Differential Targeting of Gr-MDSCs, T Cells and Prostate Cancer Cells by Dactolisib and Dasatinib
Abstract
1. Introduction
2. Results
2.1. Distinct Protein Expression Pattern by PCa Cells, T Cells and Gr-MDSCs in a Mouse CRPC Model
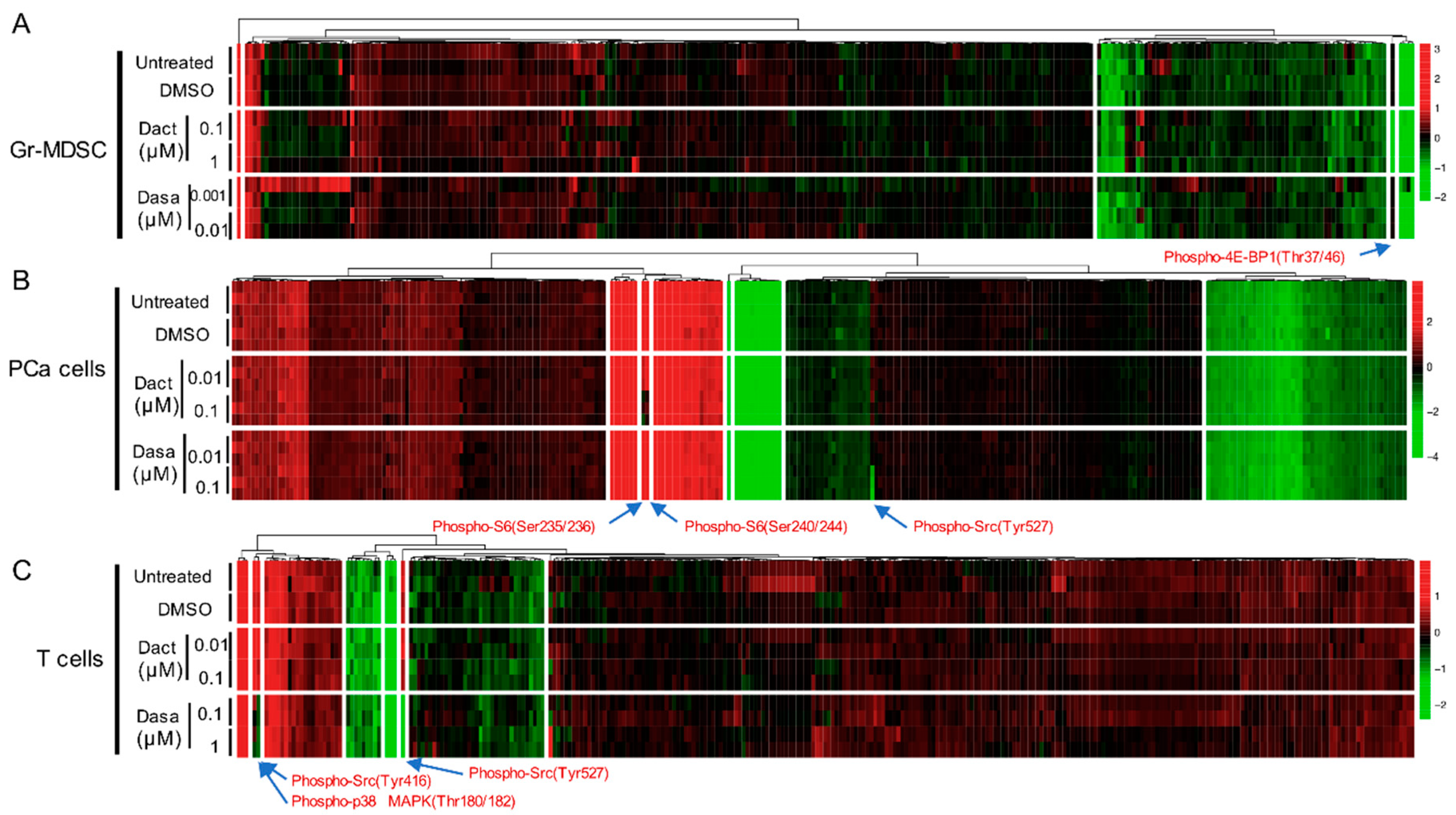
2.2. Differential Effect of Dactolisib and Dasatinib on Protein Phosphorylation in PCa Cells, T cells and Gr-MDSCs
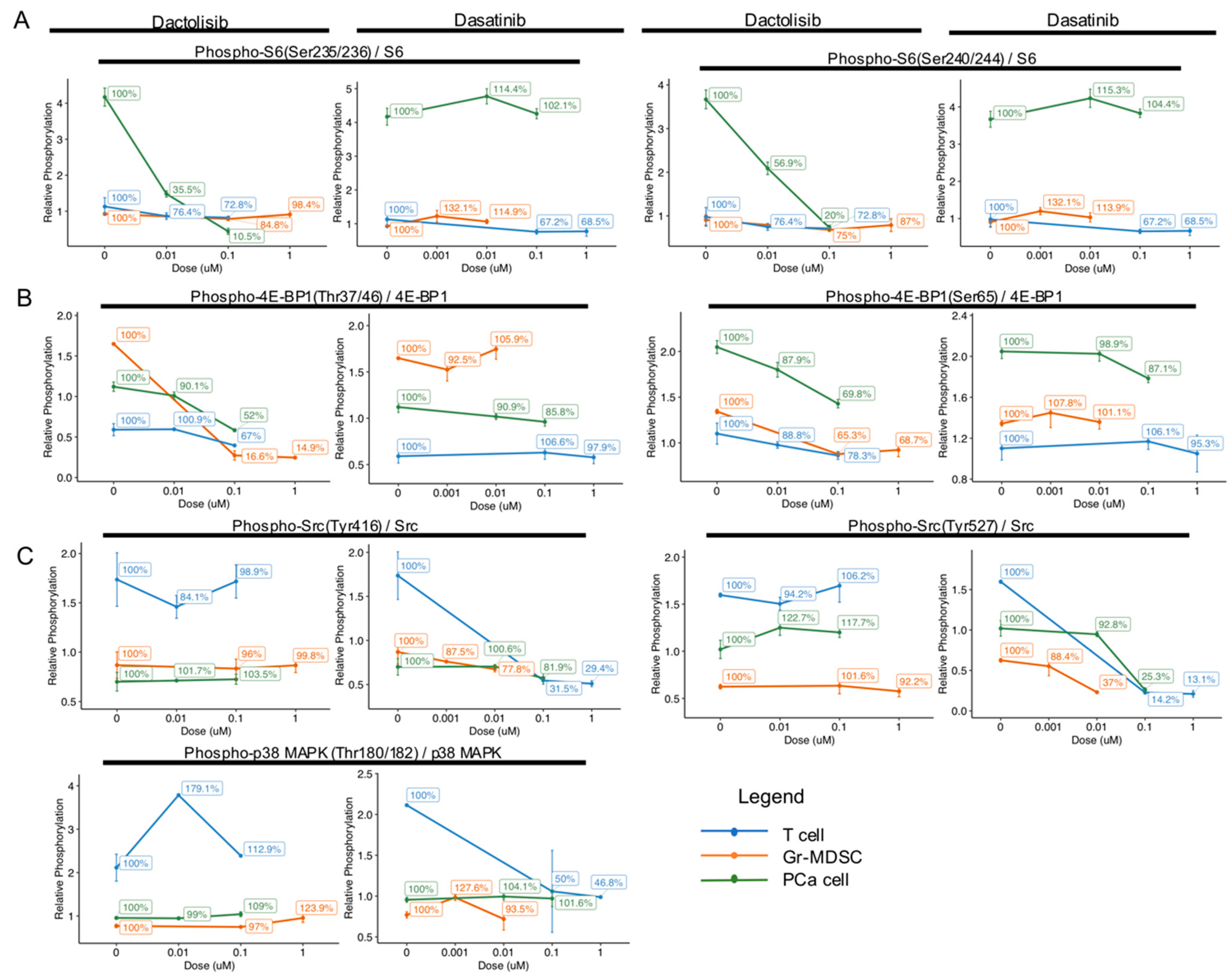
2.3. Cell Type-Specific Downregulation of Phosphorylation Levels of S6, 4E-BP1, Src and p38 MAPK by Dactolisib and Dasatinib
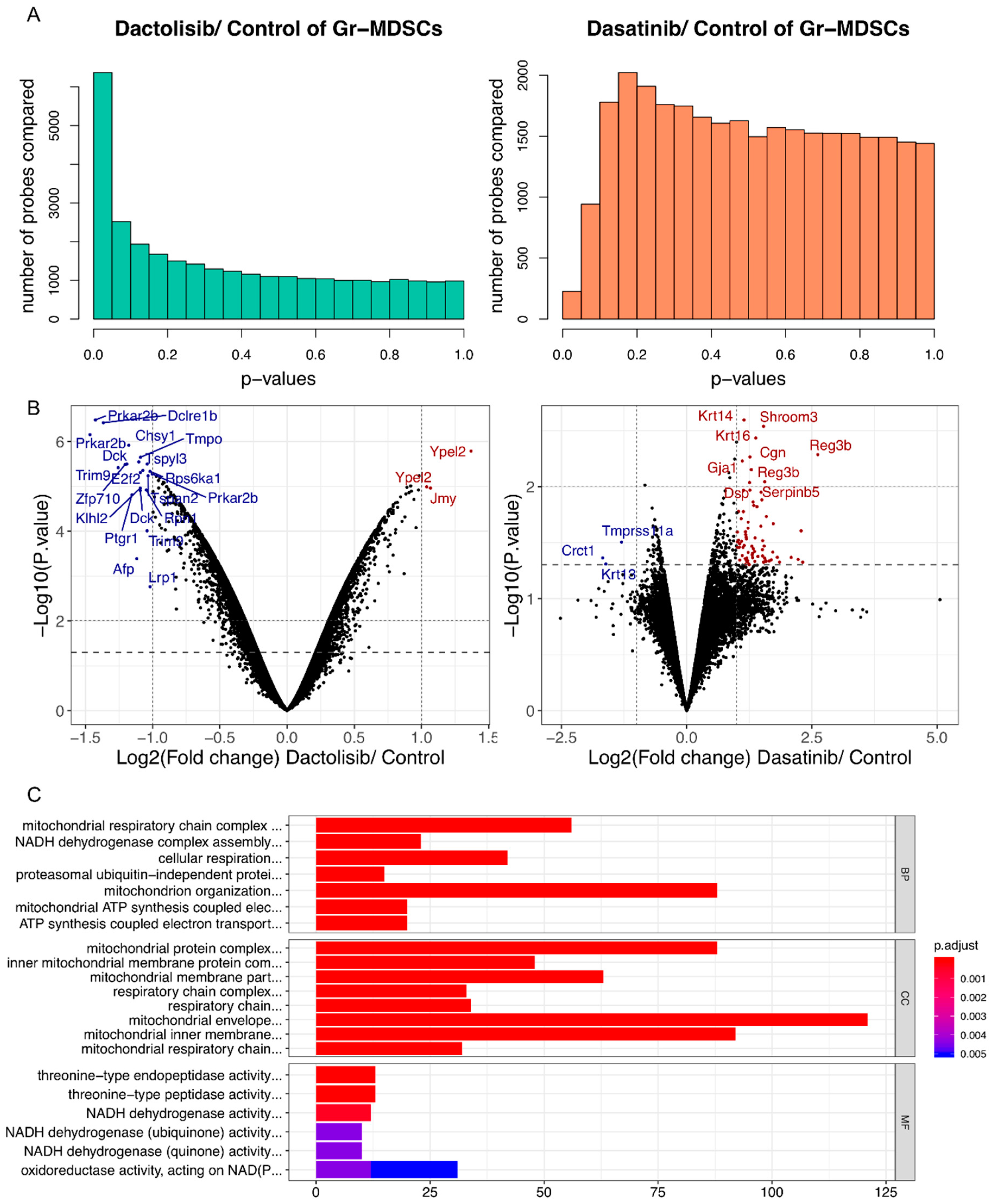
2.4. Dactolisib, but not Dasatinib, Elicited Substantial Transcriptomic Alteration of Gr-MDSCs and Downregulated Genes Associated with Mitochondrial Respiration
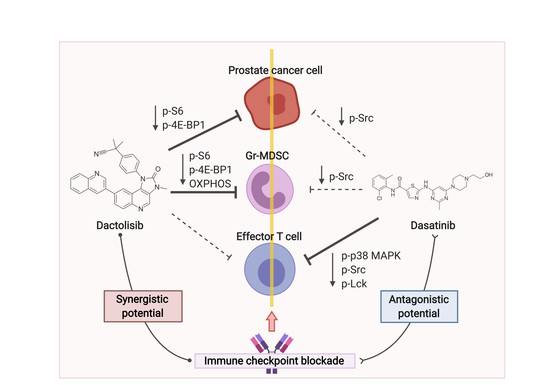
3. Discussion
4. Materials and Methods
4.1. Mouse Model
4.2. Isolation of PCa Cells, Gr-MDSCs and T Cells
4.3. Inhibitor Treatment
4.4. Reverse Phase Protein Array (RPPA)
4.5. Western Blot
4.6. Microarray and Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 4E-BP1 | eukaryotic translation initiation factor 4E binding protein 1 |
| ARID1A | AT rich interactive domain 1A |
| ATP | adenosine triphosphate |
| CD3ε | T-cell surface glycoprotein CD3 epsilon chain |
| CPPSML | PB-Cre+ PtenL/L p53L/L Smad4L/L mTmGL/+ LSL-LucL/+ |
| CRPC | castration-resistant prostate cancer |
| Csk | C-terminal Src kinase |
| CTL | cytotoxic T lymphocyte |
| CTLA4 | cytotoxic-T-lymphocyte-associated protein 4 |
| eIF4G | eukaryotic translation initiation factor 4G |
| FACS | fluorescence-activated cell sorting |
| Fyn | tyrosine-protein kinase Fyn |
| GO | Gene Ontology |
| Gr-MDSC | granulocytic myeloid-derived suppressor cell |
| ICB | immune checkpoint blockade |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| Lck | lymphocyte protein tyrosine kinase |
| MACS | magnetic-activated cell sorting |
