The Cell Wall PAC (Proline-Rich, Arabinogalactan Proteins, Conserved Cysteines) Domain-Proteins Are Conserved in the Green Lineage
Abstract
1. Introduction
2. Results and Discussion
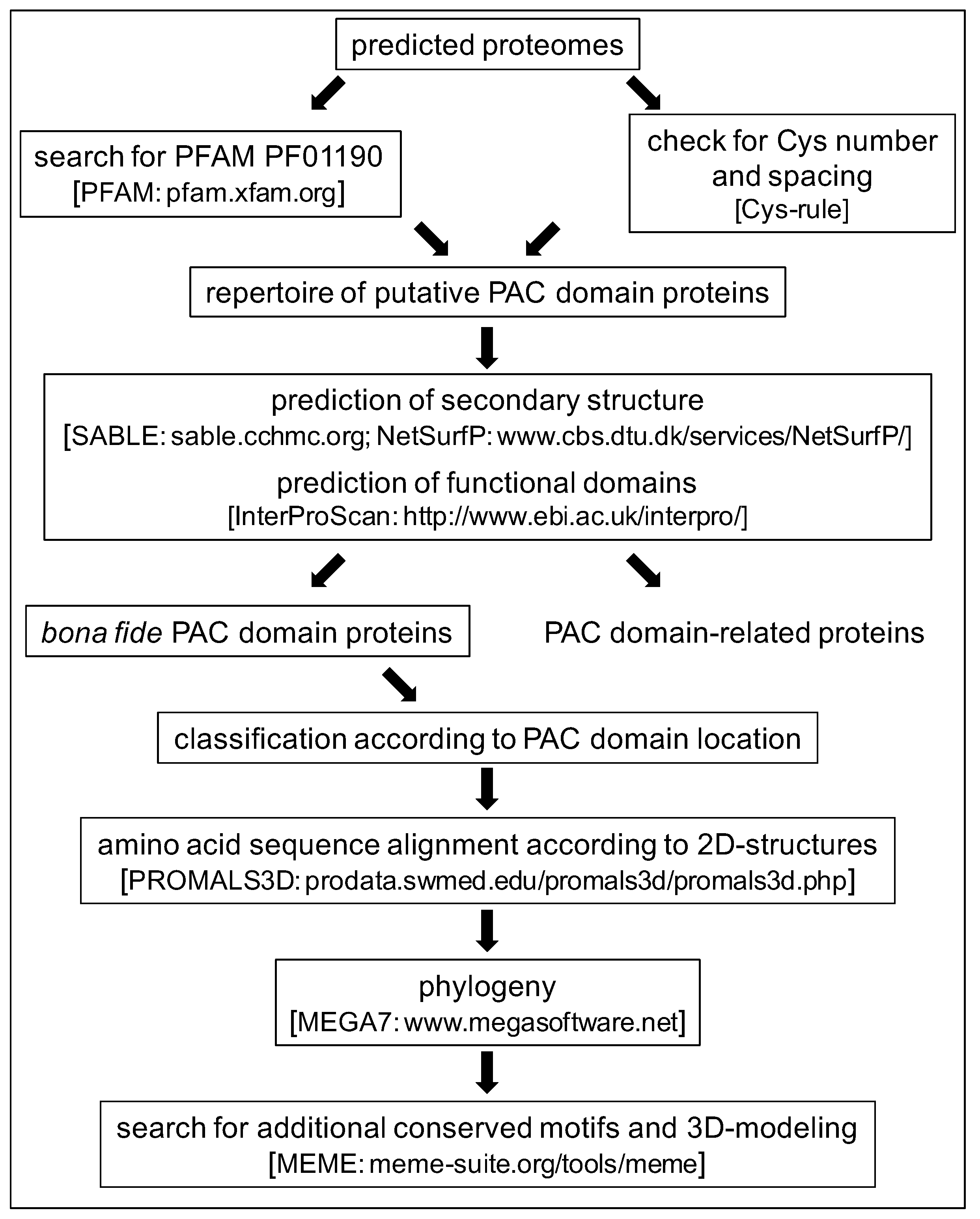
2.1. Characteristics of the PAC Domain and Search for New PDP Candidates
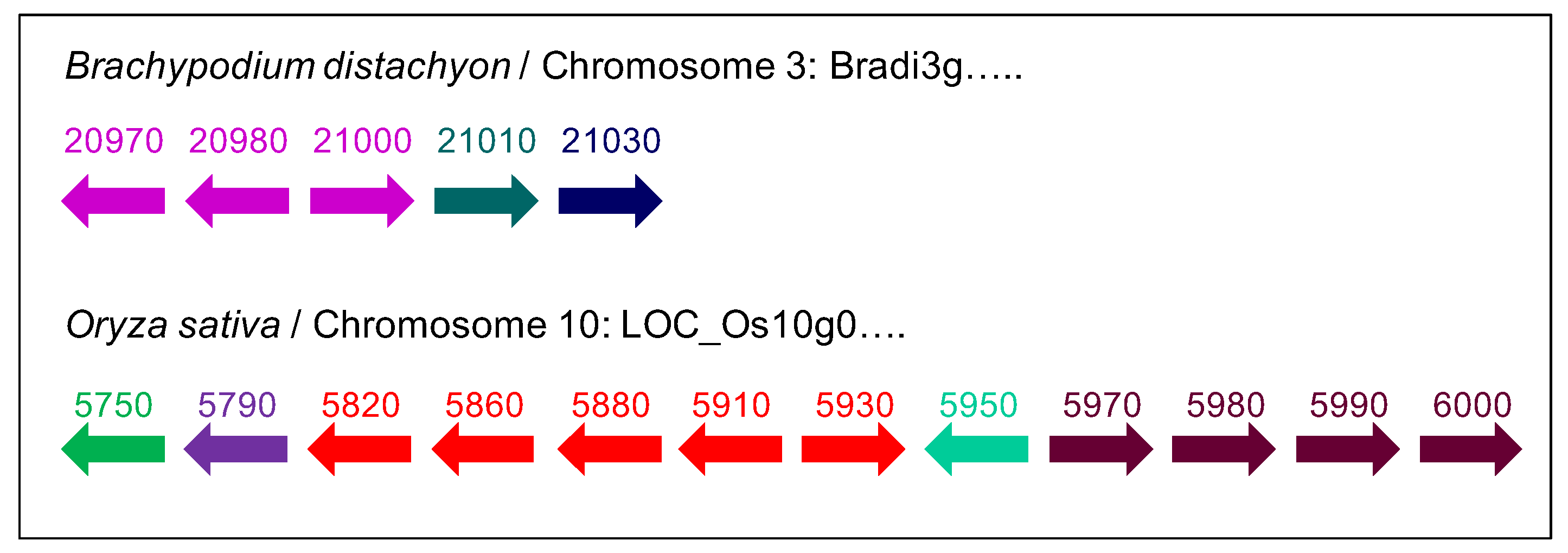
2.2. The Number and the Diversity of PAC Domain Proteins Increase Along the Green Lineage
2.3. A Possible Origin for the PAC Domain
2.4. Three-Dimensional-Modeling of PAC Domain Proteins
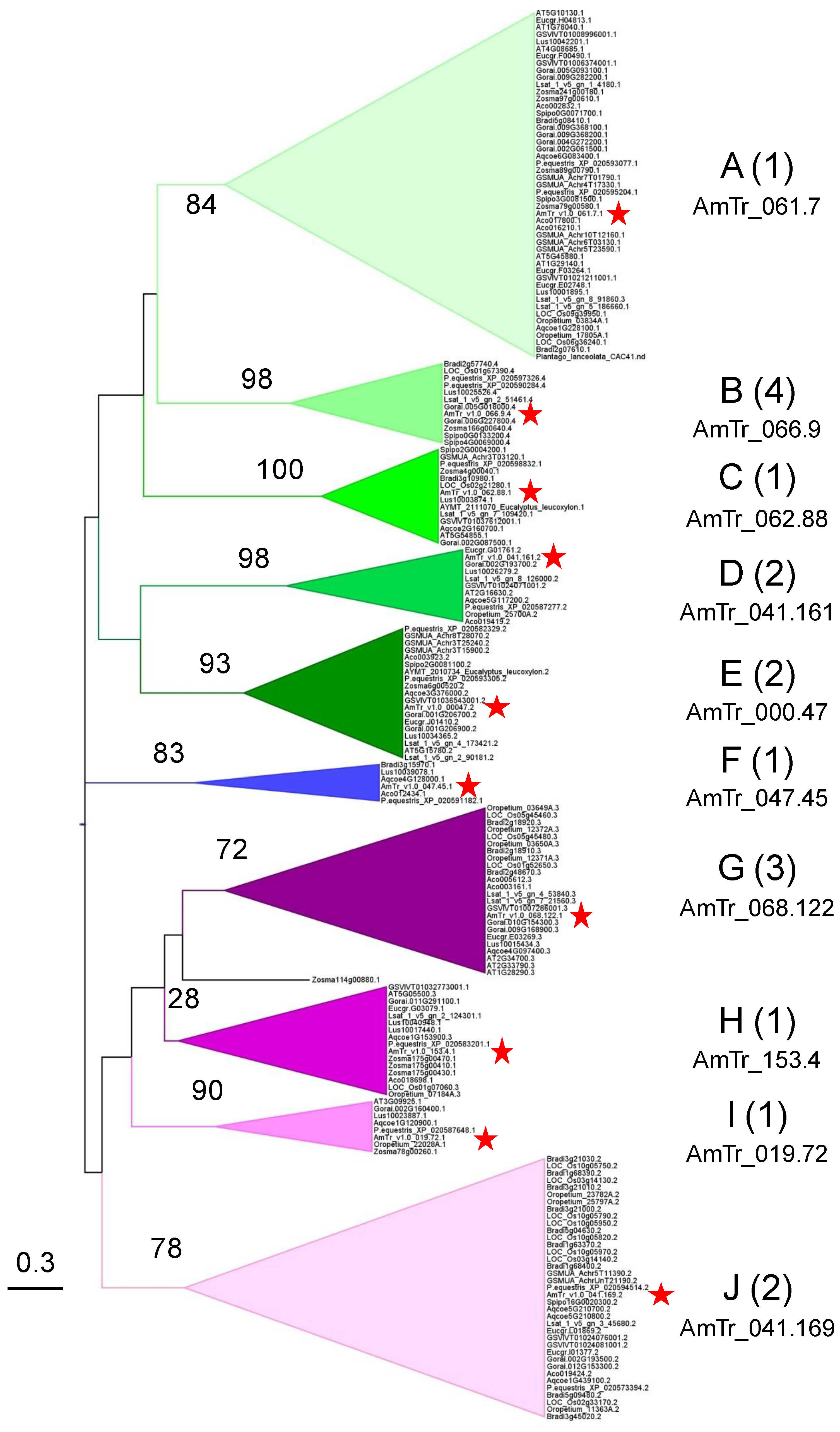
2.5. Phylogenetic Analyses Reveal the Presence of a Few Clades Grouping the PAC Domain Proteins According to their Associated Domains
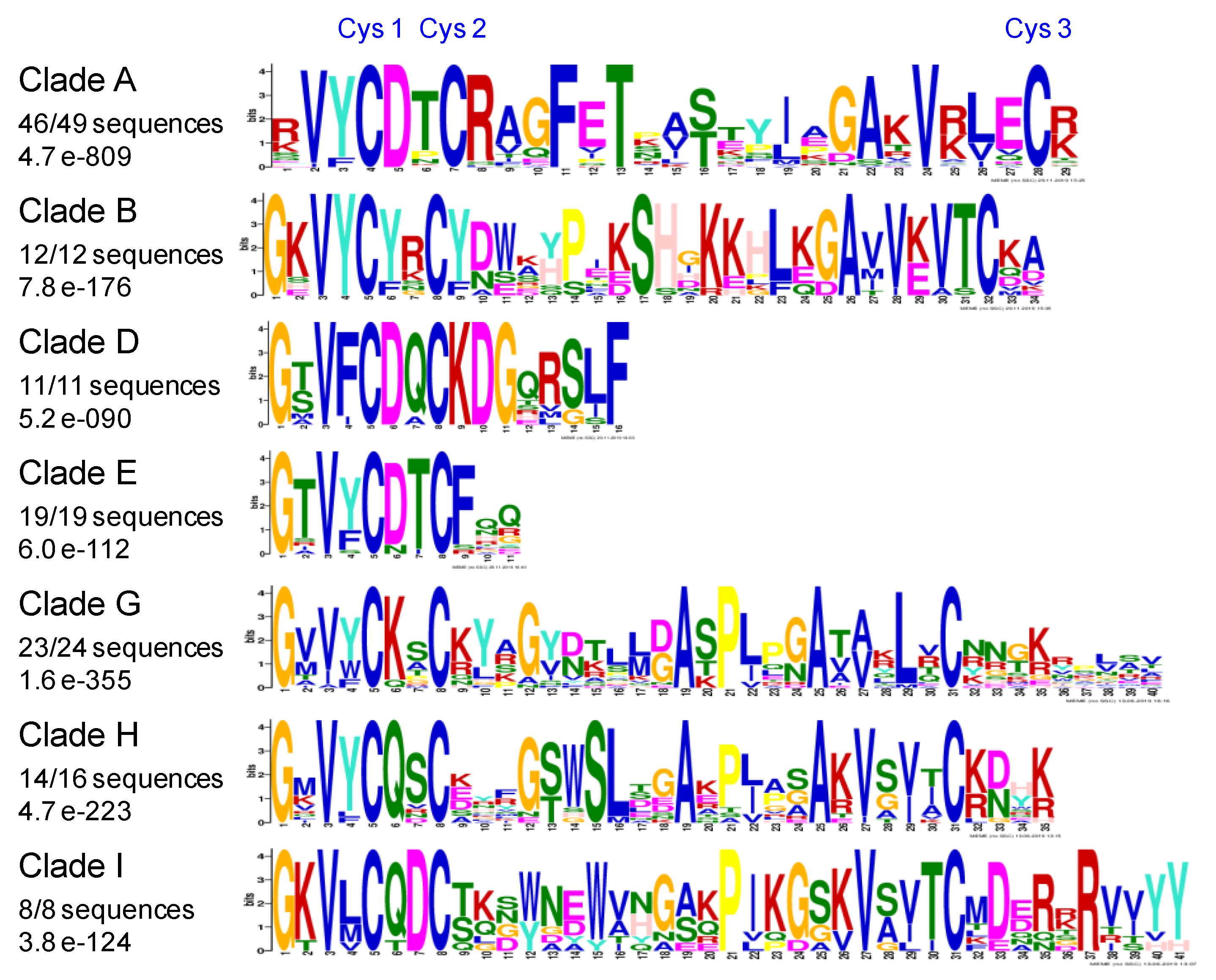
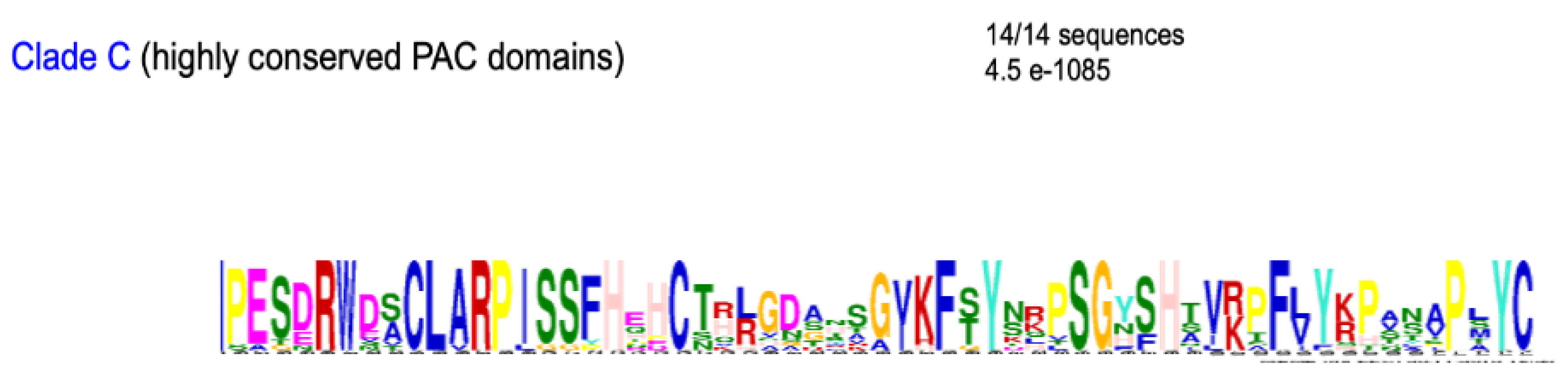
2.6. Conserved Amino Acids Motifs Inside Clades
3. Materials and Methods
3.1. Databases
3.2. Comparisons and Alignment of PAC Domains
3.3. Three-Dimensional Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PAC | Proline-rich, Arabinogalactan protein, conserved Cysteines |
| PDP | PAC Domain Protein |
References
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants, consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Franková, L.; Fry, S.C. Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J. Exp. Bot. 2013, 64, 3519–3550. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Jamet, E.; Albenne, C.; Boudart, G.; Irshad, M.; Canut, H.; Pont-Lezica, R. Recent advances in plant cell wall proteomics. Proteomics 2008, 8, 893–908. [Google Scholar] [CrossRef]
