Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances
Abstract
1. Introduction
2. Results
2.1. Isolation, Identification and Typing of Vaginal LAB
2.2. Antibiotic Susceptibility
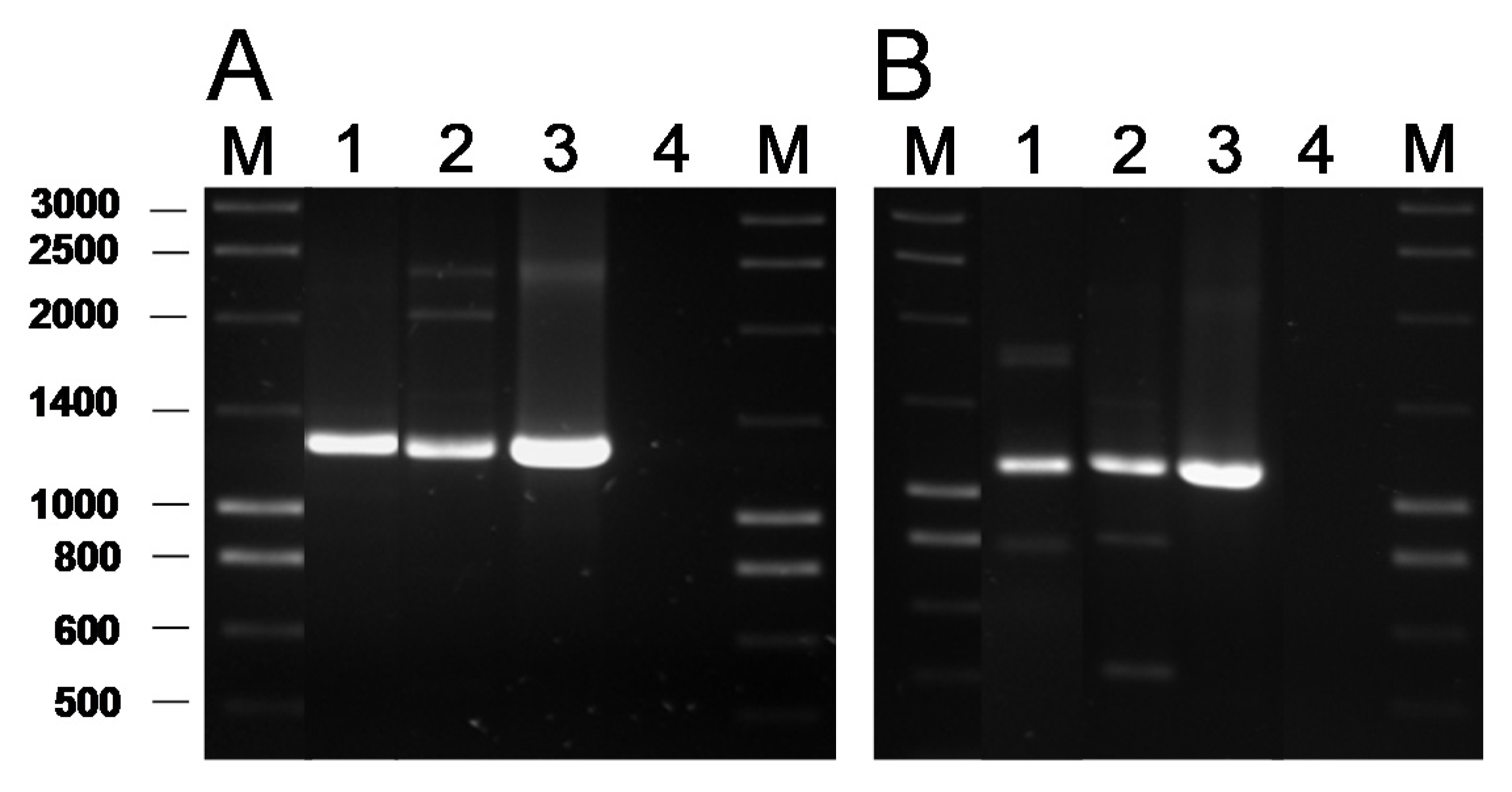
2.3. Detection of AR Genes by PCR
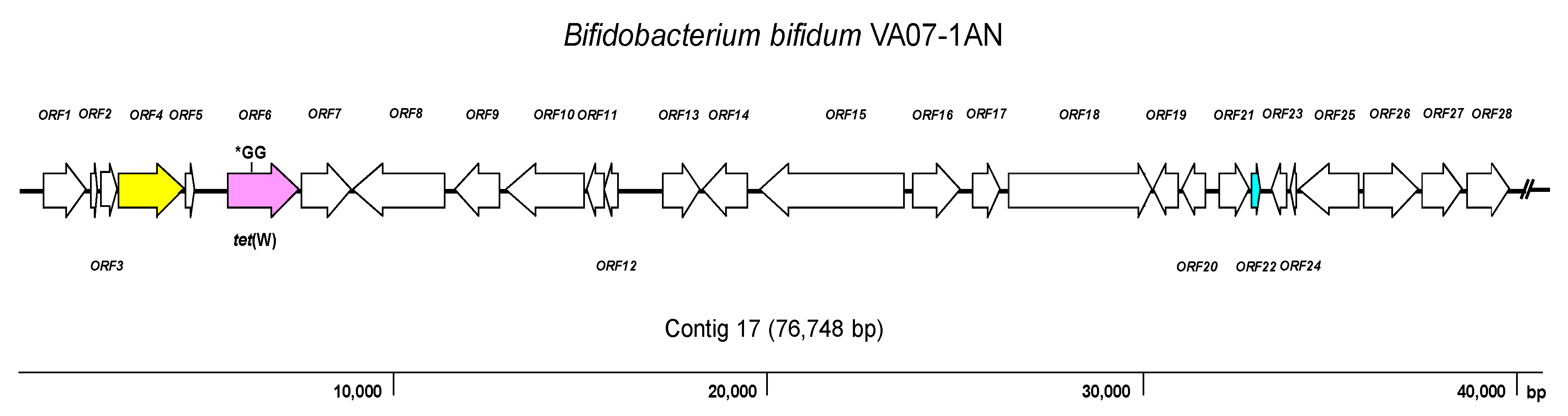
2.4. Genome Analysis for AR Genes
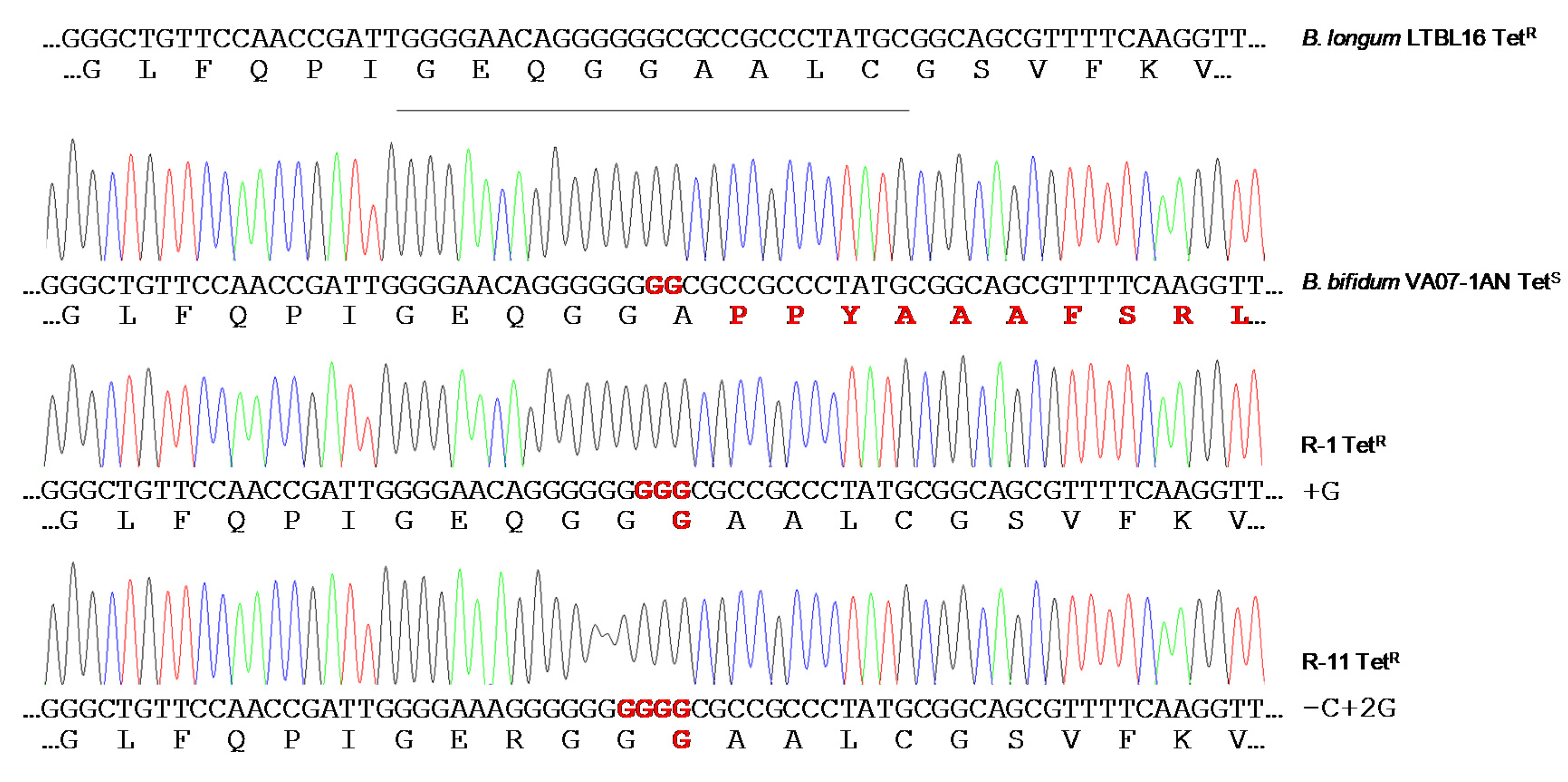
2.5. Restoration of the Tetracycline Resistance Phenotype in B. Bifidum VA07-1AN
3. Discussion
4. Materials and Methods
4.1. Sample Selection and Collection
4.2. Isolation of Lactic Acid Bacteria (LAB)
4.3. Identification of Bacteria by 16S rRNA Gene Sequencing
4.4. Molecular PCR Fingerprinting
4.5. Antibiotic Susceptibility Testing
4.6. PCR Detection and Identification of AR Genes
4.7. Genome Sequencing, Annotation, and Analysis
4.8. Stability of the Disrupted tet(W) Gene of B. Bifidum
4.9. GenBank Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance: Global Report on Surveillance. 2014. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 6 December 2019).
- von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar]
- Sáez-Lara, M.J.; Gómez-Llorente, C.; Plaza-Díaz, J.; Gil, A. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: A systematic review of randomized human clinical trials. BioMed Res. Int. 2015. [Google Scholar] [CrossRef]
- EFSA. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 7: Suitability of taxonomic units notified to EFSA until September 2017. EFSA J. 2018, 16, e05131. [Google Scholar]
- FDA. Generally Recognized as Safe (GRAS). Notifications FDA. 2010. Available online: https://www.fda.gov/food/generally-recognized-safe-gras/microorganisms-microbial-derived-ingredients-used-food-partial-list (accessed on 1 October 2019).
