A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization
Abstract
1. Introduction
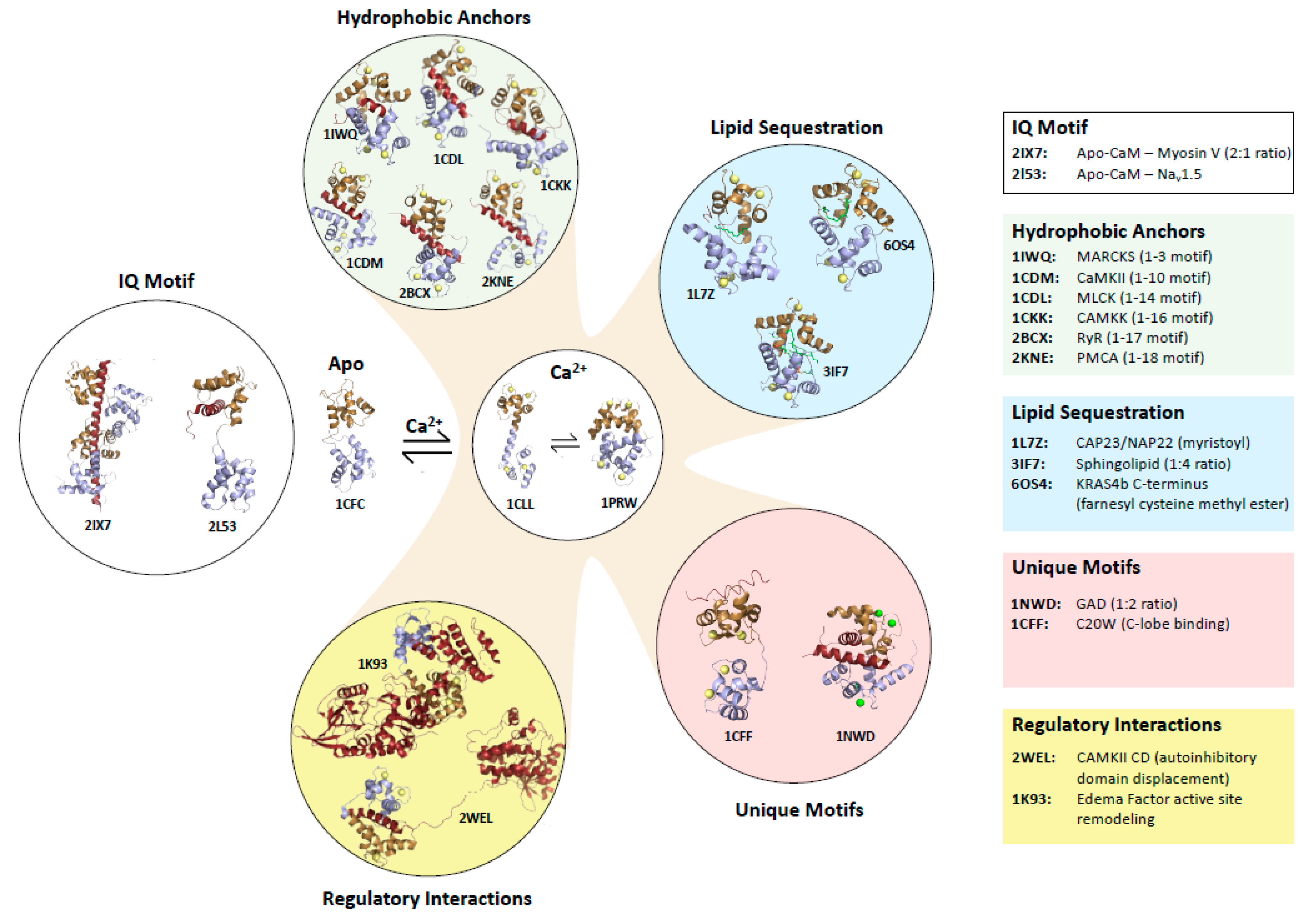
1.1. Target Protein Binding Motifs of Ca2+-Bound and apo-CaM
1.2. Mechanisms by Which Ca2+-CaM Regulates Target Proteins
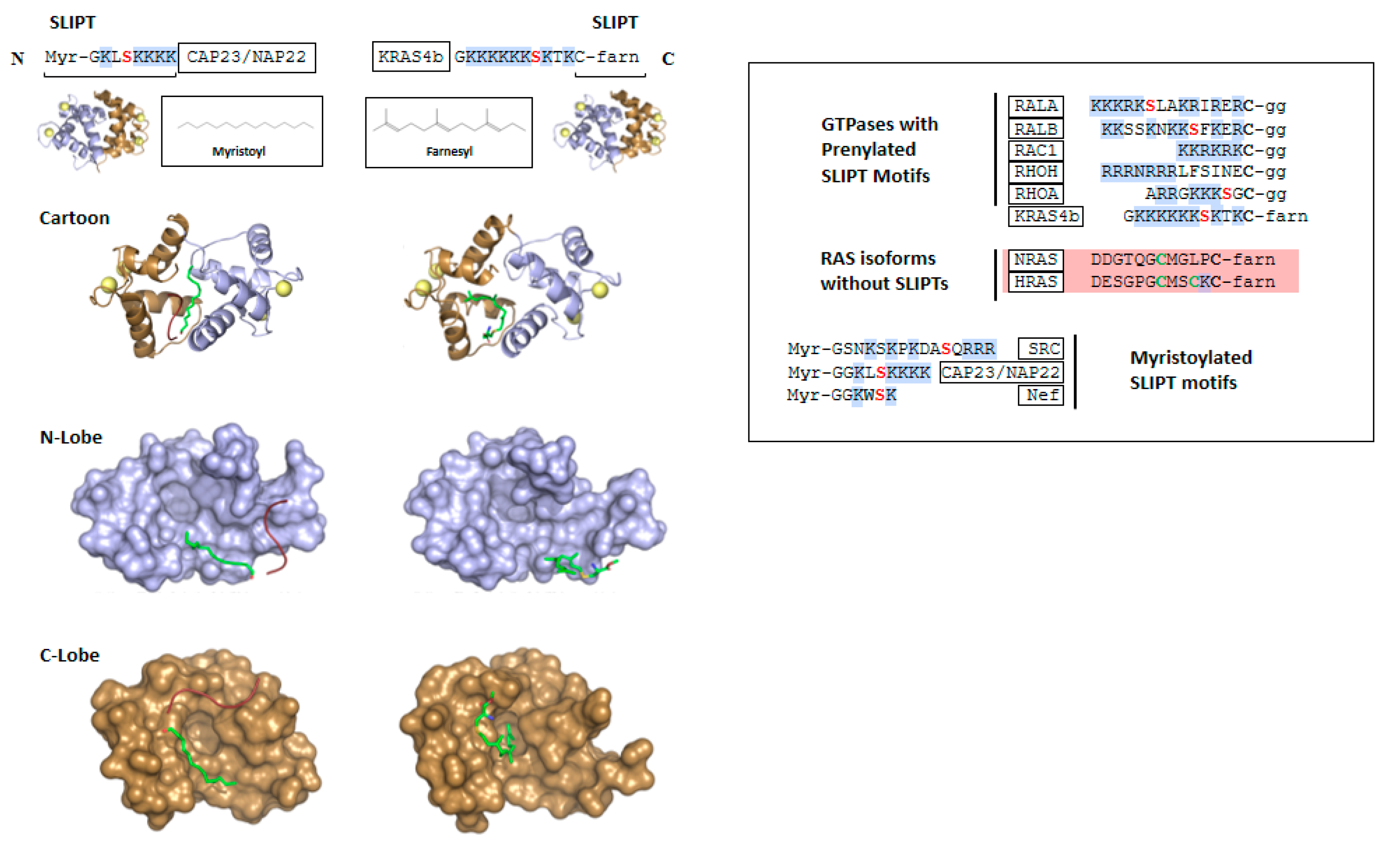
2. CaM Binding to Myristoylated Proteins
3. KRAS4b
Structural Characterization of CaM Binding to KRAS4b
4. Comparison of CaM Bound to Myristoylated versus Farnesylated Moieties
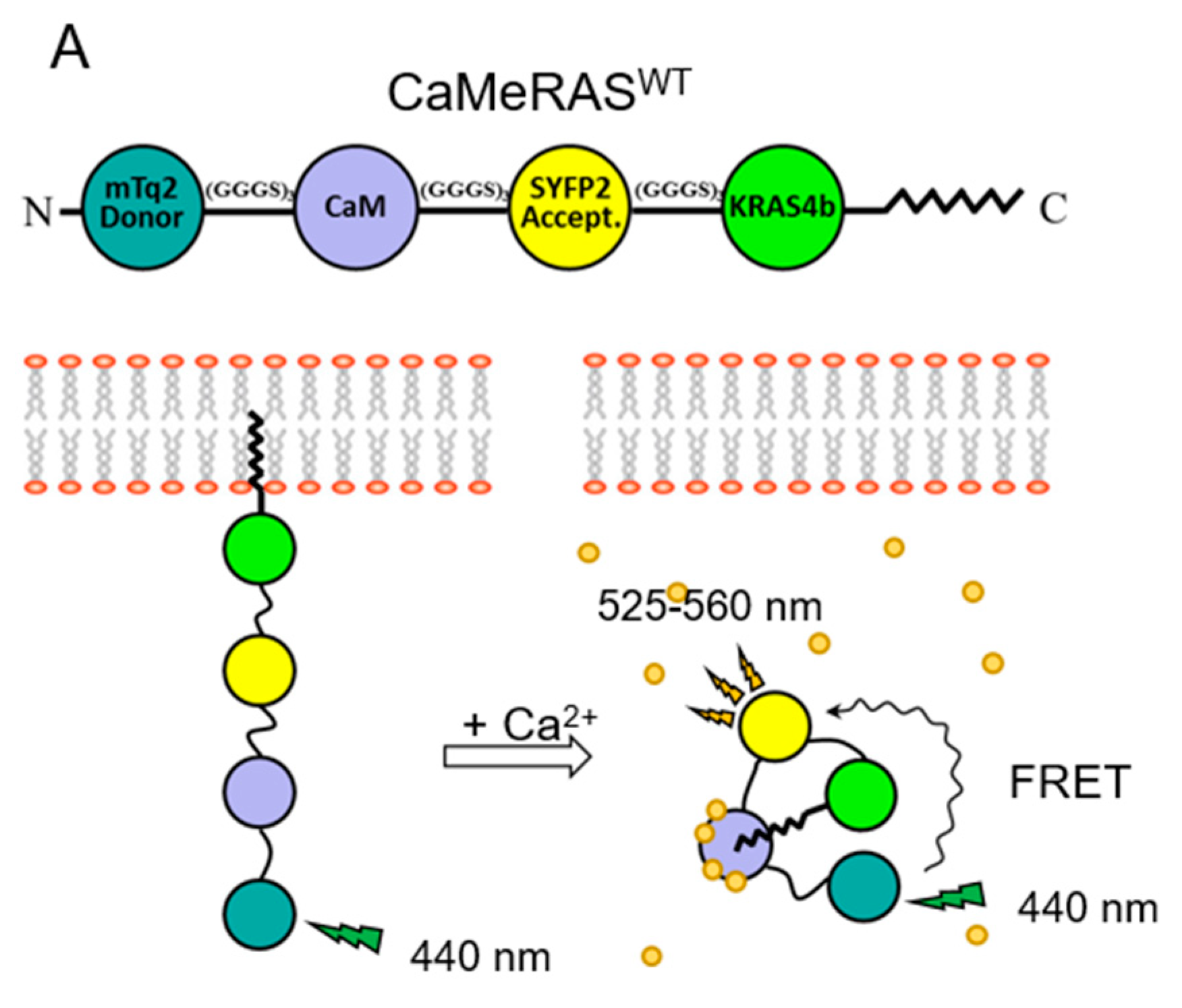
5. CaM Regulates KRAS4b Localization in Cells
6. Other RAS Isoforms and GTPases
7. A Plant CaM Isoform with a SLIPT
8. Regulation of CaM-Lipid Interactions by Phosphorylation of SLIPTs
9. Other Lipo-Peptide Sequestering Proteins
10. Complex Interplay between CaM and Lipidated Targets, and Crosstalk between Ca2+ and Oncogenic Signaling
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tidow, H.; Nissen, P. Structural diversity of calmodulin binding to its target sites. FEBS J. 2013, 280, 5551–5565. [Google Scholar] [CrossRef]
- Zhang, M.; Tanaka, T.; Ikura, M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995, 2, 758–767. [Google Scholar] [CrossRef]
- Kuboniwa, H.; Tjandra, N.; Grzesiek, S.; Ren, H.; Klee, C.B.; Bax, A. Solution structure of calcium-free calmodulin. Nat. Struct. Biol. 1995, 2, 768–776. [Google Scholar] [CrossRef]
- Vogel, H.J.; Zhang, M. Protein engineering and NMR studies of calmodulin. Mol. Cell. Biochem. 1995, 149–150, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Ames, J.B. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: Two ways to promote multifunctionality. Proc. Natl. Acad. Sci. USA 2006, 103, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, A.; Taglialatela, M.; Bernardo-Seisdedos, G.; Alaimo, A.; Agirre, J.; Alberdi, A.; Gomis-Perez, C.; Soldovieri, M.V.; Ambrosino, P.; Malo, C.; et al. The ever changing moods of calmodulin: How structural plasticity entails transductional adaptability. J. Mol. Biol. 2014, 426, 2717–2735. [Google Scholar] [CrossRef] [PubMed]
- Hoeflich, K.P.; Ikura, M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 2002, 108, 739–742. [Google Scholar] [CrossRef]
- Crivici, A.; Ikura, M. Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 85–116. [Google Scholar] [CrossRef]
- Yamniuk, A.P.; Vogel, H.J. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004, 27, 33–57. [Google Scholar] [CrossRef]
- Rhoads, A.R.; Friedberg, F. Sequence motifs for calmodulin recognition. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 1997, 11, 331–340. [Google Scholar] [CrossRef]
- Ikura, M.; Clore, G.M.; Gronenborn, A.M.; Zhu, G.; Klee, C.B.; Bax, A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 1992, 256, 632–638. [Google Scholar] [CrossRef]