- Fich, E.A.; Fegerson, N.A.; Rose, J.K.C. The plant polyester cutin: Biosynthesis, structure, and biological roles. Ann. Rev. Plant Biol. 2016, 76, 207–233. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef]
- Schaller, A.; Stintzi, A.; Rivas, S.; Serrano, I.; Chichkova, N.V.; Vartapetian, A.B.; Martınez, D.; Guiamet, J.J.; Sueldo, D.J.; van der Hoorn, R.A.L.; et al. From structure to function—A family portrait of plant subtilases. New Phytol. 2018, 218, 901–915. [Google Scholar] [CrossRef]
- Wolf, S.; Hématy, K.; Höfte, H. Growth Control and Cell Wall Signaling in Plants. Annu. Rev. Plant Boil. 2012, 63, 381–407. [Google Scholar] [CrossRef]
- Baldwin, T.C.; van Hengel, A.; Roberts, K. The C-terminal PAC domain of a secreted arabinogalactan protein from carrot defines a family of basic proline-rich proteins. In Cell and Developmental Biology of Arabinogalactan Proteins; Kluwer Academic Publishers: New York, NY, USA, 2000; pp. 43–50. [Google Scholar]
- Du, H.; Simpson, R.; Clarke, A.E.; Bacic, A. Molecular characterization of a stigma-specific gene encoding an arabinogalactan-protein (AGP) from Nicotiana alata. Plant J. 1996, 9, 313–323. [Google Scholar] [CrossRef]
- Hijazi, M.; Roujol, D.; Nguyen-Kim, H.; del Rocio Cisneros Castillo, L.; Saland, E.; Jamet, E.; Albenne, C. Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls? Ann. Bot. 2014, 114, 1087–1097. [Google Scholar] [CrossRef]
- Baldwin, T.C.; Domingo, C.; Schindler, T.; Seetharaman, G.; Stacey, N.; Roberts, K.J. DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol. Boil. 2001, 45, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Van Hengel, A.J.; Roberts, K.J. AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J. 2003, 36, 256–270. [Google Scholar] [CrossRef]