- EFSA. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar]
- Flórez, A.B.; Campedelli, I.; Delgado, S.; Alegría, A.; Salvetti, E.; Felis, G.E.; Mayo, B.; Torriani, S. Antibiotic susceptibility profiles of dairy Leuconostoc, analysis of the genetic basis of atypical resistances and transfer of genes in vitro and in a food matrix. PLoS ONE 2016, 11, e0145203. [Google Scholar]
- Martin, D.H. The microbiota of the vagina and its influence on women’s health and disease. Am. J. Med. Sci. 2012, 343, 2–9. [Google Scholar] [CrossRef]
- Sirichoat, A.; Buppasiri, P.; Engchanil, C.; Namwat, W.; Faksri, K.; Sankuntaw, N.; Pasomsub, E.; Chantratita, W.; Lulitanond, V. Characterization of vaginal microbiota in Thai women. PeerJ 2018, 6, e5977. [Google Scholar] [CrossRef]
- Smith, S.B.; Ravel, J. The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 2017, 595, 451–463. [Google Scholar] [CrossRef]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- Gad, G.F.; Abdel-Hamid, A.M.; Farag, Z.S. Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz. J. Microbiol. 2014, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Fraqueza, M.J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Ngu, D.Y.; Dan, L.A.; Ooi, A.; Lim, R.L. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Park, I.S.; Walsh, C.T. D-Alanyl-D-lactate and D-alanyl-D-alanine synthesis by D-alanyl-D-alanine ligase from vancomycin-resistant Leuconostoc mesenteroides. Effects of a phenylalanine 261 to tyrosine mutation. J. Biol. Chem. 1997, 272, 9210–9214. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 1–37. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Milani, C.; Mangifesta, M.; Ferrario, C.; Anzalone, R.; Viappiani, A.; van Sinderen, D.; et al. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl. Environ. Microbiol. 2017, 83, e02894-16. [Google Scholar] [CrossRef]
- Štšepetova, J.; Taelma, H.; Smidt, I.; Hutt, P.; Lapp, E.; Aotäht, E.; Mändar, R. Assessment of phenotypic and genotypic antibiotic susceptibility of vaginal Lactobacillus sp. J. Appl. Microbiol. 2017, 123, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2018, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed]
- Rozman, V.; Lorbeg, P.M.; Accetto, T.; Matijašić, B.B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food. Microbiol. 2020, 314, 108388. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; O’Toole, P.W. When regulation challenges innovation: The case of the genus Lactobacillus. Trends Food Sci. Technol. 2017, 66, 187–194. [Google Scholar] [CrossRef]
- Delgado, S.; Flórez, A.B.; Mayo, B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 2005, 50, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Casado Munoz, M.D.C.; Lavilla Lerma, L.; Perez Montoro, B.; Bockelmann, W.; Pichner, R.; Kabisch, J.; Cho, G.S.; Franz, C.M.A.P.; Gálvez, A.; et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015, 78, 465–481. [Google Scholar] [CrossRef]
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside resistance: The emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
