TmPGRP-SA regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio molitor
Abstract
1. Introduction
2. Results
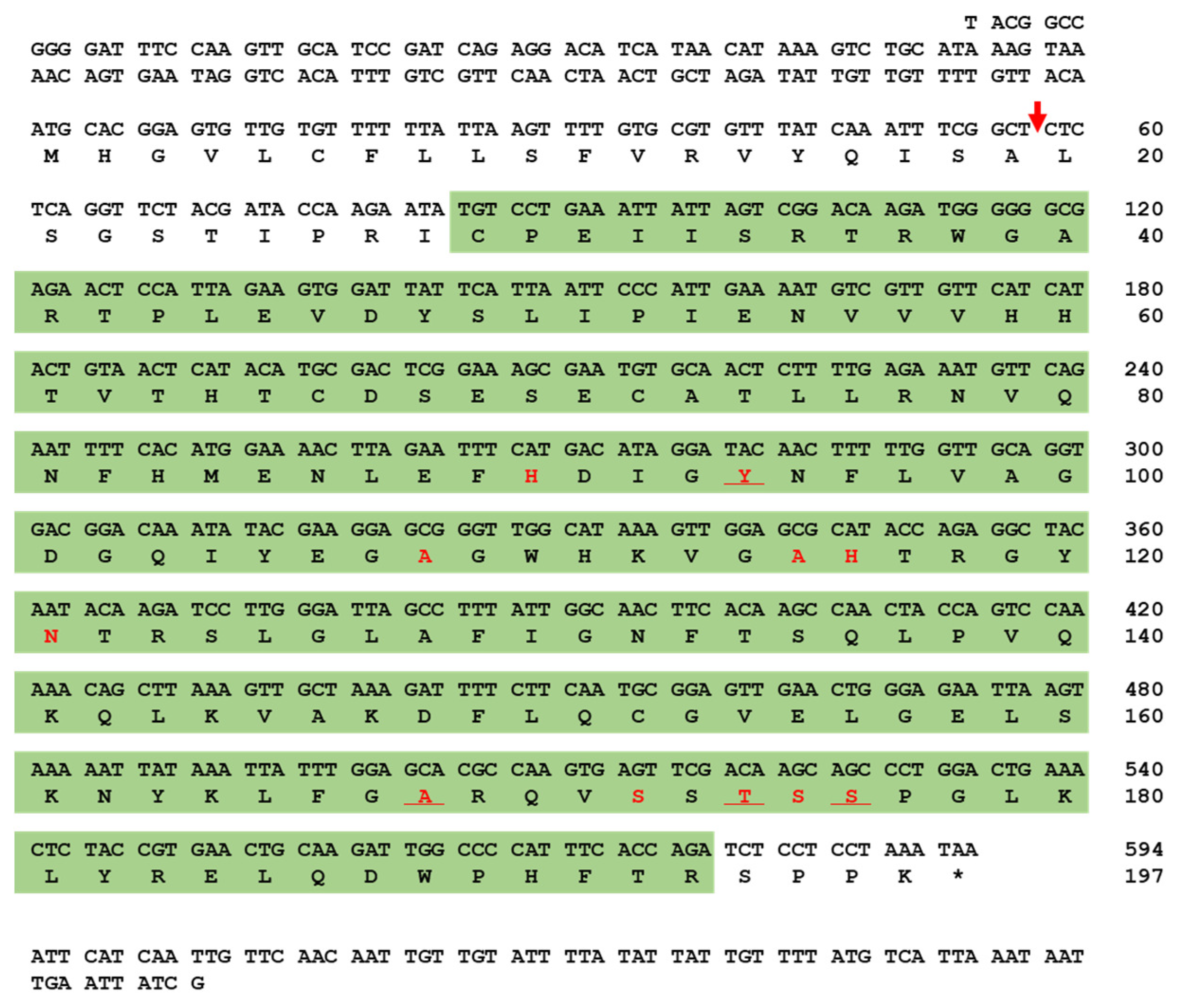
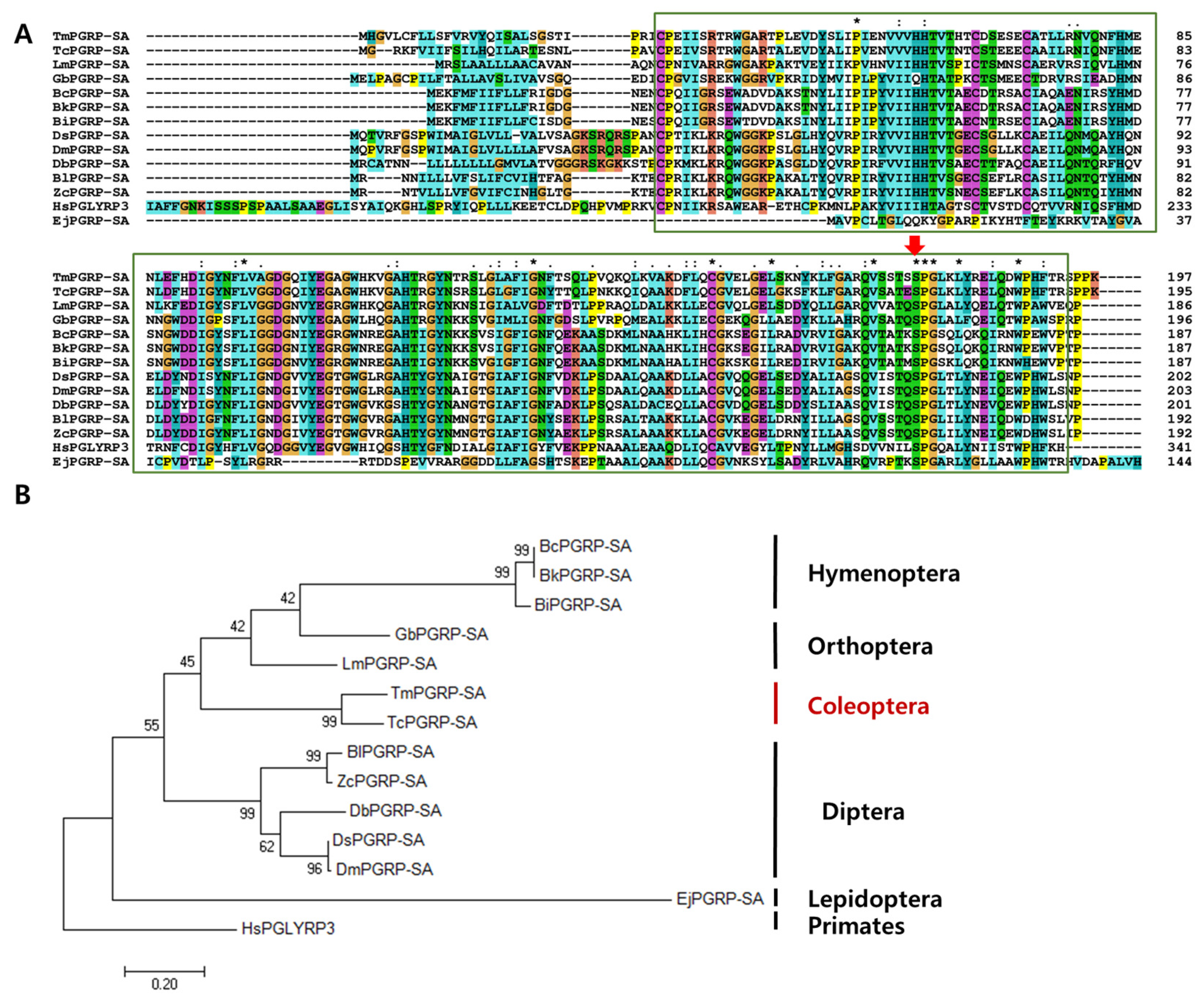
2.1. Gene Organization, cDNA Analysis, and Phylogenetic Tree of TmPGRP-SA
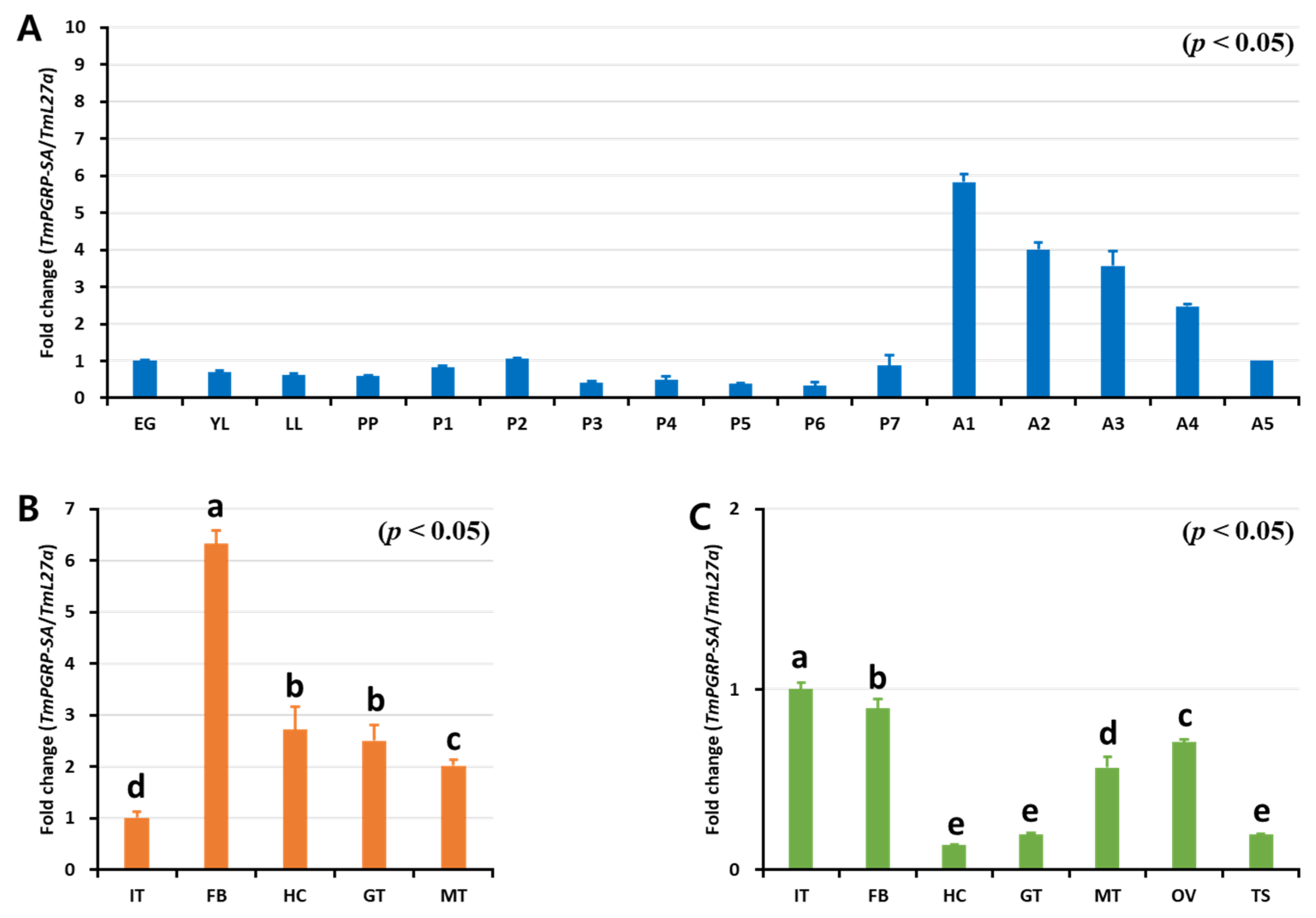
2.2. Developmental and Tissue Distribution of TmPGRP-SA
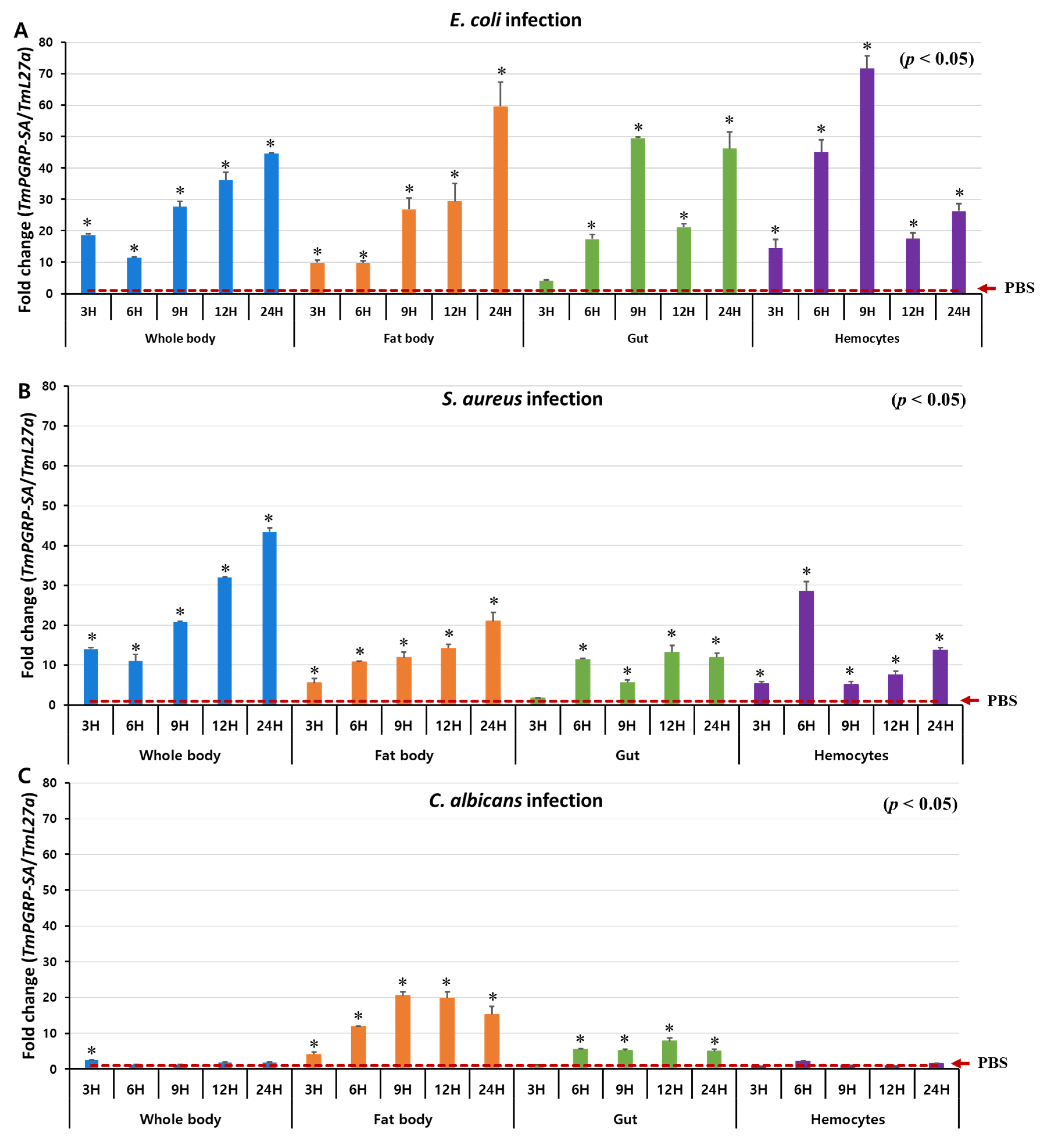
2.3. TmPGRP-SA is Upregulated following Microbial Infection in vivo
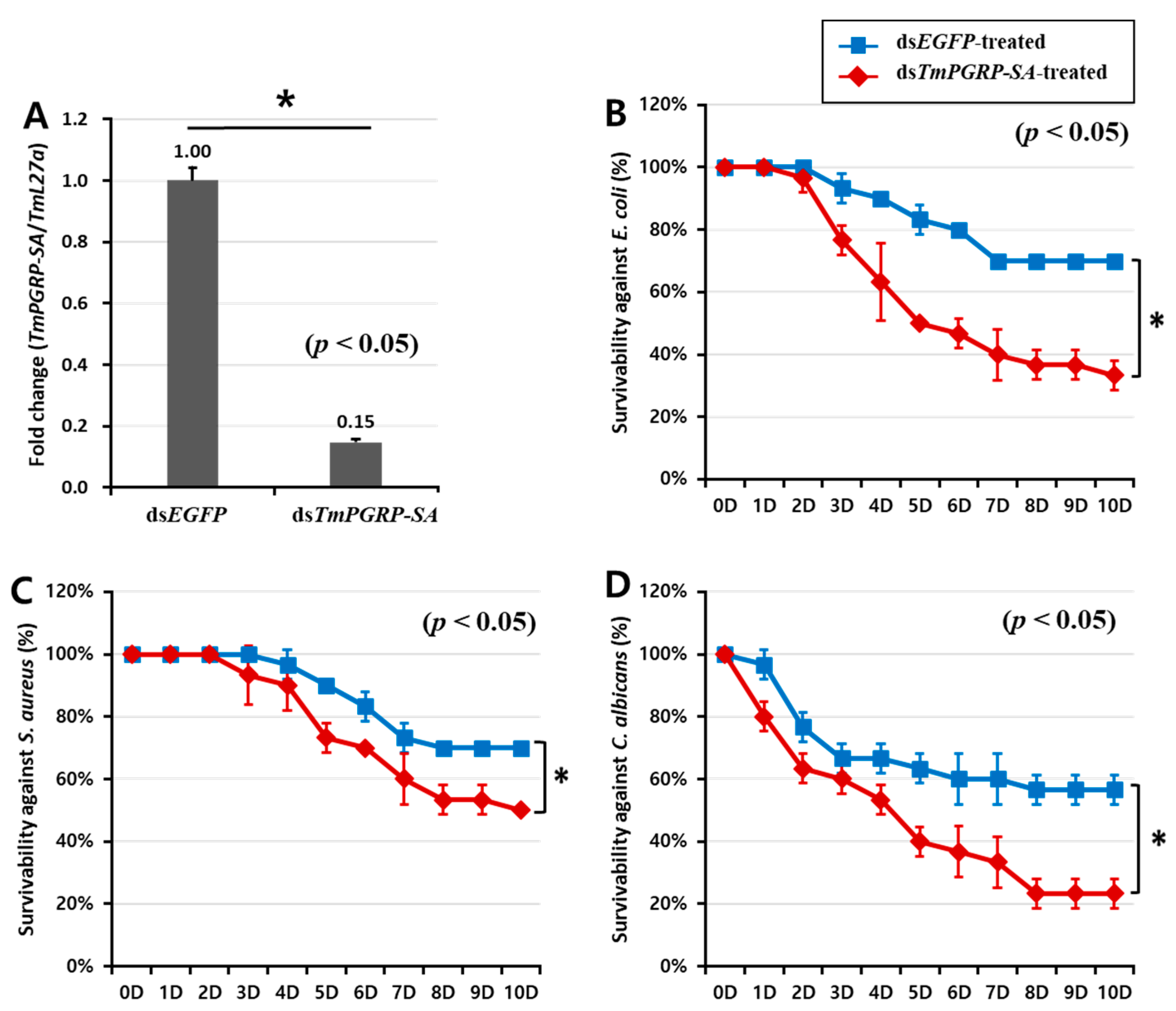
2.4. TmPGRP-SA Silenced Larvae are Susceptible to Microbial Infections
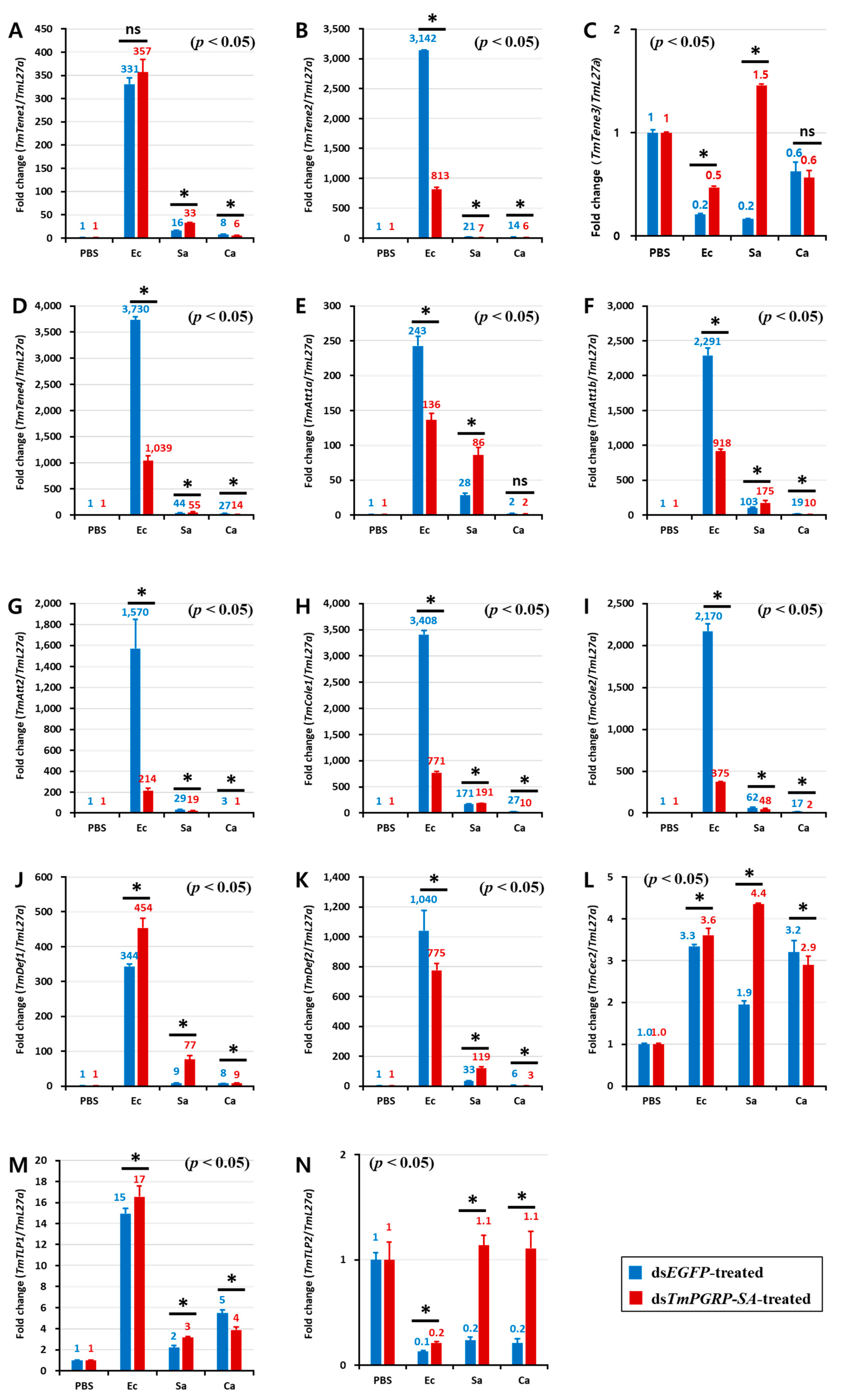
2.5. Induction of AMPs is Regulated by TmPGRP-SA in Immunocompetent Tissues
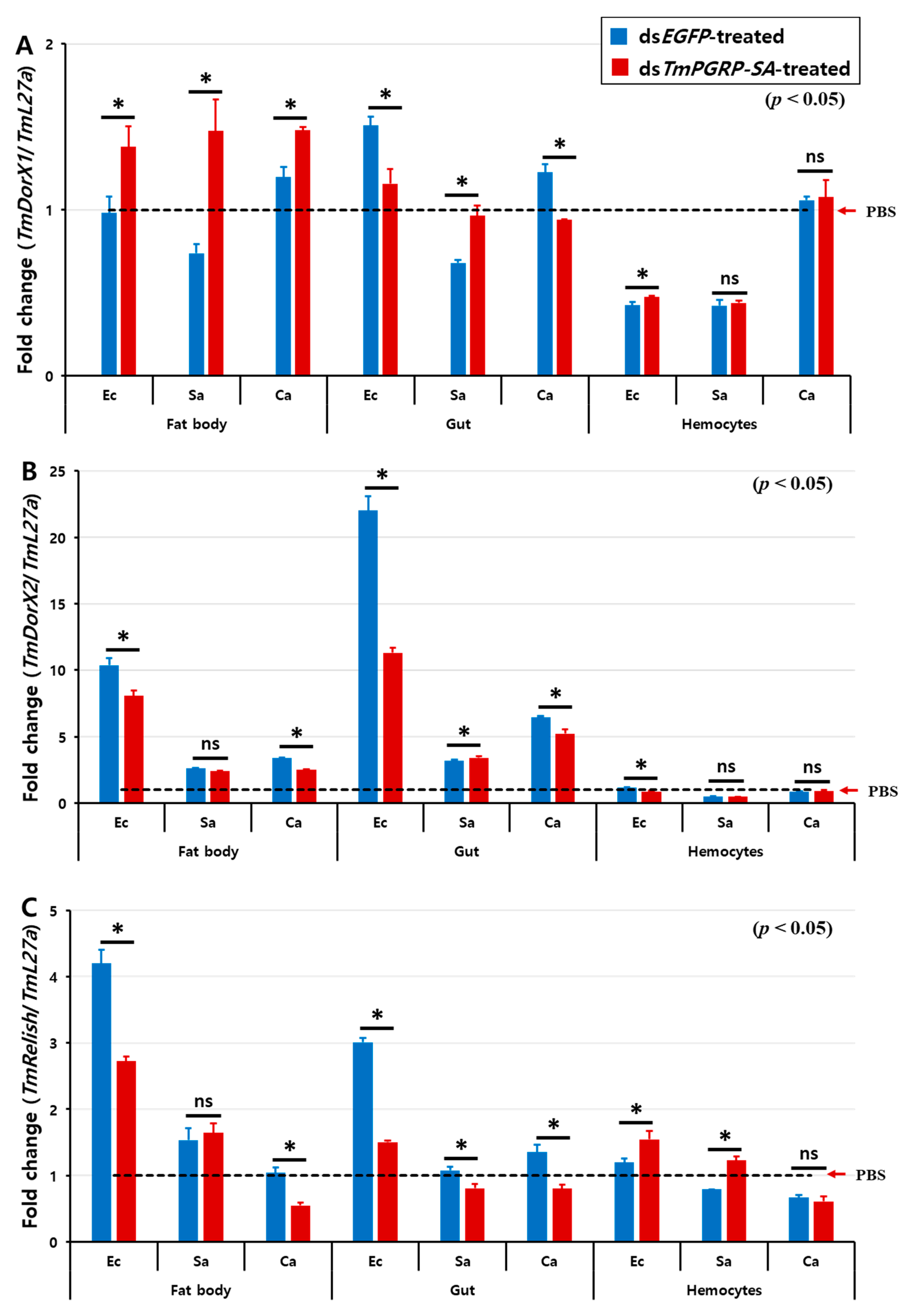
2.6. Effect of TmPGRP-SA Knockdown on NF-κB Gene Expression
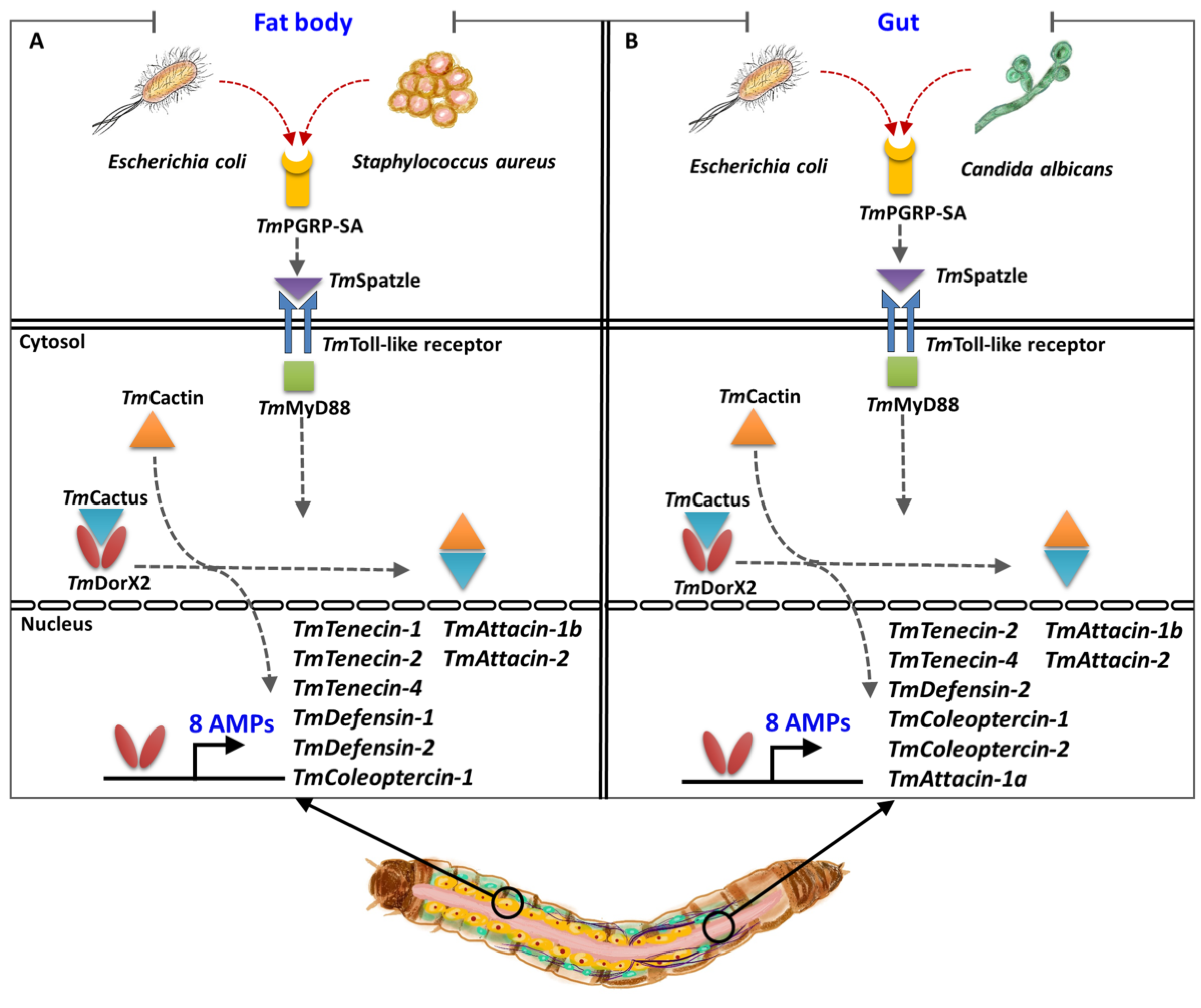
3. Discussion
4. Materials and Methods
4.1. Rearing Stock of T. Molitor
4.2. Bioinformatics Analysis for Identification and Sequence Characterization of TmPGRP-SA
4.3. Gene Expression Analysis of TmPGRP-SA in Multiple Tissues and in Different Developmental Stages
4.4. RNA Isolation and cDNA Synthesis
4.5. Quantitative Reverse-Transcription PCR (RT-qPCR) Analysis
4.6. Microbial Strains
4.7. TmPGRP-SA mRNA Expression Following Microbial Challenges
4.8. Preparation of Double-Stranded TmPGRP-SA
4.9. T. molitor Survival Bioassay
4.10. Analysis of the Effect of TmPGRP-SA Silencing on AMP and NF-κB Gene Expression Post-Microbial Challenge
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef]
- Nürnberger, T.; Brunner, F. Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef]
- Kusumoto, S.; Fukase, K.; Shiba, T. Key structures of bacterial peptidoglycan and lipopolysaccharide triggering the innate immune system of higher animals: Chemical synthesis and functional studies. Proc. Jpn. Acad. Ser. B 2010, 86, 322–337. [Google Scholar] [CrossRef]