- Liu, C.; Mehdy, M.C. A Nonclassical Arabinogalactan Protein Gene Highly Expressed in Vascular Tissues, AGP31, is Transcriptionally Repressed by Methyl Jasmonic Acid in Arabidopsis1 [OA]. Plant Physiol. 2007, 145, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Mang, H.G.; Lee, J.-H.; Park, J.-A.; Pyee, J.; Pai, H.-S.; Lee, J.; Kim, W.T. The CaPRP1 gene encoding a putative proline-rich glycoprotein is highly expressed in rapidly elongating early roots and leaves in hot pepper (Capsicum annuum L. cv. Pukang). Biochim. Biophys. Acta 2004, 1674, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.-Y.; Huang, G.-Q.; Sun, X.; Li, P.; Zhao, L.L.; Zhang, D.J.; Li, X.B. GhAGP31, a cotton non-classical arabinogalactan protein, is involved in responsae to cold stress during early seedling development. Plant Biol. 2012, 14, 447–457. [Google Scholar] [CrossRef]
- Twomey, M.C.; Brooks, J.K.; Corey, J.M.; Singh-Cundy, A. Characterization of PhPRP1, a histidine domain arabinogalactan protein from Petunia hybrida pistils. J. Plant Physiol. 2013, 170, 1384–1388. [Google Scholar] [CrossRef]
- Hijazi, M.; Durand, J.; Pichereaux, C.; Pont, F.; Jamet, E.; Albenne, C. Characterization of the arabinogalactan protein 31 (AGP31) of Arabidopsis thaliana: New advances on the Hyp-O-glycosylation of the Pro-rich domain. J. Biol. Chem. 2012, 287, 9623–9632. [Google Scholar] [CrossRef]
- Nguyen-Kim, H. Recherche de la Fonction de Protéines Riches en Hydroxyproline Dans les Parois Végétales. Ph.D. Thesis, Toulouse University, Toulouse, France, 2015. [Google Scholar]
