Recombination Pattern Characterization via Simulation Using Different Maize Populations
Abstract
1. Introduction
2. Results
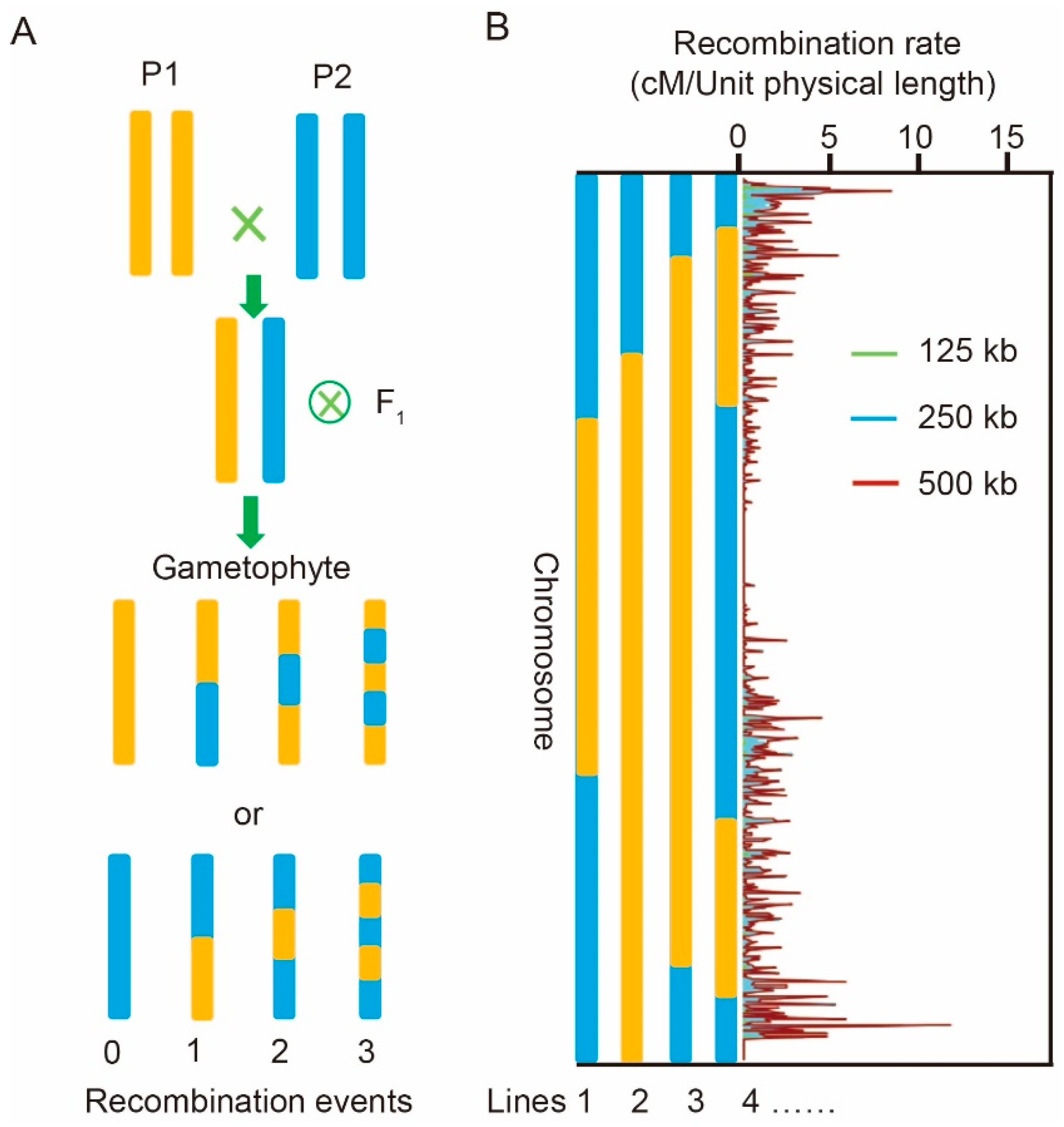
2.1. Variance of Two Key Recombination Parameters
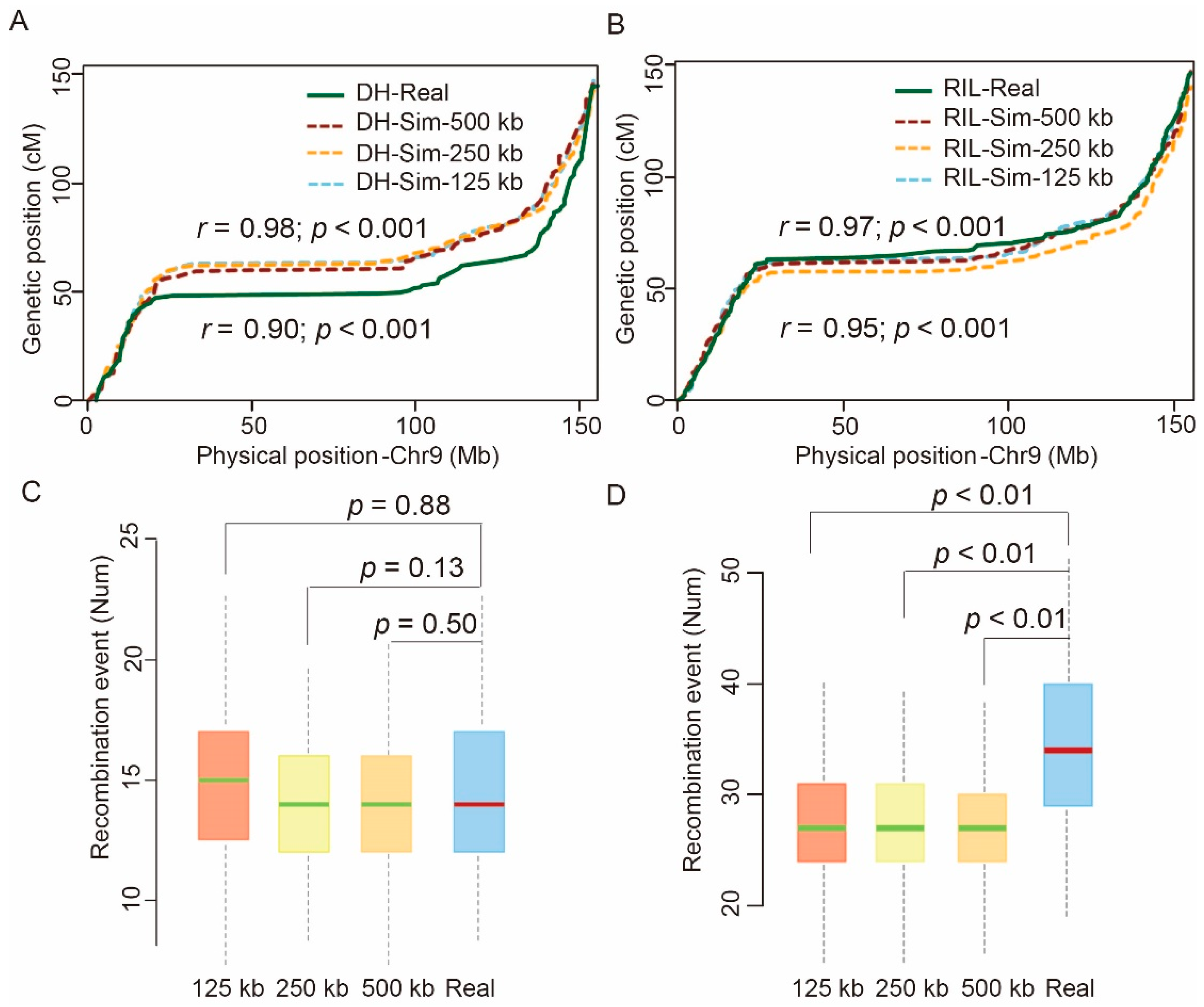
2.2. Comparison between Simulated and Real Genotypic Data
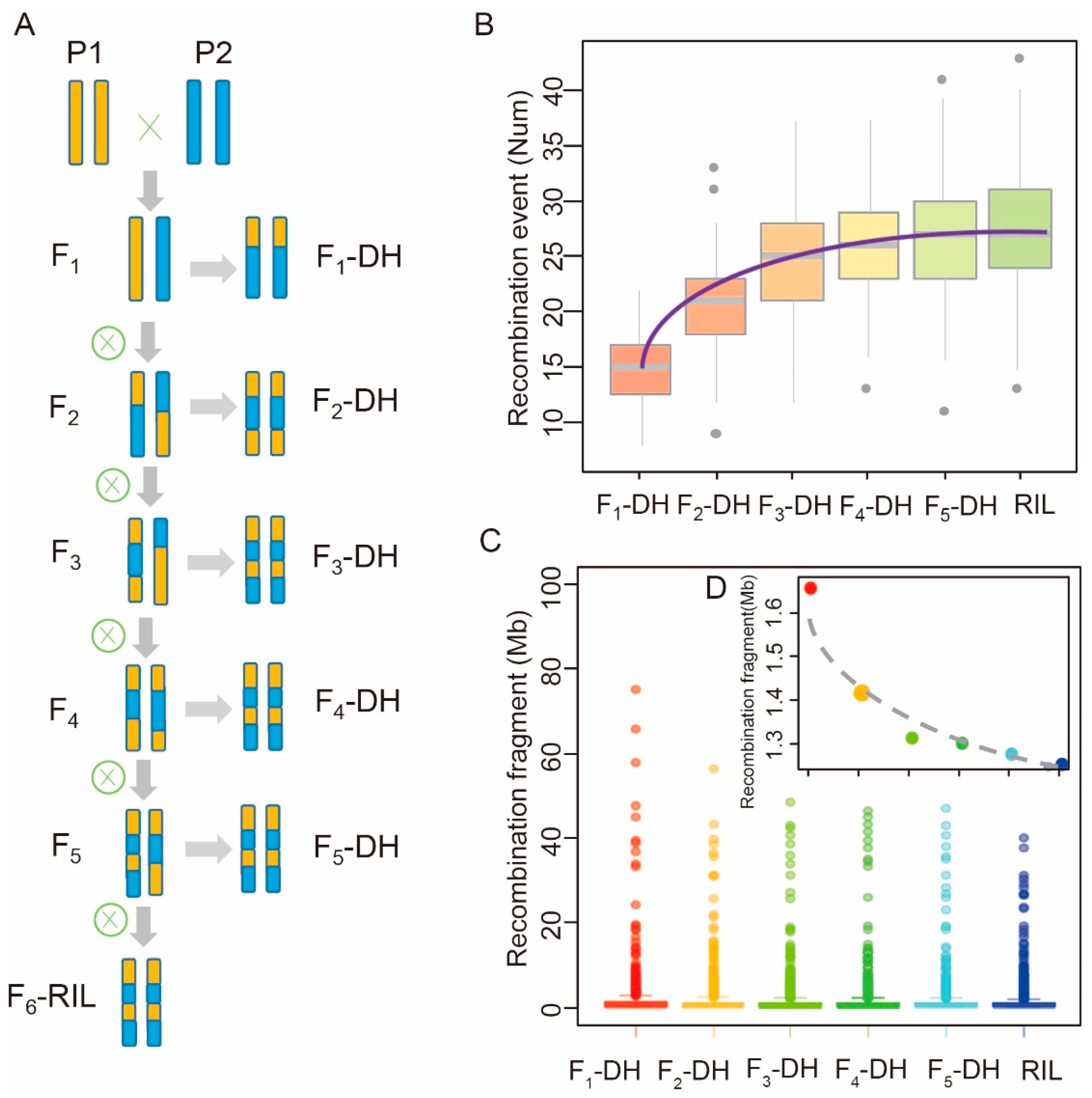
2.3. Recombination Pattern in DH Populations Induced at Different Generations
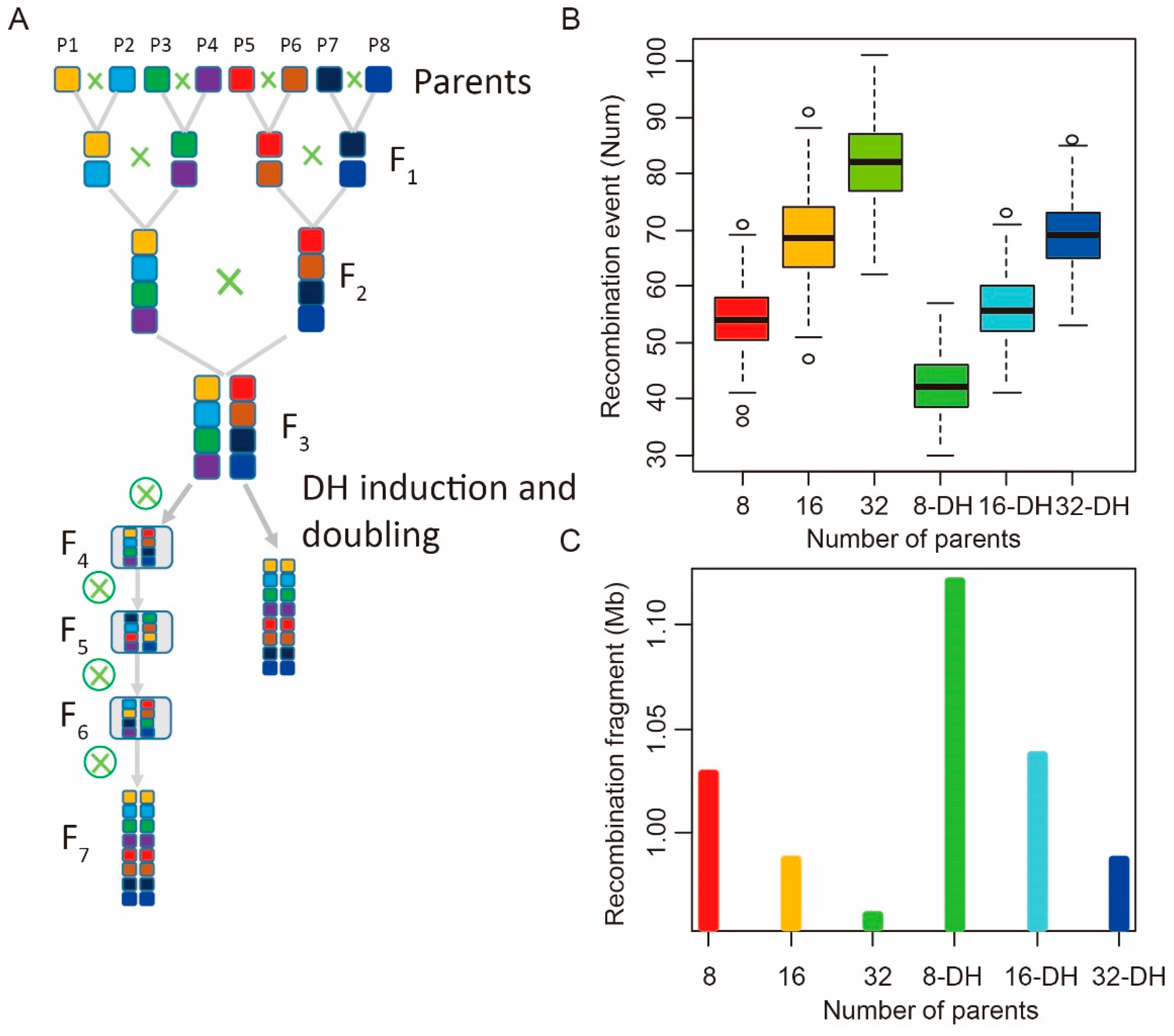
2.4. Recombination Pattern Comparison of MAGIC Populations Constructed by DH Induction and Continuous Self-Crossing
3. Discussion
3.1. Feasibility and Influencing Factors of the Simulation Method
3.2. Recombination Simulation Is a Fast and Cost-Effective Approach for Genetic Studies and Crop Improvement
3.3. The Simulation Method Has Broader Implications
4. Materials and Methods
4.1. Determination of the Two Recombination Parameters
4.2. Estimation of the Number of Recombination Events and Fragment Length
4.3. Comparison of the Linkage Maps of the Simulated and Real Populations
4.4. Analysis of the Recombination Pattern in Different MAGIC Populations
4.5. Code Available
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | Bulk segregant analysis |
| BSR-seq | Bulked segregant RNA-seq |
| CO | The number of crossover events |
| CS | Self-crossing |
| DSB | Double-strand breaks |
| DH | Doubled haploid |
| GCA | General combining ability |