- Werner, T.; Liu, G.; Kang, D.; Ekengren, S.; Steiner, H.; Hultmark, D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13772–13777. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 2003, 278, 49044–49052. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef]
- Montaño, A.M.; Tsujino, F.; Takahata, N.; Satta, Y. Evolutionary origin of peptidoglycan recognition proteins in vertebrate innate immune system. BMC Evol. Biol. 2011, 11, 79. [Google Scholar] [CrossRef]
- Mellroth, P.; Karlsson, J.; Steiner, H. A scavenger function for a DrosophilaPeptidoglycan recognition protein. J. Biol. Chem. 2003, 278, 7059–7064. [Google Scholar] [CrossRef]
- Guan, R.; Roychowdhury, A.; Ember, B.; Kumar, S.; Boons, G.-J.; Mariuzza, R.A. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17168–17173. [Google Scholar] [CrossRef]
- Guan, R.; Mariuzza, R.A. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007, 15, 127–134. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, M.; Liu, X.; Xia, H.; Chen, K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 2019, 106, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef]
- Werner, T. Peptidoglycan Recognition Proteins in Drosophila Melanogaster. Ph.D. Thesis, Umeå Centrum för Molekylär Patogenes (UCMP Medicinska fakulteten), Umea, Sweden, 2004. [Google Scholar]
- Michel, T.; Reichhart, J.-M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756. [Google Scholar] [CrossRef]
- Gobert, V.; Gottar, M.; Matskevich, A.A.; Rutschmann, S.; Royet, J.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 2003, 302, 2126–2130. [Google Scholar] [CrossRef]
- Pili-Floury, S.; Leulier, F.; Takahashi, K.; Saigo, K.; Samain, E.; Ueda, R.; Lemaitre, B. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 2004, 279, 12848–12853. [Google Scholar] [CrossRef]
- Wang, L.; Weber, A.N.; Atilano, M.L.; Filipe, S.R.; Gay, N.J.; Ligoxygakis, P. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006, 25, 5005–5014. [Google Scholar] [CrossRef]