- Boron, A.K.; Van Orden, J.; Markakis, M.N.; Mouille, G.; Adriaensen, D.; Verbelen, J.-P.; Höfte, H.; Vissenberg, K. Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 5485–5495. [Google Scholar] [CrossRef]
- Hunt, L.; Amsbury, S.; Baillie, A.; Movahedi, M.; Mitchell, A.; Afsharinafar, M.; Swarup, K.; Denyer, T.; Hobbs, J.K.; Swarup, R.; et al. Formation of the Stomatal Outer Cuticular Ledge Requires a Guard Cell Wall Proline-Rich Protein. Plant Physiol. 2017, 174, 689–699. [Google Scholar] [CrossRef]
- van Hengel, A.; Barber, C.; Roberts, K. The expression patterns of arabinogalactan-protein AtAGP30 and GLABRA2 reveal a role for abcisic acid in the early stages of root epidermal patterning. Plant J. 2004, 39, 70–83. [Google Scholar] [CrossRef]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.F.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Boil. 2008, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Stemeseder, T.; Freier, R.; Wildner, S.; Fuchs, J.E.; Briza, P.; Lang, R.; Batanero, E.; Lidholm, J.; Liedl, K.R.; Campo, P.; et al. Crystal structure of Pla l 1 reveals both structural similarity and allergenic divergence within the Ole e 1–like protein family. J. Allergy Clin. Immunol. 2016, 140, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Amborella Genome Project. The Amborella genome and the evolution of flowering plants. Science 2013, 242, 1241089. [Google Scholar]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef] [PubMed]