- Meador, W.E.; Means, A.R.; Quiocho, F.A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 1992, 257, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Meador, W.E.; Means, A.R.; Quiocho, F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 1993, 262, 1718–1721. [Google Scholar] [CrossRef] [PubMed]
- Osawa, M.; Tokumitsu, H.; Swindells, M.B.; Kurihara, H.; Orita, M.; Shibanuma, T.; Furuya, T.; Ikura, M. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 1999, 6, 819–824. [Google Scholar] [PubMed]
- Maximciuc, A.A.; Putkey, J.A.; Shamoo, Y.; Mackenzie, K.R. Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure 2006, 14, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Juranic, N.; Atanasova, E.; Filoteo, A.G.; Macura, S.; Prendergast, F.G.; Penniston, J.T.; Strehler, E.E. Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif. J. Biol. Chem. 2010, 285, 4015–4024. [Google Scholar] [CrossRef]
- Ishida, H.; Vogel, H.J. The solution structure of a plant calmodulin and the CaM-binding domain of the vacuolar calcium-ATPase BCA1 reveals a new binding and activation mechanism. J. Biol. Chem. 2010, 285, 38502–38510. [Google Scholar] [CrossRef]
- Yamauchi, E.; Nakatsu, T.; Matsubara, M.; Kato, H.; Taniguchi, H. Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca2+-calmodulin. Nat. Struct. Biol. 2003, 10, 226–231. [Google Scholar] [CrossRef]
- Yap, K.L.; Yuan, T.; Mal, T.K.; Vogel, H.J.; Ikura, M. Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J. Mol. Biol. 2003, 328, 193–204. [Google Scholar] [CrossRef]
- Dunlap, T.B.; Guo, H.F.; Cook, E.C.; Holbrook, E.; Rumi-Masante, J.; Lester, T.E.; Colbert, C.L.; Vander Kooi, C.W.; Creamer, T.P. Stoichiometry of the calcineurin regulatory domain-calmodulin complex. Biochemistry 2014, 53, 5779–5790. [Google Scholar] [CrossRef]
- Yuan, T.; Walsh, M.P.; Sutherland, C.; Fabian, H.; Vogel, H.J. Calcium-dependent and -independent interactions of the calmodulin-binding domain of cyclic nucleotide phosphodiesterase with calmodulin. Biochemistry 1999, 38, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Houdusse, A.; Gaucher, J.F.; Krementsova, E.; Mui, S.; Trybus, K.M.; Cohen, C. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci. USA 2006, 103, 19326–19331. [Google Scholar] [CrossRef]
- Feldkamp, M.D.; Yu, L.; Shea, M.A. Structural and energetic determinants of apo calmodulin binding to the IQ motif of the Na(V)1.2 voltage-dependent sodium channel. Structure 2011, 19, 733–747. [Google Scholar] [CrossRef]
- Chagot, B.; Chazin, W.J. Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 2011, 406, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Drum, C.L.; Yan, S.Z.; Bard, J.; Shen, Y.Q.; Lu, D.; Soelaiman, S.; Grabarek, Z.; Bohm, A.; Tang, W.J. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 2002, 415, 396–402. [Google Scholar] [CrossRef]