- Huys, G.; D’Haene, K.; Cnockaert, M.; Tosi, L.; Danielsen, M.; Flórez, A.B.; Mättö, J.; Axelsson, L.; Korhonen, J.; Mayrhofer, S.; et al. Intra- and interlaboratory performances of two commercial antimicrobial susceptibility testing methods for bifidobacteria and nonenterococcal lactic acid bacteria. Antimicrob. Agents Chemother. 2010, 54, 2567–2574. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; van Hoek, A.H.; de Los Reyes-Gavilán, C.G.; Aarts, H.J.; Margolles, A.; Mayo, B. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J. Mol. Microbiol. Biotechnol. 2008, 14, 6–15. [Google Scholar] [CrossRef]
- Condon, S. Aerobic metabolism of lactic acid bacteria. Irish J. Food Sci. Technol. 1983, 7, 15–25. [Google Scholar]
- Elkins, C.A.; Mullis, L.B. Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 2004, 70, 7200–7209. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Demirci, H.; Murphy, F.V.t.; Murphy, E.L.; Connetti, J.L.; Dahlberg, A.E.; Jogl, G.; Gregory, S.T. Structural analysis of base substitutions in Thermus thermophilus 16S rRNA conferring streptomycin resistance. Antimicrob. Agents Chemother. 2014, 58, 4308–4317. [Google Scholar] [CrossRef] [PubMed]
- Kiwaki, M.; Sato, T. Antimicrobial susceptibility of Bifidobacterium breve strains and genetic analysis of streptomycin resistance of probiotic B. breve strain Yakult. Int. J. Food Microbiol. 2009, 134, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.; Tyrrell, K.L.; Citron, D.M. Lactobacillus species: Taxonomic complexity and controversial susceptibilities. Clin. Infect. Dis. 2015, 60, S98–S107. [Google Scholar] [CrossRef] [PubMed]
- van Pijkeren, J.P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef]
- Katla, A.K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Castro, W.; Navarro, M.; Biot, C. Medicinal potential of ciprofloxacin and its derivatives. Future Med. Chem. 2013, 5, 81–96. [Google Scholar] [CrossRef]
- Rosander, A.; Connolly, E.; Roos, S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032–6040. [Google Scholar] [CrossRef]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Alvarez-Martín, P.; Margolles, A.; Mayo, B. Analysis of tetracycline resistance tet(W) genes and their flanking sequences in intestinal Bifidobacterium species. J. Antimicrob. Chemother. 2008, 62, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Flórez, A.B.; van Hoek, A.H.; Stuer-Lauridsen, B.; Strøman, P.; de los Reyes-Gavilán, C.G.; Margolles, A. Genetic basis of tetracycline resistance in Bifidobacterium animalis subsp. lactis. Appl. Environ. Microbiol. 2010, 76, 3364–3369. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, L.; Giraffa, G. Rapid identification of dairy lactic acid bacteria by M13-generated, RAPD-PCR fingerprint databases. J. Microbiol. Methods 2005, 63, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Mättö, J.; Malinen, E.; Suihko, M.L.; Alander, M.; Palva, A.; Saarela, M. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J. Appl. Microbiol. 2004, 97, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Koeuth, T.; Versalovic, J.; Lupski, J.R. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome. Res. 1995, 5, 408–418. [Google Scholar] [CrossRef]
- IDF. Milk and Milk Products: Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-enterococcal Lactic Acid Bacteria. ISO Standard 10932:2010. 2010. Available online: https://www.iso.org/obp/ui/#iso:std:iso:10932:ed-1:v1:en (accessed on 8 April 2020).