| MAPK | mitogen-activated protein kinase |
| mTOR | mechanistic target of rapamycin kinase |
| mTORC1 | mTOR Complex I |
| NDRG1 | N-myc downstream regulated gene 1 |
| PCa | prostate cancer |
| PD1 | programmed cell death 1 |
| PD-L1 | programmed cell death 1 ligand 1 |
| PI3K | phosphoinositide-3-kinase |
| PTM | post-translational modification |
| RPPA | Reverse Phase Protein Array |
| S6K1 | ribosomal protein S6 kinase |
| Src | Rous sarcoma oncogene |
| TAZ | tafazzin |
| TKi | tyrosine kinase inhibitor |
| TNF-α | tumor necrosis factor alpha |
| YAP | yes-associated protein |
References
- Hanahan, D.; Coussens, L. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef] [PubMed]
- Veglia, F.; Perego, M.; Gabrilovich, D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018, 19, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I. Myeloid-Derived Suppressor Cells. Cancer Immunol. Res. 2017, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cheng, X.; Zhang, L.; Lu, X.; Chaudhary, S.; Teng, R.; Frederickson, C.; Champion, M.; Zhao, R.; Cheng, L.; et al. Myeloid-Derived Suppressor Cells Inhibit T Cell Activation through Nitrating LCK in Mouse Cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 10094–10099. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef]
- Beer, T.M.; Kwon, E.D.; Drake, C.G.; Fizazi, K.; Logothetis, C.; Gravis, G.; Ganju, V.; Polikoff, J.; Saad, F.; Humanski, P.; et al. Randomized, Double-Blind, Phase III Trial of Ipilimumab Versus Placebo in Asymptomatic or Minimally Symptomatic Patients With Metastatic Chemotherapy-Naive Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2017, 35, 40–47. [Google Scholar] [CrossRef]