- Filipe, S.R.; Tomasz, A.; Ligoxygakis, P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005, 6, 327–333. [Google Scholar] [CrossRef]
- Chang, C.-I.; Pili-Floury, S.; Hervé, M.; Parquet, C.; Chelliah, Y.; Lemaitre, B.; Mengin-Lecreulx, D.; Deisenhofer, J. A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual L, D-carboxypeptidase activity. PLoS Biol. 2004, 2, e277. [Google Scholar] [CrossRef]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.-H.; Caroff, M.; Lee, W.-J.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003, 4, 478. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, X.; Huang, J.; Chen, M.; An, J. Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans. J. Immunol. 2019, 202, 249–259. [Google Scholar] [CrossRef]
- Canteri de Souza, P.; Custódio Caloni, C.; Wilson, D.; Sergio Almeida, R. An invertebrate host to study fungal infections, mycotoxins and antifungal drugs: Tenebrio molitor. J. Fungi 2018, 4, 125. [Google Scholar] [CrossRef]
- de Souza, P.C.; Morey, A.T.; Castanheira, G.M.; Bocate, K.P.; Panagio, L.A.; Ito, F.A.; Furlaneto, M.C.; Yamada-Ogatta, S.F.; Costa, I.N.; Mora-Montes, H.M. Tenebrio molitor (Coleoptera: Tenebrionidae) as an alternative host to study fungal infections. J. Microbiol. Methods 2015, 118, 182–186. [Google Scholar] [CrossRef]
- Park, S.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Kim, C.E.; Bae, Y.M.; Kim, B.; Jun, S.A.; Bang, I.S.; Lee, Y.S. TmToll-7 plays a crucial role in innate immune responses against Gram-negative bacteria by regulating 5 AMP genes in Tenebrio molitor. Front. Immunol. 2019, 10, 310. [Google Scholar] [CrossRef]
- Park, J.-W.; Kim, C.-H.; Kim, J.-H.; Je, B.-R.; Roh, K.-B.; Kim, S.-J.; Lee, H.-H.; Ryu, J.-H.; Lim, J.-H.; Oh, B.-H. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 2007, 104, 6602–6607. [Google Scholar] [CrossRef]