- Voigt, J.; Frank, R.; Wöstemeyer, J. The chaotrope-soluble glycoprotein GP1 is a constituent of the insoluble glycoprotein framework of the Chlamydomonascell wall. FEMS Microbiol. Lett. 2009, 291, 209–215. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Voigt, J.; Woestemeyer, J.; Frank, R.; Voigt, J. The Chaotrope-soluble Glycoprotein GP2 is a Precursor of the Insoluble Glycoprotein Framework of the Chlamydomonas Cell Wall. J. Boil. Chem. 2007, 282, 30381–30392. [Google Scholar] [CrossRef]
- Lamport, D.T.A.; Tan, L.; Held, M.; Kieliszewski, M.J. The Role of the Primary Cell Wall in Plant Morphogenesis. Int. J. Mol. Sci. 2018, 19, 2674. [Google Scholar] [CrossRef]
- Barre, A.; Simplicien, M.; Benoist, H.; Van Damme, E.J.M.; Rougé, P. Mannose-Specific Lectins from Marine Algae: Diverse Structural Scaffolds Associated to Common Virucidal and Anti-Cancer Properties. Mar. Drugs 2019, 17, 440. [Google Scholar] [CrossRef]
- Koharudin, L.M.I.; Furey, W.; Gronenborn, A.M. Novel Fold and Carbohydrate Specificity of the Potent Anti-HIV Cyanobacterial Lectin from Oscillatoria agardhii. J. Boil. Chem. 2010, 286, 1588–1597. [Google Scholar] [CrossRef]
- Ruhfel, B.R.; A Gitzendanner, M.; Soltis, P.S.; Soltis, D.; Burleigh, J.G. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Boil. 2014, 14, 23. [Google Scholar] [CrossRef]
- Garau, G.; Di Guilmi, A.-M.; Hall, B.G. Structure-based phylogeny of the metallo-lactamases. Antimicrob. Agents Chemother. 2005, 49, 2778–2784. [Google Scholar] [CrossRef] [PubMed]
- Kakarala, K.K.; Jamil, K. Sequence-structure based phylogeny of GRCR classs A rhodopsin receptors. Mol. Phylogenetics Evol. 2014, 74, 66–96. [Google Scholar] [CrossRef]