- Watson, G.M.; Wilce, J.A. Direct Interaction between Calmodulin and the Grb7 RA-PH Domain. Int. J. Mol. Sci. 2020, 21, 1336. [Google Scholar] [CrossRef]
- Li, H.; Sanchez-Torres, J.; del Carpio, A.F.; Nogales-Gonzalez, A.; Molina-Ortiz, P.; Moreno, M.J.; Torok, K.; Villalobo, A. The adaptor Grb7 is a novel calmodulin-binding protein: Functional implications of the interaction of calmodulin with Grb7. Oncogene 2005, 24, 4206–4219. [Google Scholar] [CrossRef][Green Version]
- Schumacher, M.A.; Rivard, A.F.; Bachinger, H.P.; Adelman, J.P. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature 2001, 410, 1120–1124. [Google Scholar] [CrossRef]
- Nunez, E.; Muguruza-Montero, A.; Villarroel, A. Atomistic Insights of Calmodulin Gating of Complete Ion Channels. Int. J. Mol. Sci. 2020, 21, 1285. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Hedman, A.C.; Ames, J.B.; Sacks, D.B. Calmodulin Lobes Facilitate Dimerization and Activation of Estrogen Receptor-alpha. J. Biol. Chem. 2017, 292, 4614–4622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Sacks, D.B.; Ames, J.B. Structural basis for Ca2+-induced activation and dimerization of estrogen receptor alpha by calmodulin. J. Biol. Chem. 2012, 287, 9336–9344. [Google Scholar] [CrossRef] [PubMed]
- York, B.; Lou, D.; Noonan, D.J. Tuberin nuclear localization can be regulated by phosphorylation of its carboxyl terminus. Mol. Cancer Res. 2006, 4, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, A.; Hayashi, N.; Matsubara, M.; Yamauchi, E.; Taniguchi, H. Identification of the calmodulin-binding domain of neuron-specific protein kinase C substrate protein CAP-22/NAP-22. Direct involvement of protein myristoylation in calmodulin-target protein interaction. J. Biol. Chem. 1999, 274, 11848–11853. [Google Scholar] [CrossRef]
- Hayashi, N.; Matsubara, M.; Jinbo, Y.; Titani, K.; Izumi, Y.; Matsushima, N. Nef of HIV-1 interacts directly with calcium-bound calmodulin. Protein. Sci. 2002, 11, 529–537. [Google Scholar] [CrossRef]
- Hayashi, N.; Nakagawa, C.; Ito, Y.; Takasaki, A.; Jinbo, Y.; Yamakawa, Y.; Titani, K.; Hashimoto, K.; Izumi, Y.; Matsushima, N. Myristoylation-regulated direct interaction between calcium-bound calmodulin and N-terminal region of pp60v-src. J. Mol. Biol. 2004, 338, 169–180. [Google Scholar] [CrossRef]
- Matsubara, M.; Nakatsu, T.; Kato, H.; Taniguchi, H. Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. EMBO J. 2004, 23, 712–718. [Google Scholar] [CrossRef]