- Yang, Y.T.; Lee, M.R.; Lee, S.J.; Kim, S.; Nai, Y.S.; Kim, J.S. Tenebrio molitor Gram-negative-binding protein 3 (TmGNBP3) is essential for inducing downstream antifungal Tenecin 1 gene expression against infection with Beauveria bassiana JEF-007. Insect Sci. 2018, 25, 969–977. [Google Scholar] [CrossRef]
- Yagi, Y.; Nishida, Y.; Ip, Y.T. Functional analysis of Toll-related genes in Drosophila. Dev. Growth Differ. 2010, 52, 771–783. [Google Scholar] [CrossRef]
- Edosa, T.T.; Jo, Y.H.; Keshavarz, M.; Bae, Y.M.; Kim, D.H.; Lee, Y.S.; Han, Y.S. TmSpz6 Is Essential for Regulating the Immune Response to Escherichia Coli and Staphylococcus Aureus Infection in Tenebrio Molitor. Insects 2020, 11, 105. [Google Scholar] [CrossRef]
- Patnaik, B.B.; Patnaik, H.H.; Seo, G.W.; Jo, Y.H.; Lee, Y.S.; Lee, B.L.; Han, Y.S. Gene structure, cDNA characterization and RNAi-based functional analysis of a myeloid differentiation factor 88 homolog in Tenebrio molitor larvae exposed to Staphylococcus aureus infection. Dev. Comp. Immunol. 2014, 46, 208–221. [Google Scholar] [CrossRef]
- Jo, Y.H.; Kim, Y.J.; Park, K.B.; Seong, J.H.; Kim, S.G.; Park, S.; Noh, M.Y.; Lee, Y.S.; Han, Y.S. TmCactin plays an important role in Gram-negative and-positive bacterial infection by regulating expression of 7 AMP genes in Tenebrio molitor. Sci. Rep. 2017, 7, 46459. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Edosa, T.T.; Lee, Y.S.; Han, Y.S. Tm DorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-J.; Xu, S.; He, Z.-H.; Shi, X.-Z.; Zhao, X.-F.; Wang, J.-X. Activation of toll pathway is different between kuruma shrimp and Drosophila. Front. Immunol. 2017, 8, 1151. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Jiang, H.; Kanost, M.R. Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010, 277, 148–162. [Google Scholar] [CrossRef]