- Simons, J.W. Prostate Cancer Immunotherapy: Beyond Immunity to Curability. Cancer Immunol. Res. 2014, 2, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Di Mitri, D.; Toso, A.; Chen, J.J.; Sarti, M.; Pinton, S.; Jost, T.R.; D’Antuono, R.; Montani, E.; Garcia-Escudero, R.; Guccini, I.; et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature 2014, 515, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lu, X.; Dey, P.; Deng, P.; Wu, C.C.; Jiang, S.; Fang, Z.; Zhao, K.; Konaparthi, R.; Hua, S.; et al. Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discov. 2016, 6, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Spataro, C.; Zagato, E.; Di Mitri, D.; Gil, V.; Crespo, M.; De Bernardis, G.; Losa, M.; Mirenda, M.; Pasquini, E.; et al. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature 2018, 559, 363–369. [Google Scholar] [CrossRef]
- Hossain, D.M.; Pal, S.K.; Moreira, D.; Duttagupta, P.; Zhang, Q.; Won, H.; Jones, J.; D’Apuzzo, M.; Forman, S.; Kortylewski, M. TLR9-Targeted STAT3 Silencing Abrogates Immunosuppressive Activity of Myeloid-Derived Suppressor Cells from Prostate Cancer Patients. Clin. Cancer Res. 2015, 21, 3771–3782. [Google Scholar] [CrossRef]
- Lu, X.; Horner, J.W.; Paul, E.; Shang, X.; Troncoso, P.; Deng, P.; Jiang, S.; Chang, Q.; Spring, D.J.; Sharma, P.; et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature 2017, 543, 728–732. [Google Scholar] [CrossRef]
- Schade, A.E.; Schieven, G.L.; Townsend, R.; Jankowska, A.M.; Susulic, V.; Zhang, R.; Szpurka, H.; Maciejewski, J.P. Dasatinib, a small-molecule protein tyrosine kinase inhibitor, inhibits T-cell activation and proliferation. Blood 2008, 111, 1366–1377. [Google Scholar] [CrossRef]
- Akbani, R.; Becker, K.-F.; Carragher, N.; Goldstein, T.; de Koning, L.; Korf, U.; Liotta, L.; Mills, G.B.; Nishizuka, S.S.; Pawlak, M.; et al. Realizing the Promise of Reverse Phase Protein Arrays for Clinical, Translational, and Basic Research: A Workshop Report: The RPPA (Reverse Phase Protein Array) Society. Mol. Cell. Proteom. 2014, 13, 1625–1643. [Google Scholar] [CrossRef]