- Perez, Y.; Maffei, M.; Igea, A.; Amata, I.; Gairi, M.; Nebreda, A.R.; Bernado, P.; Pons, M. Lipid binding by the Unique and SH3 domains of c-Src suggests a new regulatory mechanism. Sci. Rep. 2013, 3, 1295. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Brunsveld, L.; Kuhlmann, J.; Alexandrov, K.; Wittinghofer, A.; Goody, R.S.; Waldmann, H. Lipidated ras and rab peptides and proteins--synthesis, structure, and function. Angew. Chem. Int. Ed. Engl. 2006, 45, 6622–6646. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.D.; Kirschmeier, P.; Bishop, W.R. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 2006, 47, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Villalonga, P.; Lopez-Alcala, C.; Bosch, M.; Chiloeches, A.; Rocamora, N.; Gil, J.; Marais, R.; Marshall, C.J.; Bachs, O.; Agell, N. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol. Cell Biol. 2001, 21, 7345–7354. [Google Scholar] [CrossRef] [PubMed]
- Fivaz, M.; Meyer, T. Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 2005, 170, 429–441. [Google Scholar] [CrossRef]
- Wang, M.T.; Holderfield, M.; Galeas, J.; Delrosario, R.; To, M.D.; Balmain, A.; McCormick, F. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell 2015, 163, 1237–1251. [Google Scholar] [CrossRef]
- Sperlich, B.; Kapoor, S.; Waldmann, H.; Winter, R.; Weise, K. Regulation of K-Ras4B Membrane Binding by Calmodulin. Biophys. J. 2016, 111, 113–122. [Google Scholar] [CrossRef]
- Saito, N.; Mine, N.; Kufe, D.W.; Von Hoff, D.D.; Kawabe, T. CBP501 inhibits EGF-dependent cell migration, invasion and epithelial-to-mesenchymal transition of non-small cell lung cancer cells by blocking KRas to calmodulin binding. Oncotarget 2017, 8, 74006–74018. [Google Scholar] [CrossRef]
- Sidhu, R.S.; Clough, R.R.; Bhullar, R.P. Ca2+/calmodulin binds and dissociates K-RasB from membrane. Biochem. Biophys. Res. Commun. 2003, 304, 655–660. [Google Scholar] [CrossRef]
- Dharmaiah, S.; Bindu, L.; Tran, T.H.; Gillette, W.K.; Frank, P.H.; Ghirlando, R.; Nissley, D.V.; Esposito, D.; McCormick, F.; Stephen, A.G.; et al. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEdelta. Proc. Natl. Acad. Sci. USA 2016, 113, E6766–E6775. [Google Scholar] [CrossRef]
- Lopez-Alcala, C.; Alvarez-Moya, B.; Villalonga, P.; Calvo, M.; Bachs, O.; Agell, N. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J. Biol. Chem. 2008, 283, 10621–10631. [Google Scholar] [CrossRef]
- Wu, L.J.; Xu, L.R.; Liao, J.M.; Chen, J.; Liang, Y. Both the C-terminal polylysine region and the farnesylation of K-RasB are important for its specific interaction with calmodulin. PLoS ONE 2011, 6, e21929. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Jang, H.; Nussinov, R.; Gaponenko, V. The disordered hypervariable region and the folded catalytic domain of oncogenic K-Ras4B partner in phospholipid binding. Curr. Opin. Struct. Biol. 2016, 36, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Banerjee, A.; Chavan, T.; Gaponenko, V.; Nussinov, R. Flexible-body motions of calmodulin and the farnesylated hypervariable region yield a high-affinity interaction enabling K-Ras4B membrane extraction. J. Biol. Chem. 2017, 292, 12544–12559. [Google Scholar] [CrossRef] [PubMed]