- Park, J.W.; Je, B.-R.; Piao, S.; Inamura, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Söderhäll, K.; Ha, N.-C.; Lee, B.L. A synthetic peptidoglycan fragment as a competitive inhibitor of the melanization cascade. J. Biol. Chem. 2006, 281, 7747–7755. [Google Scholar] [CrossRef]
- Yu, Y.; Park, J.-W.; Kwon, H.-M.; Hwang, H.-O.; Jang, I.-H.; Masuda, A.; Kurokawa, K.; Nakayama, H.; Lee, W.-J.; Dohmae, N. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem. 2010, 285, 32937–32945. [Google Scholar] [CrossRef]
- Kikkawa, H.S.; Ueda, T.; Suzuki, S.-I.; Yasuda, J. Characterization of the catalytic activity of the γ-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol. Lett. 2008, 286, 236–240. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Zhao, X.; Naeem, M.; An, J. Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae). Protein Sci. 2018, 27, 893–897. [Google Scholar] [CrossRef]
- Consortium, H.G.S. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931. [Google Scholar]
- Dziarski, R. Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 2004, 40, 877–886. [Google Scholar] [CrossRef]
- Kang, D.; Liu, G.; Lundström, A.; Gelius, E.; Steiner, H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 1998, 95, 10078–10082. [Google Scholar] [CrossRef]
- You, H.; Wan, H.; Li, J.; Jin, B.R. Molecular cloning and characterization of a short peptidoglycan recognition protein (PGRP-S) with antibacterial activity from the bumblebee Bombus ignitus. Dev. Comp. Immunol. 2010, 34, 977–985. [Google Scholar] [CrossRef]
- Dai, L.-S.; Qian, C.; Wang, L.; Wei, G.-Q.; Liu, Q.-N.; Sun, Y.; Zhang, C.-F.; Li, J.; Liu, D.-R.; Zhu, B.-J. Characterization and function of a short peptidoglycan recognition protein from the Chinese oak silkworm, Antheraea pernyi. J. Asia Pac. Entomol. 2015, 18, 701–707. [Google Scholar] [CrossRef]
- Kurata, S. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 2014, 42, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, X.; Cai, S.; Zhang, S.; Luo, H.; Wu, C.; Zhang, R.; Zhang, J. A novel peptidoglycan recognition protein involved in the prophenoloxidase activation system and antimicrobial peptide production in Antheraea pernyi. Dev. Comp. Immunol. 2018, 86, 78–85. [Google Scholar] [CrossRef]