- Lakshmi, B.; Mishra, M.; Srinivasan, N.; Archunan, G. Structure-Based Phylogenetic Analysis of the Lipocalin Superfamily. PLoS ONE 2015, 10, e0135507. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.; Puttick, M.; Clark, J.; Edwards, D.; Kenrick, P.; Pressel, S.; Wellman, C.; Yang, Z.; Schneider, H.; Donoghue, P. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 2018, 115, E2274–E2283. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.D.; Soltis, U.E.; Soltis, P.S. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 2010, 97, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Patthy, L. Modular assembly of genes and the evolution of new functions. Genetica 2003, 118, 217–231. [Google Scholar] [CrossRef]
- Chao, Y.-T.; Yen, S.-H.; Yeh, J.-H.; Chen, W.-C.; Shih, M.-C. Orchidstra 2.0—A Transcriptomics Resource for the Orchid Family. Plant Cell Physiol. 2017, 58, 9. [Google Scholar] [CrossRef]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef]
- Goodstein, D.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Carpenter, E.J.; Matasci, N.; Ayyampalayam, S.; Wu, S.; Sun, J.; Yu, J.; Vieira, F.R.J.; Bowler, C.; Dorrell, R.G.; A Gitzendanner, M.; et al. Access to RNA-sequencing data from 1,173 plant species: The 1000 Plant transcriptomes initiative (1KP). GigaScience 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, R.; Porollo, A.; Meller, J.; Porollo, A. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins Struct. Funct. Bioinform. 2005, 59, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.S.; Jespersen, M.C.; Nielsen, H.; Jensen, K.K.; Jurtz, V.I.; Sønderby, C.K.; Sommer, M.O.A.; Winther, O.; Nielsen, M.; Petersen, B.; et al. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins Struct. Funct. Bioinform. 2019, 87, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.