- Agamasu, C.; Ghirlando, R.; Taylor, T.; Messing, S.; Tran, T.H.; Bindu, L.; Tonelli, M.; Nissley, D.V.; McCormick, F.; Stephen, A.G. KRAS Prenylation Is Required for Bivalent Binding with Calmodulin in a Nucleotide-Independent Manner. Biophys. J. 2019, 116, 1049–1063. [Google Scholar] [CrossRef]
- Grant, B.M.M.; Enomoto, M.; Back, S.I.; Lee, K.Y.; Gebregiworgis, T.; Ishiyama, N.; Ikura, M.; Marshall, C. Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety. Sci. Signal. 2020, 13, eaaz0344. [Google Scholar] [CrossRef] [PubMed]
- Gillette, W.K.; Esposito, D.; Abreu Blanco, M.; Alexander, P.; Bindu, L.; Bittner, C.; Chertov, O.; Frank, P.H.; Grose, C.; Jones, J.E.; et al. Farnesylated and methylated KRAS4b: High yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 2015, 5, 15916. [Google Scholar] [CrossRef]
- Bhagatji, P.; Leventis, R.; Rich, R.; Lin, C.J.; Silvius, J.R. Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys. J. 2010, 99, 3327–3335. [Google Scholar] [CrossRef]
- Hayashi, N.; Titani, K. N-myristoylated proteins, key components in intracellular signal transduction systems enabling rapid and flexible cell responses. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 494–508. [Google Scholar] [CrossRef]
- Kovacs, E.; Harmat, V.; Toth, J.; Vertessy, B.G.; Modos, K.; Kardos, J.; Liliom, K. Structure and mechanism of calmodulin binding to a signaling sphingolipid reveal new aspects of lipid-protein interactions. FASEB J. 2010, 24, 3829–3839. [Google Scholar] [CrossRef]
- Rocks, O.; Peyker, A.; Kahms, M.; Verveer, P.J.; Koerner, C.; Lumbierres, M.; Kuhlmann, J.; Waldmann, H.; Wittinghofer, A.; Bastiaens, P.I. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 2005, 307, 1746–1752. [Google Scholar] [CrossRef]
- Rocks, O.; Peyker, A.; Bastiaens, P.I. Spatio-temporal segregation of Ras signals: One ship, three anchors, many harbors. Curr. Opin. Cell Biol. 2006, 18, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Elsaraj, S.M.; Bhullar, R.P. Regulation of platelet Rac1 and Cdc42 activation through interaction with calmodulin. Biochim. Biophys. Acta 2008, 1783, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Quadras, M.; Gelabert-Baldrich, M.; Soriano-Castell, D.; Llado, A.; Rentero, C.; Calvo, M.; Pol, A.; Enrich, C.; Tebar, F. Rac1 and calmodulin interactions modulate dynamics of ARF6-dependent endocytosis. Traffic 2011, 12, 1879–1896. [Google Scholar] [CrossRef]
