Heterogenic Origin of Micro RNAs in Atlantic Salmon (Salmo salar) Seminal Plasma
Abstract
1. Introduction
2. Results
2.1. Atlantic Salmon Sperm Small RNA Sequencing and Annotation
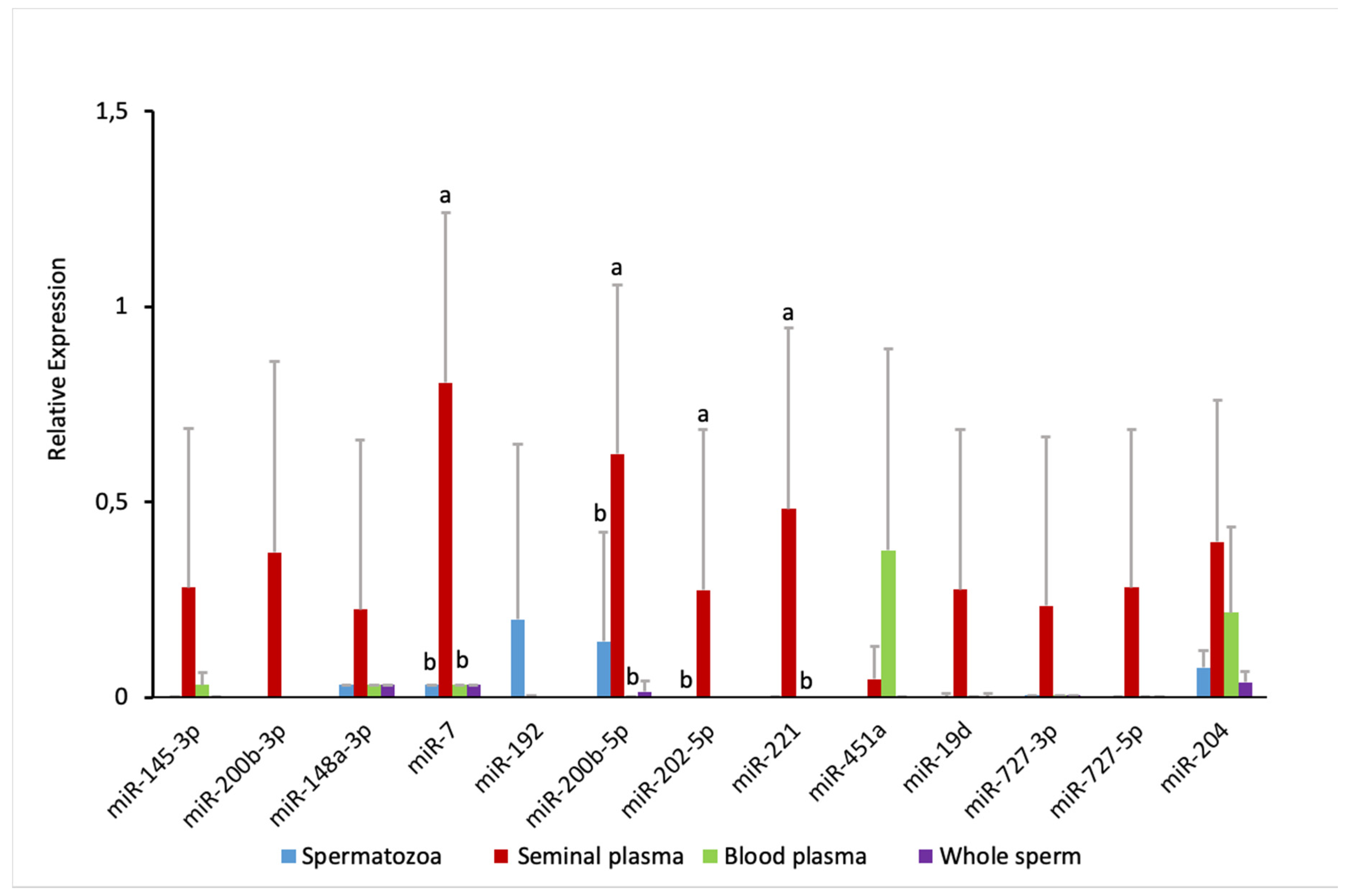
2.2. miRNA Profiling in the Whole Sperm and Its Fractions
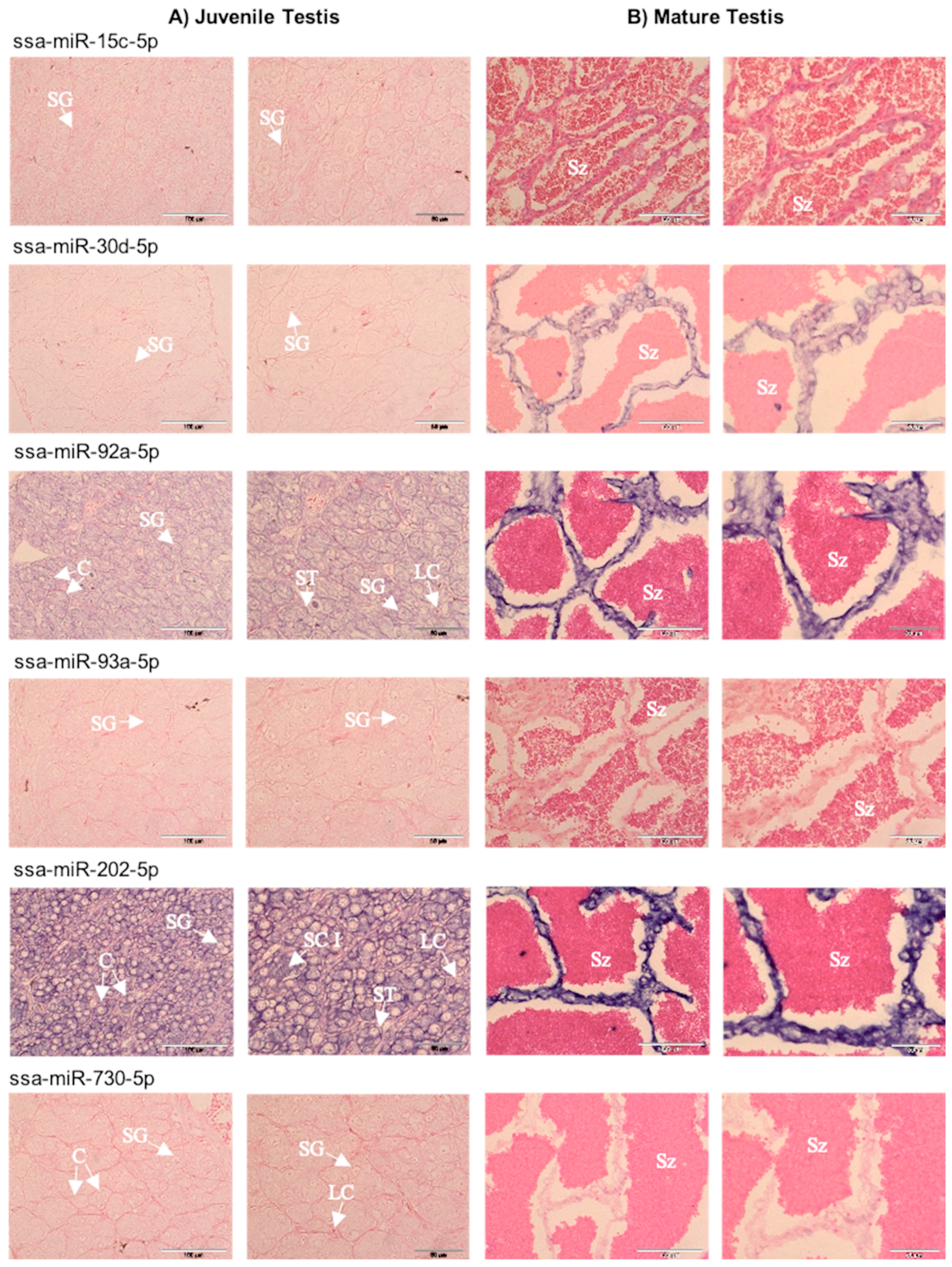
2.3. Spatial Expression of Selected miRNAs in Gonads
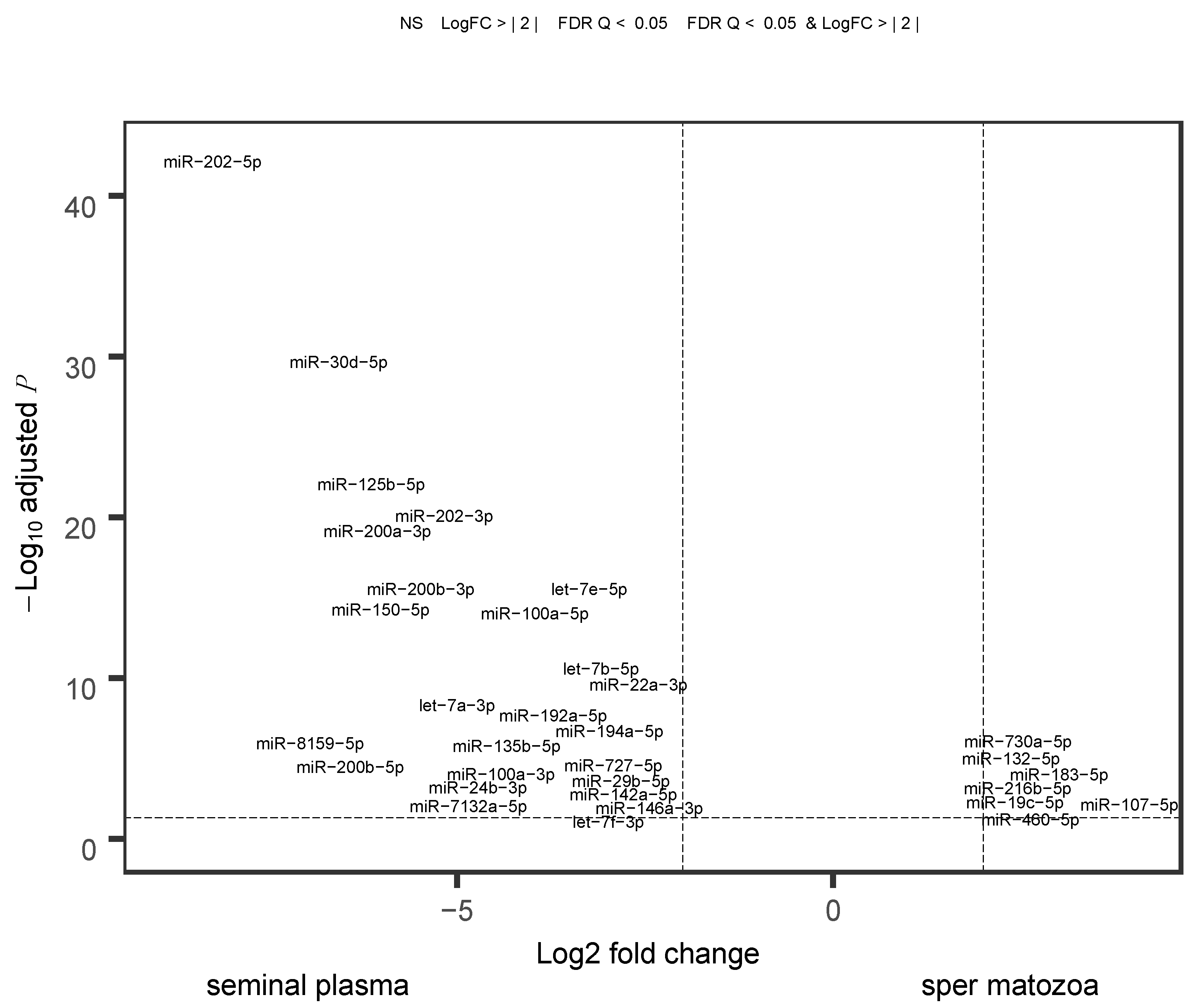
2.4. Comparison of Sperm and Blood Plasma miRNAs
2.5. GO Terms Annotation of the Differentially Expressed miRNA Targets
3. Discussion
3.1. miRNA Heterogeneity in Sperm Fractions
3.2. miRNA Accumulation in Atlantic Salmon Sperm
3.3. Plausible Sources and Functions of Seminal Plasma miRNAs
4. Materials and Methods
4.1. Ethics
4.2. Animals, Sperm Collection, and Fractionation
4.3. RNA Extraction and Small RNA Sequencing
4.4. Data Analysis
4.5. In Situ Hybridization
4.6. Quantitative Reverse Transcription PCR (RT-qPCR)
4.7. Assessment of Extracellular miRNAs Function
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GO | Gene ontology |
| ISH | In situ hybridization |
| LNA | Locked nucleic acid |
| miRNA | microRNA |
| PCR | Polymerase chain reaction |
| piRNA | Piwi-interacting RNA |
| RNA | Ribonucleic acid |
| RT-qPCR | Quantitative reverse transcription polymerase chain reaction |
| snRNA | small nuclear RNA |
| tRFs | transfer RNA-derived fragments |
| UTR | Untranslated region |
References
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.M.; Pang, R.T.K.; Chiu, P.C.N.; Wong, B.P.C.; Lao, K.; Lee, K.F.; Yeung, W.S.B. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. PNAS 2012, 109, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.; Mansuy, I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Sendler, E.; Johnson, G.D.; Mao, S.; Goodrich, R.J.; Diamond, M.P.; Hauser, R.; Krawetz, S.A. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013, 41, 4104–4117. [Google Scholar] [CrossRef] [PubMed]
- Siklenka, K.; Erkek, S.; Godmann, M.; Lambrot, R.; McGraw, S.; Lafleur, C.; Cohen, T.; Xia, J.; Suderman, M.; Hallett, M.; et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015, 350, aab2006. [Google Scholar] [CrossRef] [PubMed]