-H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Boil. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; De Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012, 41, D344–D347. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.; Qureshi, M.; Richardson, L.; A Salazar, G.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Boil. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Laimer, J.; Hofer, H.; Fritz, M.; Wegenkittl, S.; Lackner, P. MAESTRO-multi agent stability prediction upon point mutations. BMC Bioinform. 2015, 16, 116. [Google Scholar] [CrossRef]
- Shen, M.-Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Tyka, M.D.; Keedy, D.; André, I.; DiMaio, F.; Song, Y.; Richardson, D.C.; Richardson, J.S.; Baker, D. Alternate States of Proteins Revealed by Detailed Energy Landscape Mapping. J. Mol. Boil. 2010, 405, 607–618. [Google Scholar] [CrossRef]
- Voss, N.; Gerstein, M. Calculation of Standard Atomic Volumes for RNA and Comparison with Proteins: RNA is Packed More Tightly. J. Mol. Boil. 2005, 346, 477–492. [Google Scholar] [CrossRef]
- Joosten, R.P.; Beek, T.A.H.T.; Krieger, E.; Hekkelman, M.; Hooft, R.; Schneider, R.; Sander, C.; Vriend, G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2010, 39, D411–D419. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
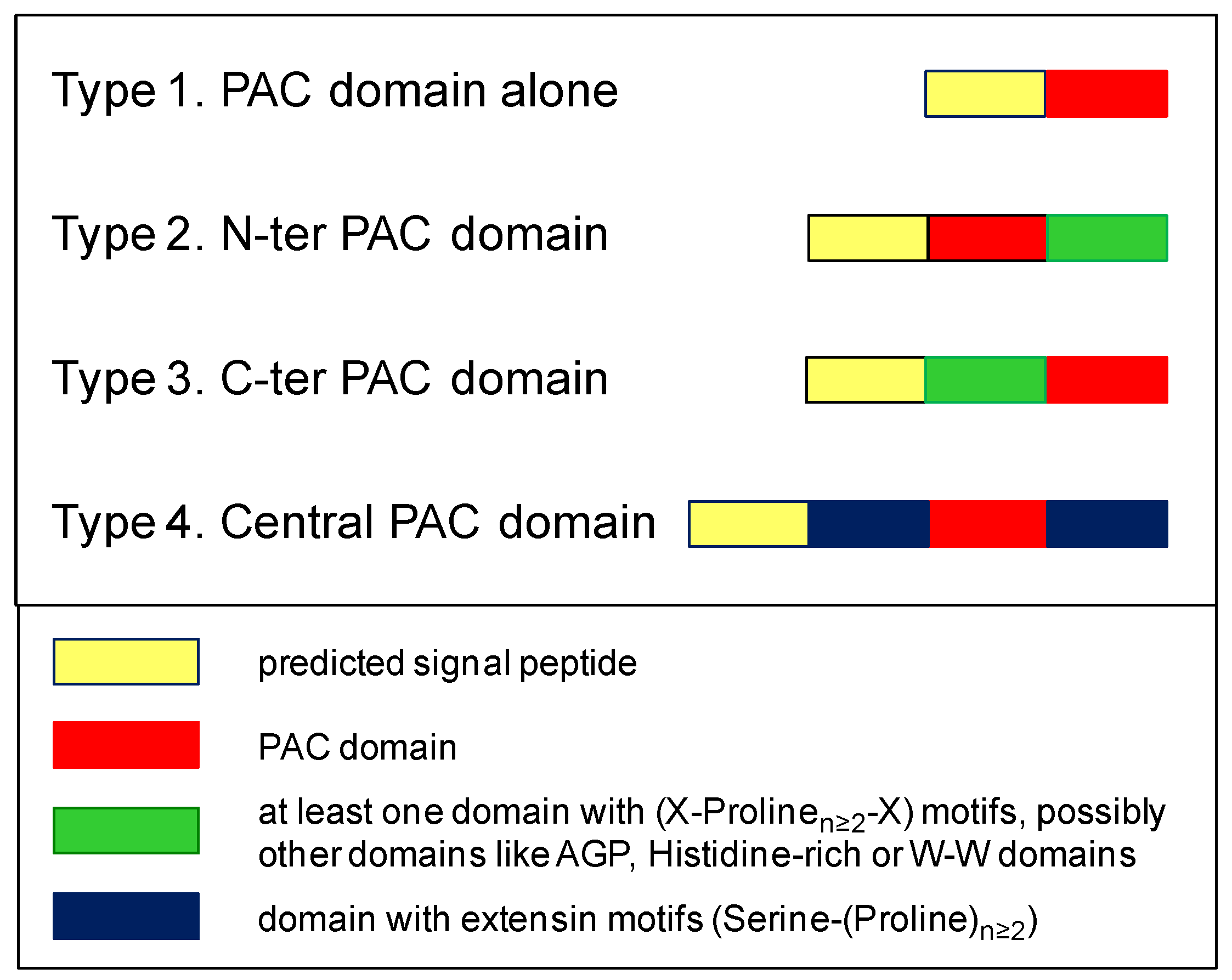


| 1. Present in a protein with a predicted signal peptide |
| 2. Presence of six Cys residues downstream a Glycine residue and with a defined spacing 1: Gly (3) Cys 1 (2) Cys 2 (10,30) Cys 3 (20,50) Cys 4 (8,20) Cys 5 (25,60) Cys 6 |