- Xu, B.; Chelikani, P.; Bhullar, R.P. Characterization and functional analysis of the calmodulin-binding domain of Rac1 GTPase. PLoS ONE 2012, 7, e42975. [Google Scholar] [CrossRef]
- Wang, K.L.; Roufogalis, B.D. Ca2+/calmodulin stimulates GTP binding to the ras-related protein ral-A. J. Biol. Chem. 1999, 274, 14525–14528. [Google Scholar] [CrossRef]
- Wang, K.L.; Khan, M.T.; Roufogalis, B.D. Identification and characterization of a calmodulin-binding domain in Ral-A, a Ras-related GTP-binding protein purified from human erythrocyte membrane. J. Biol. Chem. 1997, 272, 16002–16009. [Google Scholar] [CrossRef]
- Clough, R.R.; Sidhu, R.S.; Bhullar, R.P. Calmodulin binds RalA and RalB and is required for the thrombin-induced activation of Ral in human platelets. J. Biol. Chem. 2002, 277, 28972–28980. [Google Scholar] [CrossRef]
- Sidhu, R.S.; Elsaraj, S.M.; Grujic, O.; Bhullar, R.P. Calmodulin binding to the small GTPase Ral requires isoprenylated Ral. Biochem. Biophys. Res. Commun. 2005, 336, 105–109. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Toledo-Ortiz, G.; Yalovsky, S.; Caldelari, D.; Gruissem, W. Carboxyl-methylation of prenylated calmodulin CaM53 is required for efficient plasma membrane targeting of the protein. Plant J. 2000, 24, 775–784. [Google Scholar] [CrossRef]
- Bush, D.S. Calcium regulation in plant cells and its role in signaling. Annu. Rev. Pant. Physiol. 1995, 46, 95–122. [Google Scholar] [CrossRef]
- Trewavas, A.J.; Malho, R. Signal Perception and Transduction: The Origin of the Phenotype. Plant Cell 1997, 9, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Bivona, T.G.; Quatela, S.E.; Bodemann, B.O.; Ahearn, I.M.; Soskis, M.J.; Mor, A.; Miura, J.; Wiener, H.H.; Wright, L.; Saba, S.G.; et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell 2006, 21, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.J.; Tsai, F.D.; Vais, H.; Court, H.; Yang, J.; Fehrenbacher, N.; Foskett, J.K.; Philips, M.R. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc. Natl. Acad. Sci. USA 2013, 110, 20593–20598. [Google Scholar] [CrossRef]
- Alvarez-Moya, B.; Barcelo, C.; Tebar, F.; Jaumot, M.; Agell, N. CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases 2011, 2, 99–103. [Google Scholar] [CrossRef][Green Version]
- Sawada, N.; Itoh, H.; Yamashita, J.; Doi, K.; Inoue, M.; Masatsugu, K.; Fukunaga, Y.; Sakaguchi, S.; Sone, M.; Yamahara, K.; et al. cGMP-dependent protein kinase phosphorylates and inactivates RhoA. Biochem. Biophys. Res. Commun. 2001, 280, 798–805. [Google Scholar] [CrossRef]
- Wang, H.; Owens, C.; Chandra, N.; Conaway, M.R.; Brautigan, D.L.; Theodorescu, D. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010, 70, 8760–8769. [Google Scholar] [CrossRef]