- Sharma, P.; Dube, D.; Sinha, M.; Yadav, S.; Kaur, P.; Sharma, S.; Singh, T.P. Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: Structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid. PLoS ONE 2013, 8, e53756. [Google Scholar] [CrossRef]
- Yang, C.; Wang, L.; Jia, Z.; Yi, Q.; Xu, Q.; Wang, W.; Gong, C.; Liu, C.; Song, L. Two short peptidoglycan recognition proteins from Crassostrea gigas with similar structure exhibited different PAMP binding activity. Dev. Comp. Immunol. 2017, 70, 9–18. [Google Scholar] [CrossRef]
- Koyama, H.; Kato, D.; Minakuchi, C.; Tanaka, T.; Yokoi, K.; Miura, K. Peptidoglycan recognition protein genes and their roles in the innate immune pathways of the red flour beetle, Tribolium castaneum. J. Invertebr. Pathol. 2015, 132, 86–100. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the basic local alignment search tool (BLAST). Cold Spring Harbor Protocols 2007, 2007, pdb.top17. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucl. Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Yaffe, H.; Buxdorf, K.; Shapira, I.; Ein-Gedi, S.; Zvi, M.M.-B.; Fridman, E.; Moshelion, M.; Levy, M. LogSpin: A simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes 2012, 5, 45. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protocols 2008, 3, 1101. [Google Scholar] [CrossRef]

| Name | Abbreviation | GenBank Number |
|---|---|---|
| Tribolium castaneum PREDICTED: peptidoglycan recognition protein 2 | TcPGRP-SA | P_008192927.1 |
| Locusta migratoria PGRP-SA | LmPGRP-SA | AFD54029.1 |
| Bombus consobrinus peptidoglycan recognition protein SA | BcPGRP-SA | ATL64828.1 |
| Bombus koreanus peptidoglycan recognition protein SA | BkPGRP-SA | ATL64813.1 |