- Teperek, M.; Simeone, A.; Gaggioli, V.; Miyamoto, K.; Allen, G.; Erkek, S.; Peters, A.; Kwon, T.; Marcotte, E.; Zegerman, P.; et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016, 26, 1034–1046. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Reproductive biology: Delivering spermatozoan RNA to the oocyte. Nature 2004, 429, 154. [Google Scholar] [CrossRef]
- Hosken, D.J.; Hodgson, D.J. Why do sperm carry RNA? Relatedness, conflict, and control. Trends Ecol. Evol. 2014, 29, 451–455. [Google Scholar] [CrossRef]
- Holman, L.; Price, T.A.R. Even more functions of sperm RNA: A response to Hosken and Hodgson. Trends Ecol. Evol. 2014, 29, 648–649. [Google Scholar] [CrossRef]
- Billard, R. Spermatogenesis and spermatology of some teleost fish species. Reprod. Nutr. Dev. 1986, 26, 877–920. [Google Scholar] [CrossRef]
- La Rocca, G.; Olejniczak, S.H.; González, A.J.; Briskin, D.; Vidigal, J.A.; Spraggon, L.; DeMatteo, R.G.; Radler, M.R.; Lindsten, T.; Ventura, A.; et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. PNAS 2015, 112, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.D.; Mackie, P.; Jodar, M.; Moskovtsev, S.; Krawetz, S.A. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res. 2015, 43, 6847–6859. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Arnold, G.J.; Nynca, J.; Fröhlich, T.; Otte, K.; Ciereszko, A. Characterization of carp seminal plasma proteome in relation to blood plasma. J. Proteom. 2014, 98, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Cosson, M.P.; Gagnon, C. Protease inhibitor and substrates block motility and microtubule sliding of sea urchin and carp spermatozoa. Cell Motil Cytoskel 1988, 10, 518–527. [Google Scholar] [CrossRef]
- Aas, G.H.; Refstie, T.; Gjerde, B. Evaluation of milt quality of Atlantic salmon. Aquaculture 1991, 95, 125–132. [Google Scholar] [CrossRef]
- Lahnsteiner, F.; Berger, B.; Weismann, T.; Patzner, R.A. Determination of semen quality of the rainbow trout, Oncorhynchus mykiss, by sperm motility, seminal plasma parameters, and spermatozoal metabolism. Aquaculture 1998, 163, 163–181. [Google Scholar] [CrossRef]