- Araujo, J.; Logothetis, C. Dasatinib: A potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat. Rev. 2010, 36, 492–500. [Google Scholar] [CrossRef]
- Vieira, A.F.; Paredes, J. P-cadherin and the journey to cancer metastasis. Mol. Cancer 2015, 14, 178. [Google Scholar] [CrossRef]
- Condamine, T.; Dominguez, G.A.; Youn, J.I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C.; et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016, 1. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Mishalian, I.; Singhal, S.; Cheng, G.; Kapoor, V.; Horng, W.; Fridlender, G.; Bayuh, R.; Worthen, G.S.; et al. Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLoS ONE 2012, 7, e31524. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Erwin, W.; Mawlawi, O.; Gonzalez-Lepera, C.; Tokura, M.; Kurman, M.; Liu, H.; Hong, D.S.; Meric-Bernstam, F.; Pant, S.; et al. Abstract CT128: Phase 1 study of FF-21101(90Y), a radioimmunotherapeutic (RIT) targeting P-cadherin, in advanced solid tumors. Cancer Res. 2018, 78, CT128. [Google Scholar] [CrossRef]
- Menezes, D.; Abrams, T.J.; Karim, C.; Tang, Y.; Ying, C.; Miller, K.; Fanton, C.; Ghoddusi, M.; Wang, Z.; Patawaran, M.; et al. Abstract 1682: Development and activity of a novel antibody-drug conjugate for the treatment of P-cadherin expressing cancers. Cancer Res. 2015, 75, 1682. [Google Scholar] [CrossRef]
- Maira, S.M.; Stauffer, F.; Brueggen, J.; Furet, P.; Schnell, C.; Fritsch, C.; Brachmann, S.; Chene, P.; De Pover, A.; Schoemaker, K.; et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 2008, 7, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Choo, A.Y.; Yoon, S.O.; Kim, S.G.; Roux, P.P.; Blenis, J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci. USA 2008, 105, 17414–17419. [Google Scholar] [CrossRef]
- Choo, A.Y.; Blenis, J. Not all substrates are treated equally: Implications for mTOR, rapamycin-resistance and cancer therapy. Cell Cycle 2009, 8, 567–572. [Google Scholar] [CrossRef]