| IBM | B73 × Mo17 maize population |
| MAGIC | Multiparent advanced generation intercross populations |
| QTG-seq | Quantitative trait gene sequencing |
| QTL | Quantitative trait locus |
| RIL | Recombination inbred line |
References
- Gaut, B.S.; Wright, S.I.; Rizzon, C.; Dvorak, J.; Anderson, L.K. Recombination: An underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 2007, 8, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.; Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Franklin, F.C. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011, 190, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Rodgers-Melnick, E.; Bradbury, P.J.; Elshire, R.J.; Glaubitz, J.C.; Acharya, C.B.; Mitchell, S.E.; Li, C.; Li, Y.; Buckler, E.S. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. USA 2015, 112, 3823–3828. [Google Scholar] [CrossRef] [PubMed]
- Wijnker, E.; de Jong, H. Managing meiotic recombination in plant breeding. Trends Plant Sci. 2008, 13, 640–646. [Google Scholar] [CrossRef]
- Guan, H.; Ali, F.; Pan, Q. Dissection of recombination attributes for multiple maize populations using a common SNP assay. Front. Plant Sci. 2017, 8, 2063. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Zegeye, W.A.; Zhang, Y.; Cao, L.; Cheng, S. Whole Genome Resequencing from bulked populations as a rapid QTL and gene identification method in rice. Int. J. Mol. Sci. 2018, 19, 4000. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Pan, Q.; Li, P.; Liu, Y.; Lu, X.; Zhong, W.; Li, M.; Han, L.; Li, J.; et al. QTG-Seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Mol. Plant 2019, 12, 426–437. [Google Scholar] [CrossRef]
- Liu, C.; Zhong, Y.; Qi, X.; Chen, M.; Liu, Z.; Chen, C.; Tian, X.; Li, J.; Jiao, Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2020, 18, 316–318. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Seymour, D.K.; Filiault, D.L.; Henry, I.M.; Monson-Miller, J.; Ravi, M.; Pang, A.; Comai, L.; Chan, S.W.; Maloof, J.N. Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc. Natl. Acad. Sci. USA 2012, 109, 4227–4232. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.E.; Verbyla, K.L.; Verbyla, A.P.; Raghavan, C.; Singh, V.K.; Gaur, P.; Leung, H.; Varshney, R.K.; Cavanagh, C.R. MAGIC populations in crops: Current status and future prospects. Theor. Appl. Genet. 2015, 128, 999–1017. [Google Scholar] [CrossRef]
- Lee, M.; Sharopova, N.; Beavis, W.D.; Grant, D.; Katt, M.; Blair, D.; Hallauer, A. Expanding the genetic map of maize with the intermated B73 x Mo17 (IBM) population. Plant Mol. Biol. 2002, 48, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, L.; Wang, J.; Li, C.; Zeng, X.; Xie, S.; Zhang, Y.; Liu, S.; Hu, S.; Wang, J.; et al. Quantitative Trait Locus Analysis for Deep-Sowing Germination Ability in the Maize IBM Syn10 DH Population. Front. Plant Sci. 2017, 