- Yadav, R.P.; Kotaja, N. Small RNAs in spermatogenesis. Mol. Cell. Endocrinol. 2014, 382, 498–508. [Google Scholar] [CrossRef]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, I. MicroRNA in Teleost Fish. Genome Biol. Evol. 2014, 6, 1911–1937. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Babiak, J.; Norberg, B.; Fernandes, J.M.O.; Johansen, S.D.; Babiak, I. Sex-Biased miRNA Expression in Atlantic Halibut (Hippoglossus hippoglossus) Brain and Gonads. Sex. Dev. 2012, 6, 257–266. [Google Scholar] [CrossRef]
- Bizuayehu, T.T.; Johansen, S.D.; Puvanendran, V.; Toften, H.; Babiak, I. Temperature during early development has long-term effects on microRNA expression in Atlantic cod. BMC Genom. 2015, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Wu, J.; Liu, W.; Xiong, S.; Ma, W.; Zhang, J.; Wang, W.; Gui, J.F.; Mei, J. Sex-Biased miRNAs in gonad and their potential roles for testis development in yellow catfish. PLoS ONE 2014, 9, e107946. [Google Scholar] [CrossRef]
- Xiao, J.; Zhong, H.; Zhou, Y.; Yu, F.; Gao, Y.; Luo, Y.; Tang, Z.; Guo, Z.; Guo, E.; Gan, X.; et al. Identification and characterization of microRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus) by using solexa sequencing technology. PLoS ONE 2014, 9, e86821. [Google Scholar] [CrossRef]
- Jia, K.-T.; Zhang, J.; Jia, P.; Zeng, L.; Jin, Y.; Yuan, Y.; Chen, J.; Hong, Y.; Yi, M. Identification of microRNAs in zebrafish spermatozoa. Zebrafish 2015, 12, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Presslauer, C.; Tilahun Bizuayehu, T.; Kopp, M.; Fernandes, J.M.O.; Babiak, I. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 2017, 7, 43850. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Sun, F.; Conine, C.C.; Reichholf, B.; Kukreja, S.; Herzog, V.A.; Ameres, S.L.; Rando, O.J. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 2018, 46, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; Van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Cheng, H.; Luo, M.; Yi, M.; Zhou, R.; Cheng, Y.; Zhao, H. Rapid Evolution of piRNA pathway in the teleost fish: Implication for an adaptation to transposon diversity. Genome Biol. Evol. 2014, 6, 1393–1407. [Google Scholar]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Uribe, M.C.; Grier, H.J.; Mejía-Roa, V. Comparative testicular structure and spermatogenesis in bony fishes. Spermatogenesis 2014, 4, e983400. [Google Scholar] [CrossRef] [PubMed]
- Vlok, W.; Van Vuren, J.H.J. Chemical composition of blood and seminal plasma of Barbus aeneus (cyprinidae). Comp. Biochem. Phys. A 1988, 90, 49–51. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Jin, Y.; Jia, P.; Jia, K.; Yi, M. MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish. Sci. Rep. 2017, 7, 7055. [Google Scholar] [CrossRef]
- Dabaja, A.A.; Mielnik, A.; Robinson, B.D.; Wosnitzer, M.S.; Schlegel, P.N.; Paduch, D.A. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin. Androl. 2015, 25, 2. [Google Scholar] [CrossRef]
- Rakoczy, J.; Fernandez-Valverde, S.L.; Glazov, E.A.; Wainwright, E.N.; Sato, T.; Takada, S.; Combes, A.N.; Korbie, D.J.; Miller, D.; Grimmond, S.M.; et al. MicroRNAs-140-5p/140-3p modulate Leydig cell numbers in the developing mouse testis. Biol. Reprod. 2013, 88, 143. [Google Scholar] [CrossRef]
- Wainwright, E.N.; Jorgensen, J.S.; Kim, Y.; Truong, V.; Bagheri-Fam, S.; Davidson, T.; Svingen, T.; Fernandez-Valverde, S.L.; McClelland, K.S.; Taft, R.J.; et al. SOX9 regulates microRNA miR-202-5p/3p expression during mouse testis differentiation. Biol. Reprod. 2013, 89, 1–12. [Google Scholar] [CrossRef]
- Gay, S.; Bugeon, J.; Bouchareb, A.; Henry, L.; Delahaye, C.; Legeai, F.; Montfort, J.; Le Cam, A.; Siegel, A.; Bobe, J.; et al. MiR-202 controls female fecundity by regulating medaka oogenesis. PLoS Genet 2018, 14, e1007593. [Google Scholar] [CrossRef]
- Trattner, S.; Vestergren, A.S. Tissue distribution of selected microRNA in Atlantic salmon. Eur. J. Lipid Sci. Technol. 2013, 115, 1348–1356. [Google Scholar] [CrossRef]