- Ding, Z.; Wu, C.J.; Jaskelioff, M.; Ivanova, E.; Kost-Alimova, M.; Protopopov, A.; Chu, G.C.; Wang, G.; Lu, X.; Labrot, E.S.; et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 2012, 148, 896–907. [Google Scholar] [CrossRef]
- Schmid, M.; Avraamides, C.; Dippold, H.; Franco, I.; Foubert, P.; Ellies, L.; Acevedo, L.; Manglicmot, J.; Song, X.; Wrasidlo, W.; et al. Receptor tyrosine kinases and TLR/IL1Rs Unexpectedly activate myeloid cell PI3Kγ, A single convergent point promoting tumor inflammation and progression. Cancer Cell 2011, 19, 715–727. [Google Scholar] [CrossRef]
- Davis, R.J.; Moore, E.C.; Clavijo, P.E.; Friedman, J.; Cash, H.; Chen, Z.; Silvin, C.; Van Waes, C.; Allen, C. Anti-PD-L1 Efficacy Can Be Enhanced by Inhibition of Myeloid-Derived Suppressor Cells with a Selective Inhibitor of PI3Kδ/γ. Cancer Res. 2017, 77, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Raught, B.; Gygi, S.P.; Niedzwiecka, A.; Miron, M.; Burley, S.K.; Polakiewicz, R.D.; Wyslouch-Cieszynska, A.; Aebersold, R.; Sonenberg, N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001, 15, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Orth, M.F.; Dallmayer, M.; Baldauf, M.; Pardo, C.; Rotblat, B.; Kirchner, T.; Leprivier, G.; Grunewald, T.G. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): A master regulator of mRNA translation involved in tumorigenesis. Oncogene 2016, 35, 4675–4688. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Gravel, S.-P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.-P.; Chénard, V.; Sikström, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C.; et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013, 18, 698–711. [Google Scholar] [CrossRef]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Valle, L.D.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef]