- Lim, K.H.; Brady, D.C.; Kashatus, D.F.; Ancrile, B.B.; Der, C.J.; Cox, A.D.; Counter, C.M. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol. Cell Biol. 2010, 30, 508–523. [Google Scholar] [CrossRef]
- Wilson, J.M.; Prokop, J.W.; Lorimer, E.; Ntantie, E.; Williams, C.L. Differences in the Phosphorylation-Dependent Regulation of Prenylation of Rap1A and Rap1B. J. Mol. Biol. 2016, 428, 4929–4945. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Schmick, M.; Vartak, N.; Papke, B.; Kovacevic, M.; Truxius, D.C.; Rossmannek, L.; Bastiaens, P.I.H. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 2014, 157, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Chandra, A.; Grecco, H.E.; Pisupati, V.; Perera, D.; Cassidy, L.; Skoulidis, F.; Ismail, S.A.; Hedberg, C.; Hanzal-Bayer, M.; Venkitaraman, A.R.; et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011, 14, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Baehr, W. Membrane protein transport in photoreceptors: The function of PDEdelta: The Proctor lecture. Investig. Ophthalmol. Vis. Sci. 2014, 55, 8653–8666. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Papke, B.; Ismail, S.; Vartak, N.; Chandra, A.; Hoffmann, M.; Hahn, S.A.; Triola, G.; Wittinghofer, A.; Bastiaens, P.I.; et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature 2013, 497, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gago, P.; Fansa, E.K.; Klein, C.H.; Murarka, S.; Janning, P.; Schurmann, M.; Metz, M.; Ismail, S.; Schultz-Fademrecht, C.; Baumann, M.; et al. A PDE6delta-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2. Angew. Chem. Int. Ed. Engl. 2017, 56, 2423–2428. [Google Scholar] [CrossRef]
- Dovas, A.; Couchman, J.R. RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef]
- Alory, C.; Balch, W.E. Molecular evolution of the Rab-escort-protein/guanine-nucleotide-dissociation-inhibitor superfamily. Mol. Biol. Cell 2003, 14, 3857–3867. [Google Scholar] [CrossRef]
- Constantine, R.; Zhang, H.; Gerstner, C.D.; Frederick, J.M.; Baehr, W. Uncoordinated (UNC)119: Coordinating the trafficking of myristoylated proteins. Vis. Res. 2012, 75, 26–32. [Google Scholar] [CrossRef]
- Ismail, S.A.; Chen, Y.X.; Miertzschke, M.; Vetter, I.R.; Koerner, C.; Wittinghofer, A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J. 2012, 31, 4085–4094. [Google Scholar] [CrossRef]
- Sztul, E.; Chen, P.W.; Casanova, J.E.; Cherfils, J.; Dacks, J.B.; Lambright, D.G.; Lee, F.S.; Randazzo, P.A.; Santy, L.C.; Schurmann, A.; et al. ARF GTPases and their GEFs and GAPs: Concepts and challenges. Mol. Biol. Cell. 2019, 30, 1249–1271. [Google Scholar] [CrossRef]
- Ames, J.B.; Tanaka, T.; Stryer, L.; Ikura, M. Portrait of a myristoyl switch protein. Curr. Opin. Struct. Biol. 1996, 6, 432–438. [Google Scholar] [CrossRef]