- Andreassen, R.; Worren, M.; Hoyheim, B. Discovery and characterization of miRNA genes in atlantic salmon (Salmo salar) by use of a deep sequencing approach. BMC Genom. 2013, 14, 482. [Google Scholar] [CrossRef]
- Bekaert, M.; Lowe, N.R.; Bishop, S.C.; Bron, J.E.; Taggart, J.B.; Houston, R.D. Sequencing and characterisation of an extensive Atlantic salmon (Salmo salar L.) microRNA repertoire. PLoS ONE 2013, 8, e70136. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef] [PubMed]
- García-López, J.; Alonso, L.; Cárdenas, D.B.; Artaza-Alvarez, H.; Hourcade, J.D.D.; Martínez, S.; Brieño-Enríquez, M.A.; Mazo, J.D. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 2015, 21, 946–962. [Google Scholar] [CrossRef] [PubMed]
- Krawetz, S.A.; Kruger, A.; Lalancette, C.; Tagett, R.; Anton, E.; Draghici, S.; Diamond, M.P. A survey of small RNAs in human sperm. Hum. Reprod. 2011, 26, 3401–3412. [Google Scholar] [CrossRef]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.P.; Krawetz, S.A. Reproductive Medicine Network. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Fagerlind, M.; Stålhammar, H.; Olsson, B.; Klinga-Levan, K. Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reprod. Domest. Anim. 2015, 50, 587–594. [Google Scholar] [CrossRef]
- Tscherner, A.; Gilchrist, G.; Smith, N.; Blondin, P.; Gillis, D.; LaMarre, J. MicroRNA-34 family expression in bovine gametes and preimplantation embryos. Reprod. Biol. Endocrinol. 2014, 12, 85. [Google Scholar] [CrossRef]
- Morisawa, M.; Suzuki, K. Osmolality and potassium ion: Their roles in initiation of sperm motility in teleosts. Science 1980, 210, 1145. [Google Scholar] [CrossRef]
- Inaba, K.; Morisawa, S.; Morisawa, M. Proteasomes regulate the motility of salmonid fish sperm through modulation of cAMP-dependent phosphorylation of an outer arm dynein light chain. J. Cell Sci. 1998, 111, 1105. [Google Scholar]
- Morisawa, M.; Okuno, M. Cyclic AMP induces maturation of trout sperm axoneme to initiate motility. Nature 1982, 295, 703–704. [Google Scholar] [CrossRef]
- Schulz, R.W.; De França, L.R.; Lareyre, J.J.; LeGac, F.; Chiarini-Garcia, H.; Nobrega, R.H.; Miura, T. Spermatogenesis in fish. Gen. Comp. Endocrinol. 2010, 165, 390–411. [Google Scholar] [CrossRef]