- Salmond, R.J.; Filby, A.; Qureshi, I.; Caserta, S.; Zamoyska, R. T-cell receptor proximal signaling via the Src-family kinases, Lck and Fyn, influences T-cell activation, differentiation, and tolerance. Immunol. Rev. 2009, 228, 9–22. [Google Scholar] [CrossRef]
- Lee, K.C.; Ouwehand, I.; Giannini, A.L.; Thomas, N.S.; Dibb, N.J.; Bijlmakers, M.J. Lck is a key target of imatinib and dasatinib in T-cell activation. Leukemia 2010, 24, 896–900. [Google Scholar] [CrossRef]
- Thomas, S.M.; Brugge, J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. [Google Scholar] [CrossRef]
- Dodeller, F.; Schulze-Koops, H. The p38 mitogen-activated protein kinase signaling cascade in CD4 T cells. Arthritis Res. Ther. 2006, 8, 205. [Google Scholar] [CrossRef]
- Schafer, P.H.; Wang, L.; Wadsworth, S.A.; Davis, J.E.; Siekierka, J.J. T Cell Activation Signals Up-Regulate p38 Mitogen-Activated Protein Kinase Activity and Induce TNF-α Production in a Manner Distinct from LPS Activation of Monocytes. J. Immunol. 1999, 162, 659–668. [Google Scholar] [PubMed]
- Heine, A.; Schilling, J.; Grunwald, B.; Kruger, A.; Gevensleben, H.; Held, S.A.; Garbi, N.; Kurts, C.; Brossart, P.; Knolle, P.; et al. The induction of human myeloid derived suppressor cells through hepatic stellate cells is dose-dependently inhibited by the tyrosine kinase inhibitors nilotinib, dasatinib and sorafenib, but not sunitinib. Cancer Immunol. Immunother. CII 2016, 65, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Christiansson, L.; Soderlund, S.; Mangsbo, S.; Hjorth-Hansen, H.; Hoglund, M.; Markevarn, B.; Richter, J.; Stenke, L.; Mustjoki, S.; Loskog, A.; et al. The tyrosine kinase inhibitors imatinib and dasatinib reduce myeloid suppressor cells and release effector lymphocyte responses. Mol. Cancer Ther. 2015, 14, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Fei, F.; Yu, Y.; Schmitt, A.; Rojewski, M.T.; Chen, B.; Greiner, J.; Gotz, M.; Guillaume, P.; Dohner, H.; Bunjes, D.; et al. Dasatinib exerts an immunosuppressive effect on CD8+ T cells specific for viral and leukemia antigens. Exp. Hematol. 