8, 813. [Google Scholar] [CrossRef] [PubMed]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding. Rice (NY) 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Falque, M.; Walter, H.; Bauland, C.; Camisan, C.; Campo, L.; Meyer, N.; Ranc, N.; Rincent, R.; Schipprack, W.; et al. Intraspecific variation of recombination rate in maize. Genome Biol. 2013, 14, R103. [Google Scholar] [CrossRef]
- Stumpf, M.P.; McVean, G.A. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 2003, 4, 959–968. [Google Scholar] [CrossRef]
- Pan, Q.; Deng, M.; Yan, J.; Li, L. Complexity of genetic mechanisms conferring nonuniformity of recombination in maize. Sci. Rep. 2017, 7, 1205. [Google Scholar] [CrossRef]
- Radding, C.M. Molecular mechanisms in genetic recombination. Annu. Rev. Genet. 1973, 7, 87–111. [Google Scholar] [CrossRef]
- Pan, Q.; Ali, F.; Yang, X.; Li, J.; Yan, J. Exploring the genetic characteristics of two recombinant inbred line populations via high-density SNP markers in maize. PLoS ONE 2012, 7, e52777. [Google Scholar] [CrossRef]
- Li, R.; Bitoun, E.; Altemose, N.; Davies, R.W.; Davies, B.; Myers, S.R. A high-resolution map of non-crossover events reveals impacts of genetic diversity on mammalian meiotic recombination. Nat. Commun. 2019, 10, 3900. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Morgan, C.; Franklin, F.C.H.; Bomblies, K. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 2018, 208, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Modliszewski, J.L.; Wang, H.; Albright, A.R.; Lewis, S.M.; Bennett, A.R.; Huang, J.; Ma, H.; Wang, Y.; Copenhaver, G.P. Elevated temperature increases meiotic crossover frequency via the interfering (Type I) pathway in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007384. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Li, X.; Zhang, Q.; Yan, J. Single gametophyte sequencing reveals that crossover events differ between sexes in maize. Nat. Commun. 2019, 10, 785. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.B.; Seguela-Arnaud, M.; Larcheveque, C.; Lloyd, A.H.; Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. USA 2018, 115, 2431–2436. [Google Scholar] [CrossRef]
- Ishii, T.; Karimi-Ashtiyani, R.; Houben, A. Haploidization via chromosome elimination: Means and mechanisms. Annu. Rev. Plant Biol. 2016, 67, 421–438. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Goff, S.A.; Ricke, D.; Lan, T.H.; Presting, G.; Wang, R.; Dunn, M.; Glazebrook, J.; Sessions, A.; Oeller, P.; Varma, H.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Hall, R.; Kingan, S.; Clavijo, B.J.; Salzberg, S.L. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Liu, S.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap-Gap: Whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zeng, Q.; Wang, Q.; Liu, S.; Yu, S.; Mu, J.; Huang, S.; Sela, H.; Distelfeld, A.; Huang, L.; et al. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018, 131, 1481–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, H.; Lyle, D.; Li, D.; Wang, G.; Pan, Q.; Wang, J.; Lübberstedt, T. Bulk pollen pollination in maize for efficient construction of introgression populations with high genome coverage. Plant Breed. 2019, 138, 252–258. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Gong, X.; Li, K.; Zhang, H.; Chen, F.; Pan, Q. Recombination Pattern Characterization via Simulation Using Different Maize Populations. Int. J. Mol. Sci. 2020, 21, 2222. https://doi.org/10.3390/ijms21062222
Ren W, Gong X, Li K, Zhang H, Chen F, Pan Q. Recombination Pattern Characterization via Simulation Using Different Maize Populations. International Journal of Molecular Sciences. 2020; 21(6):2222. https://doi.org/10.3390/ijms21062222
Chicago/Turabian StyleRen, Wei, Xiaoping Gong, Kun Li, Hongwei Zhang, Fanjun Chen, and Qingchun Pan. 2020. "Recombination Pattern Characterization via Simulation Using Different Maize Populations" International Journal of Molecular Sciences 21, no. 6: 2222. https://doi.org/10.3390/ijms21062222
APA StyleRen, W., Gong, X., Li, K., Zhang, H., Chen, F., & Pan, Q. (2020). Recombination Pattern Characterization via Simulation Using Different Maize Populations. International Journal of Molecular Sciences, 21(6), 2222. https://doi.org/10.3390/ijms21062222