| 3. Prediction of β-sheets according to the crystal structure of the Plantago lanceolata (www.rcsb.org/structure/4Z8W) PAC domain protein |
| 4. Possibly associated to AGP, extensin, X(Prolinen≥2) X-rich, Histidine-rich, or W-W domains |
| 5. No prediction of additional functional domains |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen-Kim, H.; San Clemente, H.; Laimer, J.; Lackner, P.; Gadermaier, G.; Dunand, C.; Jamet, E. The Cell Wall PAC (Proline-Rich, Arabinogalactan Proteins, Conserved Cysteines) Domain-Proteins Are Conserved in the Green Lineage. Int. J. Mol. Sci. 2020, 21, 2488. https://doi.org/10.3390/ijms21072488
Nguyen-Kim H, San Clemente H, Laimer J, Lackner P, Gadermaier G, Dunand C, Jamet E. The Cell Wall PAC (Proline-Rich, Arabinogalactan Proteins, Conserved Cysteines) Domain-Proteins Are Conserved in the Green Lineage. International Journal of Molecular Sciences. 2020; 21(7):2488. https://doi.org/10.3390/ijms21072488
Chicago/Turabian StyleNguyen-Kim, Huan, Hélène San Clemente, Josef Laimer, Peter Lackner, Gabriele Gadermaier, Christophe Dunand, and Elisabeth Jamet. 2020. "The Cell Wall PAC (Proline-Rich, Arabinogalactan Proteins, Conserved Cysteines) Domain-Proteins Are Conserved in the Green Lineage" International Journal of Molecular Sciences 21, no. 7: 2488. https://doi.org/10.3390/ijms21072488
APA StyleNguyen-Kim, H., San Clemente, H., Laimer, J., Lackner, P., Gadermaier, G., Dunand, C., & Jamet, E. (2020). The Cell Wall PAC (Proline-Rich, Arabinogalactan Proteins, Conserved Cysteines) Domain-Proteins Are Conserved in the Green Lineage. International Journal of Molecular Sciences, 21(7), 2488. https://doi.org/10.3390/ijms21072488