2008, 36, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Tang, L.C.; Krug, K.; Clark, D.J.; Gritsenko, M.A.; Chen, L.; Clauser, K.R.; Clauss, T.R.; Shah, P.; Gillette, M.A.; et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat. Protoc. 2018, 13, 1632–1661. [Google Scholar] [CrossRef]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics (Oxford, England) 2010, 26, 2363–2367. [Google Scholar] [CrossRef]
- Kauffmann, A.; Gentleman, R.; Huber, W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef]
- Kauffmann, A.; Rayner, T.F.; Parkinson, H.; Kapushesky, M.; Lukk, M.; Brazma, A.; Huber, W. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics 2009, 25, 2092–2094. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Jin, Z.; Lu, X. Differential Targeting of Gr-MDSCs, T Cells and Prostate Cancer Cells by Dactolisib and Dasatinib. Int. J. Mol. Sci. 2020, 21, 2337. https://doi.org/10.3390/ijms21072337
Liu G, Jin Z, Lu X. Differential Targeting of Gr-MDSCs, T Cells and Prostate Cancer Cells by Dactolisib and Dasatinib. International Journal of Molecular Sciences. 2020; 21(7):2337. https://doi.org/10.3390/ijms21072337
Chicago/Turabian StyleLiu, Guoqiang, Zhijian Jin, and Xin Lu. 2020. "Differential Targeting of Gr-MDSCs, T Cells and Prostate Cancer Cells by Dactolisib and Dasatinib" International Journal of Molecular Sciences 21, no. 7: 2337. https://doi.org/10.3390/ijms21072337
APA StyleLiu, G., Jin, Z., & Lu, X. (2020). Differential Targeting of Gr-MDSCs, T Cells and Prostate Cancer Cells by Dactolisib and Dasatinib. International Journal of Molecular Sciences, 21(7), 2337. https://doi.org/10.3390/ijms21072337