. Am. J. Surg. Pathol. 2017, 41, 326–333. [Google Scholar] [CrossRef]
- Mills, A.M.; Dill, E.A.; Moskaluk, C.A.; Dziegielewski, J.; Bullock, T.N.; Dillon, P.M. The Relationship Between Mismatch Repair Deficiency and PD-L1 Expression in Breast Carcinoma. Am. J. Surg. Pathol. 2018, 42, 183–191. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Bi, W.L.; Greenwald, N.F.; Du, Z.; Agar, N.Y.; Kaiser, U.B.; Woodmansee, W.W.; Reardon, D.A.; Freeman, G.J.; Fecci, P.E.; et al. Increased expression of programmed death ligand 1 (PD-L1) in human pituitary tumors. Oncotarget 2016, 7, 76565–76576. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Wang, T.J.; Yang, Y.K.; Yao, K.; Li, Z.; Li, Y.M.; Yan, C.X. The expression profile of PD-L1 and CD8+ lymphocyte in pituitary adenomas indicating for immunotherapy. J. Neurooncol. 2018, 139, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.E.; Shen, Z.J.; Kanchwala, M.; Xing, C.; Filatenkov, A.; Shang, P.; Barnett, S.; Abedin, Z.; Malter, J.S.; Raisanen, J.M.; et al. Aggressive Behavior in Silent Subtype III Pituitary Adenomas May Depend on Suppression of Local Immune Response: A Whole Transcriptome Analysis. J. Neuropathol. Exp. Neurol. 2017, 76, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Akbay, E.A.; Koyama, S.; Carretero, J.; Altabef, A.; Tchaicha, J.H.; Christensen, C.L.; Mikse, O.R.; Cherniack, A.D.; Beauchamp, E.M.; Pugh, T.J.; et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013, 3, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Azuma, K.; Kawahara, A.; Hattori, S.; Iwama, E.; Tanizaki, J.; Harada, T.; Matsumoto, K.; Takayama, K.; Takamori, S.; et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 4014–4021. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Morita, S.; Villalta, S.A.; Feldman, H.C.; Register, A.C.; Rosenthal, W.; Hoffmann-Petersen, I.T.; Mehdizadeh, M.; Ghosh, R.; Wang, L.; Colon-Negron, K.; et al. Targeting ABL-IRE1α Signaling Spares ER-Stressed Pancreatic β Cells to Reverse Autoimmune Diabetes. Cell Metab. 2017, 25, 883–897. [Google Scholar] [CrossRef]
| Pathological Phenotype | Number |
| Gonadotroph adenoma | 32 |
| Silent corticotroph adenoma | 23 |
| Silent thyrotroph adenoma | 2 |
| Silent lactotroph adenoma | 4 |
| Silent plurihormonal (PRL, GH) adenoma | 4 |
| Null cell adenoma | 8 |
| Total | 73 |
| Clinical, Radiological, and Histopathological Data | |
| Male/Female | 36/37 |
| Age | 52.2 ± 15.7 years * |
| Micro/Macro | 0/73 |
| MIB-1 LI (%) | 1.6% ** (0.1–4.1) |
| <3%: 65, ≧3%: 8 | |
| Knosp grade | grade0: 3, grade1: 19, grade2: 15, grade3: 22, grade4: 14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uraki, S.; Ariyasu, H.; Doi, A.; Takeshima, K.; Morita, S.; Inaba, H.; Furuta, H.; Fukuhara, N.; Inoshita, N.; Nishioka, H.; et al. MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes. Int. J. Mol. Sci. 2020, 21, 2831. https://doi.org/10.3390/ijms21082831
Uraki S, Ariyasu H, Doi A, Takeshima K, Morita S, Inaba H, Furuta H, Fukuhara N, Inoshita N, Nishioka H, et al. MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes. International Journal of Molecular Sciences. 2020; 21(8):2831. https://doi.org/10.3390/ijms21082831
Chicago/Turabian StyleUraki, Shinsuke, Hiroyuki Ariyasu, Asako Doi, Ken Takeshima, Shuhei Morita, Hidefumi Inaba, Hiroto Furuta, Noriaki Fukuhara, Naoko Inoshita, Hiroshi Nishioka, and et al. 2020. "MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes" International Journal of Molecular Sciences 21, no. 8: 2831. https://doi.org/10.3390/ijms21082831
APA StyleUraki, S., Ariyasu, H., Doi, A., Takeshima, K., Morita, S., Inaba, H., Furuta, H., Fukuhara, N., Inoshita, N., Nishioka, H., Nakao, N., Yamada, S., & Akamizu, T. (2020). MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes. International Journal of Molecular Sciences, 21(8), 2831. https://doi.org/10.3390/ijms21082831