- Lanes, C.F.C.; Sampaio, L.A.; Marins, L.F. Evaluation of DNase activity in seminal plasma and uptake of exogenous DNA by spermatozoa of the Brazilian flounder Paralichthys orbignyanus. Theriogenology 2009, 71, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Zani, M.; Lavitrano, M.; French, D.; Lulli, V.; Maione, B.; Sperandio, S.; Spadafora, C. The mechanism of binding of exogenous DNA to sperm cells: Factors controlling the DNA uptake. Exp. Cell Res. 1995, 217, 57–64. [Google Scholar] [CrossRef]
- Carballada, R.; Esponda, P. Regulation of foreign DNA uptake by mouse spermatozoa. Exp. Cell Res. 2001, 262, 104–113. [Google Scholar] [CrossRef]
- Xin, N.; Liu, T.; Zhao, H.; Wang, Z.; Liu, J.; Zhang, Q.; Qi, J. The impact of exogenous DNA on the structure of sperm of olive flounder (Paralichthys olivaceus). Anim. Reprod. Sci. 2014, 149, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Spadafora, C. Sperm cells and foreign DNA: A controversial relation. Bioessays 1998, 20, 955–964. [Google Scholar] [CrossRef]
- Giordano, R.; Magnano, A.R.; Zaccagnini, G.; Pittoggi, C.; Moscufo, N.; Lorenzini, R.; Spadafora, C. Reverse transcriptase activity in mature spermatozoa of mouse. J. Cell Biol. 2000, 148, 1107–1114. [Google Scholar] [CrossRef]
- Sin, F.Y.T.; Walker, S.P.; Symonds, J.E.; Sin, I.L. Sperm-mediated gene transfer in chinook salmon, high performance fish. In Proceedings of the an International Fish Physiology Symposium at the University of British Columbia in Vancouver, Vancouver, BC, Canada, 16–21 July 1994; pp. 360–365. [Google Scholar]
- Sin, F.Y.T.; Bartley, A.L.; Walker, S.P.; Sin, I.L.; Symonds, J.E.; Hawke, L.; Hopkins, C.L. Gene transfer in chinook salmon (Oncorhynchus tshawytscha) by electroporating sperm in the presence of pRSV-lacZ DNA. Aquaculture 1993, 117, 57–69. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating MicroRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circul. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Liu, S.; Da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef]