| Gryllus bimaculatus peptidoglycan recognition protein SA | GbPGRP-SA | BBG28438.1 |
| Drosophila simulans PGRP-SA | DsPGRP-SA | XP_002106687.1 |
| Drosophila busckii PGRP-SA | DbPGRP-SA | ALC48229.1 |
| Eumeta japonica Peptidoglycan recognition protein SA | EjPGRP-SA | GBP17419.1 |
| Drosophila melanogaster peptidoglycan recognition protein SA | DmPGRP-SA | CAD89124.1 |
| Bactrocera latifrons Peptidoglycan recognition protein SA | BlPGRP-SA | JAI23539.1 |
| Zeugodacus cucurbita Peptidoglycan recognition protein SA | ZcPGRP-SA | JAD13283.1 |
| Bombus ignites peptidoglycan recognition protein SA | BiPGRP-SA | ATL64812.1 |
| Homo sapiens Peptidoglycan recognition protein 3 | HsPGLYRP3 | AAI28116.1 |
| Primer | Sequence (5′–3′) |
|---|---|
| TmPGRP_SA_qPCR_Fw TmPGRP_SA_qPCR_Rv | 5′-TCAATGCGGAGTTGAACTGGGAGA-3′ 5′-TAGAGTTTCAGTCCAGGGCTGCTT-3′ |
| dsTmPGRP-SA_Fw dsTmPGRP-SA_Rv | 5′-TAATACGACTCACTATAGGG AGAGACTCGGAAAGCGAATGTGC-3′ 5′-TAATACGACTCACTATAGGG AGACTCCCAGTTCAACTCCGCAT-3′ |
| dsEGFP_Fw dsEGFP_Rv | 5′-TAATACGACTCACTATAGGG TACGTAAACGGCCACAAGTTC-3′ 5′-TAATACGACTCACTATAGGG TTGCTCAGGTAGTGTTGTCG-3′ |
| TmTenecin-1_Fw TmTenecin-1_Rv | 5′-CAGCTGAAGAAATCGAACAAGG-3′ 5′-CAGACCCTCTTTCCGTTACAGT-3′ |
| TmTenecin-2_Fw TmTenecin-2_Rv | 5′-CAGCAAAACGGAGGATGGTC-3′ 5′-CGTTGAAATCGTGATCTTGTCC-3′ |
| TmTenecin-3_Fw TmTenecin-3_Rv | 5′-GATTTGCTTGATTCTGGTGGTC-3′ 5′-CTGATGGCCTCCTAAATGTCC-3′ |
| TmTenecin-4_Fw TmTenecin-4_Rv | 5′-GGACATTGAAGATCCAGGAAAG-3′ 5′-CGGTGTTCCTTATGTAGAGCTG-3′ |
| TmDefensin-1_Fw TmDefencin-1_Rv | 5′-AAATCGAACAAGGCCAACAC-3′ 5′-GCAAATGCAGACCCTCTTTC-3′ |
| TmDefensin-2_Fw TmDefensin-2_Rv | 5′-GGGATGCCTCATGAAGATGTAG-3′ 5′-CCAATGCAAACACATTCGTC-3′ |
| TmColeoptericin-1_Fw TmColeoptericin-1_Rv | 5′-GGACAGAATGGTGGATGGTC-3′ 5′-CTCCAACATTCCAGGTAGGC-3′ |
| TmColeoptericin-2_Fw TmColeoptericin-2_Rv | 5′-GGACGGTTCTGATCTTCTTGAT-3′ 5′-CAGCTGTTTGTTTGTTCTCGTC-3′ |
| TmAttacin-1a_Fw TmAttacin-1a_Rv | 5′-GAAACGAAATGGAAGGTGGA-3′ 5′-TGCTTCGGCAGACAATACAG-3′ |