- Anguita, E.; Villalobo, A. Src-family tyrosine kinases and the Ca(2+) signal. Biochim. Biophys. Acta. Mol. Cell Res. 2017, 1864, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Benaim, G.; Villalobo, A. Phosphorylation of calmodulin. Functional implications. Eur. J. Biochem. 2002, 269, 3619–3631. [Google Scholar] [CrossRef] [PubMed]
- Kano, Y.; Gebregiworgis, T.; Marshall, C.B.; Radulovich, N.; Poon, B.P.K.; St-Germain, J.; Cook, J.D.; Valencia-Sama, I.; Grant, B.M.M.; Herrera, S.G.; et al. Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation. Nat. Commun. 2019, 10, 224. [Google Scholar] [CrossRef]
- Bunda, S.; Heir, P.; Srikumar, T.; Cook, J.D.; Burrell, K.; Kano, Y.; Lee, J.E.; Zadeh, G.; Raught, B.; Ohh, M. Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation. Proc. Natl. Acad. Sci. USA 2014, 111, E3785–E3794. [Google Scholar] [CrossRef]
- Bosch, M.; Gil, J.; Bachs, O.; Agell, N. Calmodulin inhibitor W13 induces sustained activation of ERK2 and expression of p21(cip1). J. Biol. Chem. 1998, 273, 22145–22150. [Google Scholar] [CrossRef]
- Cullen, P.J.; Lockyer, P.J. Integration of calcium and Ras signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 339–348. [Google Scholar] [CrossRef]
- Agell, N.; Bachs, O.; Rocamora, N.; Villalonga, P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca(2+), and calmodulin. Cell Signal 2002, 14, 649–654. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol. Rev. 2018, 98, 559–621. [Google Scholar] [CrossRef]
- Pierro, C.; Zhang, X.; Kankeu, C.; Trebak, M.; Bootman, M.D.; Roderick, H.L. Oncogenic KRAS suppresses store-operated Ca(2+) entry and ICRAC through ERK pathway-dependent remodelling of STIM expression in colorectal cancer cell lines. Cell Calcium 2018, 72, 70–80. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grant, B.M.M.; Enomoto, M.; Ikura, M.; Marshall, C.B. A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization. Int. J. Mol. Sci. 2020, 21, 2751. https://doi.org/10.3390/ijms21082751
Grant BMM, Enomoto M, Ikura M, Marshall CB. A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization. International Journal of Molecular Sciences. 2020; 21(8):2751. https://doi.org/10.3390/ijms21082751
Chicago/Turabian StyleGrant, Benjamin M. M., Masahiro Enomoto, Mitsuhiko Ikura, and Christopher B. Marshall. 2020. "A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization" International Journal of Molecular Sciences 21, no. 8: 2751. https://doi.org/10.3390/ijms21082751
APA StyleGrant, B. M. M., Enomoto, M., Ikura, M., & Marshall, C. B. (2020). A Non-Canonical Calmodulin Target Motif Comprising a Polybasic Region and Lipidated Terminal Residue Regulates Localization. International Journal of Molecular Sciences, 21(8), 2751. https://doi.org/10.3390/ijms21082751