- Klare, I.; Konstabel, C.; Müller-Bertling, S.; Reissbrodt, R.; Huys, G.; Vancanneyt, M.; Swings, J.; Goossens, H.; Witte, W. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 2005, 71, 8982–8986. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Supplement M100S; CLSI: Wayne, NJ, USA, 2016. [Google Scholar]
- Clermont, D.; Chesneau, O.; De Cespédès, G.; Horaud, T. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 1997, 41, 112–116. [Google Scholar] [CrossRef]
- Barbosa, T.M.; Scott, K.P.; Flint, H.J. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1999, 1, 53–64. [Google Scholar] [CrossRef]
- Gevers, D.; Danielsen, M.; Huys, G.; Swings, J. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 2003, 69, 1270–1275. [Google Scholar] [CrossRef]
- Scott, K.P.; Melville, C.M.; Barbosa, T.M.; Flint, H.J. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 2000, 44, 775–777. [Google Scholar] [CrossRef]
- Rizzotti, L.; Simeoni, D.; Cocconcelli, P.; Gazzola, S.; Dellaglio, F.; Torriani, S. Contribution of enterococci to the spread of antibiotic resistance in the production chain of swine meat commodities. J. Food Prot. 2005, 68, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Chung, W.O.; Roe, D.; Xia, M.; Marquez, C.; Borthagaray, G.; Whittington, W.L.; Holmes, K.K. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob. Agents Chemother. 1999, 43, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Luna, V.A.; Cousin, S., Jr.; Whittington, W.L.; Roberts, M.C. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 2000, 44, 2503–2506. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hummel, A.S.; Hertel, C.; Holzapfel, W.H.; Franz, C.M. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl. Environ. Microbiol. 2007, 73, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Rojo-Bezares, B.; Sáenz, Y.; Poeta, P.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006, 111, 234–240. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Werner, G.; Huys, G.; Vankerckhoven, V.; Kahlmeter, G.; Hildebrandt, B.; Müller-Bertling, S.; Witte, W.; Goossens, H. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 2007, 59, 900–912. [Google Scholar] [CrossRef]


| Species | Strain | Antibiotic a (MIC as µg mL−1) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | KAN | STR | NEO | TET | ERY | CLI | CHL | AMP | PEN | VAN | QDA | LIN | TMP | CIP | RIF | ||
| L. crispatus | VA20-32AN b | 2 | 16 | 2 | 16 | 2 | 0.06 | 0.25 | 4 | 1 | 0.5 | 0.5 | 1 | 4 | >64 | 16 | 1 |
| VA27-7 | 4 | 32 | 64 | 8 | 1 | 1 | 2 | 8 | 2 | 2 | 1 | 1 | 4 | 32 | 64 | 4 | |
| VA27-9 | 1 | 16 | 2 | 2 | 2 | 0.03 | 0.5 | 4 | 2 | 0.5 | 0.5 | 1 | 4 | 64 | 32 | 2 | |
| VA28-12 | 1 | 16 | 2 | 2 | 2 | 0.06 | 0.5 | 4 | 2 | 0.5 | 0.5 | 2 | 4 | 64 | 32 | 2 | |
| VA32-17 | 2 | 64 | 2 | 8 | 4 | 0.03 | 0.5 | 2 | 1 | 1 | 0.5 | 1 | 2 | >64 | 64 | 8 | |
| VA32-17AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 1 | 0.5 | 1 | 1 | 2 | 16 | 32 | 4 | |
| VA50-4AN c | ≤0.5 | 32 | 1 | 2 | 4 | 0.12 | 0.12 | 4 | 4 | 1 | 0.5 | 1 | 4 | >64 | 32 | 4 | |
| L. jensenii | VA04-1AN | ≤0.5 | 4 | 2 | 1 | 0.25 | ≤0.016 | 0.12 | 4 | 0.25 | 0.12 | 1 | 0.5 | 1 | >64 | 8 | 0.25 |
| VA04-2AN | ≤0.5 | 4 | 4 | 1 | 0.5 | 0.03 | 0.12 | 2 | 0.5 | 1 | 1 | 0.5 | 2 | >64 | 8 | 0.25 | |
| VA15-2AN | ≤0.5 | ≤2 | 1 | ≤0.5 | 1 | ≤0.016 | ≤0.03 | 2 | 0.06 | 0.06 | 0.5 | 0.5 | 0.5 | >64 | 8 | 0.25 | |