| TmAttacin-1b_Fw TmAttacin-1b_Rv | 5′-GAGCTGTGAATGCAGGACAA-3′ 5′-CCCTCTGATGAAACCTCCAA-3′ |
| TmAttacin-2_Fw TmAttacin-2_Rv | 5′-AACTGGGATATTCGCACGTC-3′ 5′-CCCTCCGAAATGTCTGTTGT-3′ |
| TmCecropin-2_Fw TmCecropin-2_Rv | 5′-TACTAGCAGCGCCAAAACCT-3′ 5′-CTGGAACATTAGGCGGAGAA-3′ |
| TmThaumatin-like protein-1_Fw TmThaumatin-like protein-1_Rv | 5′-CTCAAAGGACACGCAGGACT-3′ 5′-ACTTTGAGCTTCTCGGGACA-3′ |
| TmThaumatin-like protein-2_Fw TmThaumatin-like protein-2_Rv | 5′-CCGTCTGGCTAGGAGTTCTG-3′ 5′-ACTCCTCCAGCTCCGTTACA-3′ |
| TmRelish_qPCR_Fw TmRelish_qPCR_Rv | 5′-AGCGTCAAGTTGGAGCAGAT-3′ 5′-GTCCGGACCTCATCAAGTGT-3′ |
| TmDorX1_qPCR_Fw TmDorX1_qPCR_Rv | 5′-AGCGTTGAGGTTTCGGTATG-3′ 5′-TCTTTGGTGACGCAAGACAC-3′ |
| TmDorX2_qPCR_Fw TmDorX2_qPCR_Rv | 5′-ACACCCCCGAAATCACAAAC-3′ 5′-TTTCAGAGCGCCAGGTTTTG-3′ |
| TmL27a_qPCR_Fw TmL27a_qPCR_Rv | 5′-TCATCCTGAAGGCAAAGCTCCAGT-3′ 5′-AGGTTGGTTAGGCAGGCACCTTTA-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keshavarz, M.; Jo, Y.H.; Edosa, T.T.; Bae, Y.M.; Han, Y.S. TmPGRP-SA regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio molitor. Int. J. Mol. Sci. 2020, 21, 2113. https://doi.org/10.3390/ijms21062113
Keshavarz M, Jo YH, Edosa TT, Bae YM, Han YS. TmPGRP-SA regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio molitor. International Journal of Molecular Sciences. 2020; 21(6):2113. https://doi.org/10.3390/ijms21062113
Chicago/Turabian StyleKeshavarz, Maryam, Yong Hun Jo, Tariku Tesfaye Edosa, Young Min Bae, and Yeon Soo Han. 2020. "TmPGRP-SA regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio molitor" International Journal of Molecular Sciences 21, no. 6: 2113. https://doi.org/10.3390/ijms21062113
APA StyleKeshavarz, M., Jo, Y. H., Edosa, T. T., Bae, Y. M., & Han, Y. S. (2020). TmPGRP-SA regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio molitor. International Journal of Molecular Sciences, 21(6), 2113. https://doi.org/10.3390/ijms21062113