- Babiak, I.; Dabrowski, K. Refrigeration of rainbow trout gametes and embryos. J. Exp. Zool. Part A 2003, 300, 140–151. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Dix, D.J.; Miller, D.; Khatri, P.; Krawetz, S.A. Spermatozoal RNA profiles of normal fertile men. Lancet 2002, 360, 772–777. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 May 2017).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Si, X.; Skogerbø, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014, bau110. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sturm, M.; Hackenberg, M.; Langenberger, D.; Frishman, D. TargetSpy: A supervised machine learning approach for microRNA target prediction. BMC Bioinform. 2010, 11, 292. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]



| Sample Type | Total Reads (min–max) | Total miRNA Reads (min–max) | Average Read/Size | ||
|---|---|---|---|---|---|
| 15–26 nt | 27–35 nt | 36–40 nt | |||
| Whole sperm | 31.2–37 | 0.6–0.9 | 48,681 | 70,808 | 50,636 |
| Spermatozoa | 29.7–37.2 | 0.4–0.6 | 52,191 | 62,011 | 51,635 |
| Seminal plasma | 20–35.5 | 0.4–2.7 | 19,026 | 17,126 | 8529 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bizuayehu, T.T.; Babiak, I. Heterogenic Origin of Micro RNAs in Atlantic Salmon (Salmo salar) Seminal Plasma. Int. J. Mol. Sci. 2020, 21, 2723. https://doi.org/10.3390/ijms21082723
Bizuayehu TT, Babiak I. Heterogenic Origin of Micro RNAs in Atlantic Salmon (Salmo salar) Seminal Plasma. International Journal of Molecular Sciences. 2020; 21(8):2723. https://doi.org/10.3390/ijms21082723
Chicago/Turabian StyleBizuayehu, Teshome Tilahun, and Igor Babiak. 2020. "Heterogenic Origin of Micro RNAs in Atlantic Salmon (Salmo salar) Seminal Plasma" International Journal of Molecular Sciences 21, no. 8: 2723. https://doi.org/10.3390/ijms21082723
APA StyleBizuayehu, T. T., & Babiak, I. (2020). Heterogenic Origin of Micro RNAs in Atlantic Salmon (Salmo salar) Seminal Plasma. International Journal of Molecular Sciences, 21(8), 2723. https://doi.org/10.3390/ijms21082723