| VA16-11 | ≤0.5 | 8 | 1 | 2 | 4 | 0.06 | 0.25 | 4 | 0.06 | ≤0.03 | 2 | 0.5 | 2 | >64 | 16 | 0.5 | |
| Breakpoint (µg mL−1) d | 16 | 16 | 16 | - | 4 | 1 | 4 | 4 | 2 | - | 2 | - | - | - | - | - | |
| L. salivarius | VA09-4 | 8 | 64 | 16 | 4 | 2 | 0.25 | 0.25 | 2 | 1 | 0.25 | 128 | 0.25 | 0.5 | ≤0.12 | 1 | 2 |
| VA16-20 | ≤0.5 | 4 | 2 | 0.5 | 1 | 0.06 | 0.06 | 2 | 0.5 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | |
| VA37-13 | ≤0.5 | 4 | ≤0.5 | ≤0.5 | 0.5 | 0.06 | 0.06 | 2 | 0.25 | 0.12 | >128 | 0.5 | 0.5 | 0.25 | ≤0.25 | 0.5 | |
| VA40-10 | 128 | >1024 | >256 | 256 | 2 | 1 | 1 | 4 | 1 | 0.25 | >128 | 1 | 1 | 1 | 4 | 0.5 | |
| VA40-12AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.25 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | 0.25 | 1 | 1 | |
| VA40-14AN | 4 | 128 | 32 | 4 | 2 | 0.25 | 0.5 | 4 | 0.5 | 0.25 | >128 | 1 | 0.5 | ≤0.12 | 1 | 1 | |
| Breakpoint (µg mL−1) | 16 | 64 | 64 | - | 8 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. paracasei | VA02-1AN | ≤0.5 | 16 | 8 | 1 | 2 | 0.12 | 0.06 | 8 | 1 | 0.25 | >128 | 1 | 4 | 0.5 | 4 | 0.5 |
| VA24-4 | 1 | 16 | 8 | 4 | 4 | 0.12 | 0.06 | 4 | 0.5 | 0.25 | >128 | 1 | 2 | 0.25 | 4 | 0.5 | |
| VA26-3 | ≤0.5 | 16 | 8 | 2 | 2 | 0.12 | 0.06 | 4 | 1 | 0.25 | >128 | 1 | 2 | 1 | 2 | 0.5 | |
| VA27-8 | 1 | 32 | 16 | 8 | 2 | 0.06 | 0.06 | 8 | 0.5 | 0.25 | >128 | 1 | 4 | 0.25 | 4 | 0.5 | |
| Breakpoint (µg mL−1) | 32 | 64 | 64 | - | 4 | 1 | 4 | 4 | 4 | - | n.r. | - | - | - | - | - | |
| L. reuteri | VA15-3 | ≤0.5 | 4 | 2 | ≤0.5 | 8 | 0.12 | ≤0.03 | 4 | 1 | 2 | >128 | 1 | 2 | >64 | 32 | 0.25 |
| VA24-5 | ≤0.5 | 16 | 4 | ≤0.5 | 16 | 0.06 | ≤0.03 | 4 | 2 | 8 | >128 | 0.5 | 4 | >64 | 32 | 0.25 | |
| Breakpoint (µg mL−1) | 8 | 64 | 64 | - | 32 | 1 | 4 | 4 | 2 | - | n.r. | - | - | - | - | - | |
| B. bifidum | VA07-1AN | 8 | 64 | >256 | 16 | 1 | ≤0.016 | 0.06 | 1 | ≤0.03 | ≤0.03 | 0.5 | 0.5 | 0.5 | 16 | 8 | 2 |
| VA07-2AN | 32 | 64 | >256 | 32 | 1 | ≤0.016 | ≤0.03 | 1 | ≤0.03 | ≤0.03 | 1 | 0.5 | 0.5 | 16 | 8 | 1 | |
| Breakpoint (µg mL−1) | 64 | - | 128 | - | 8 | 1 | 1 | 4 | 2 | - | 2 | - | - | - | - | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirichoat, A.; Flórez, A.B.; Vázquez, L.; Buppasiri, P.; Panya, M.; Lulitanond, V.; Mayo, B. Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances. Int. J. Mol. Sci. 2020, 21, 2594. https://doi.org/10.3390/ijms21072594
Sirichoat A, Flórez AB, Vázquez L, Buppasiri P, Panya M, Lulitanond V, Mayo B. Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances. International Journal of Molecular Sciences. 2020; 21(7):2594. https://doi.org/10.3390/ijms21072594
Chicago/Turabian StyleSirichoat, Auttawit, Ana Belén Flórez, Lucía Vázquez, Pranom Buppasiri, Marutpong Panya, Viraphong Lulitanond, and Baltasar Mayo. 2020. "Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances" International Journal of Molecular Sciences 21, no. 7: 2594. https://doi.org/10.3390/ijms21072594
APA StyleSirichoat, A., Flórez, A. B., Vázquez, L., Buppasiri, P., Panya, M., Lulitanond, V., & Mayo, B. (2020). Antibiotic Susceptibility Profiles of Lactic Acid Bacteria from the Human Vagina and Genetic Basis of Acquired Resistances. International Journal of Molecular Sciences, 21(7), 2594. https://doi.org/10.3390/ijms21072594