Combined Chibby and β-Catenin Predicts Clinical Outcomes in Patients with Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
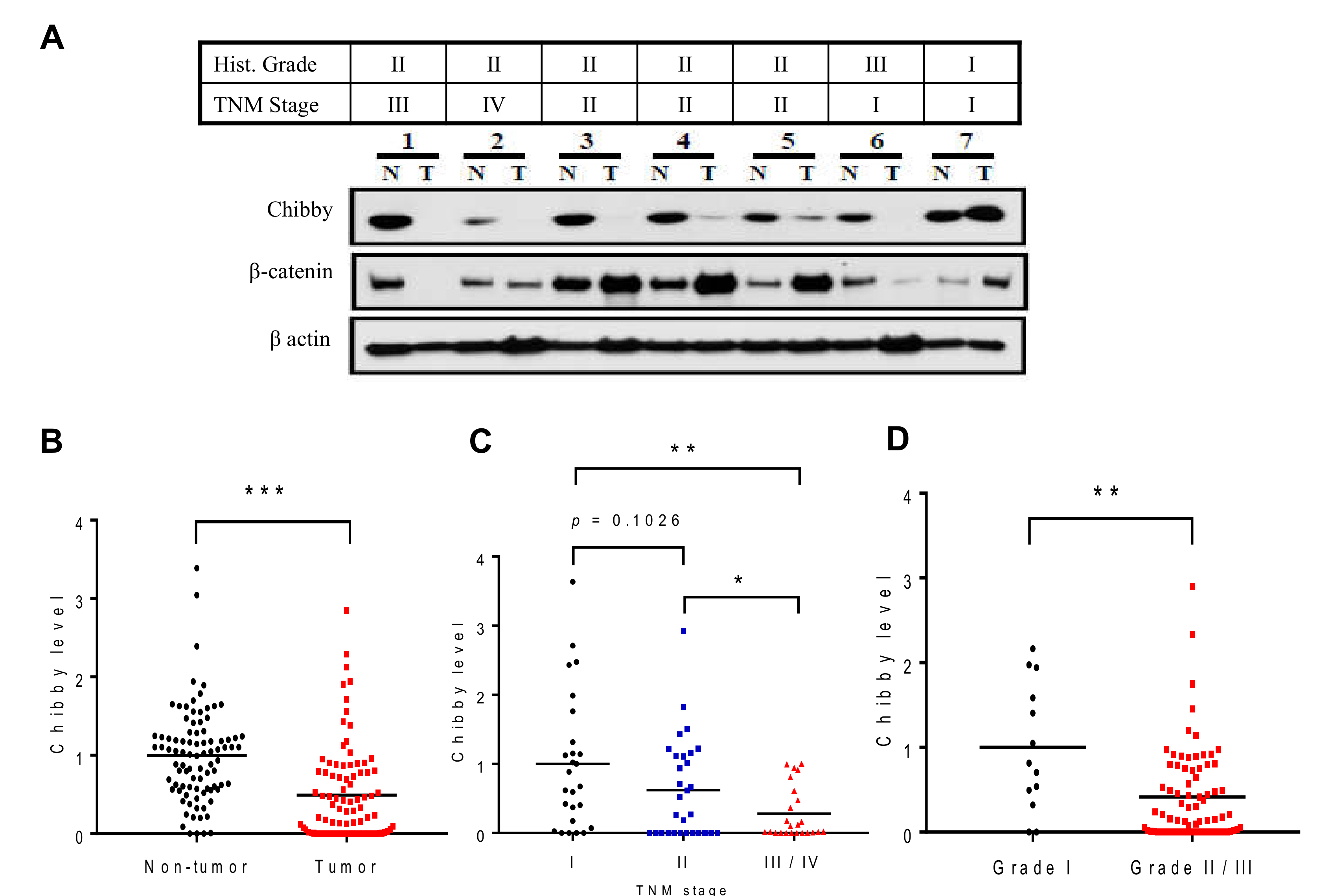
2.1. Low Expression of Chibby Correlates with High Stage of HCC
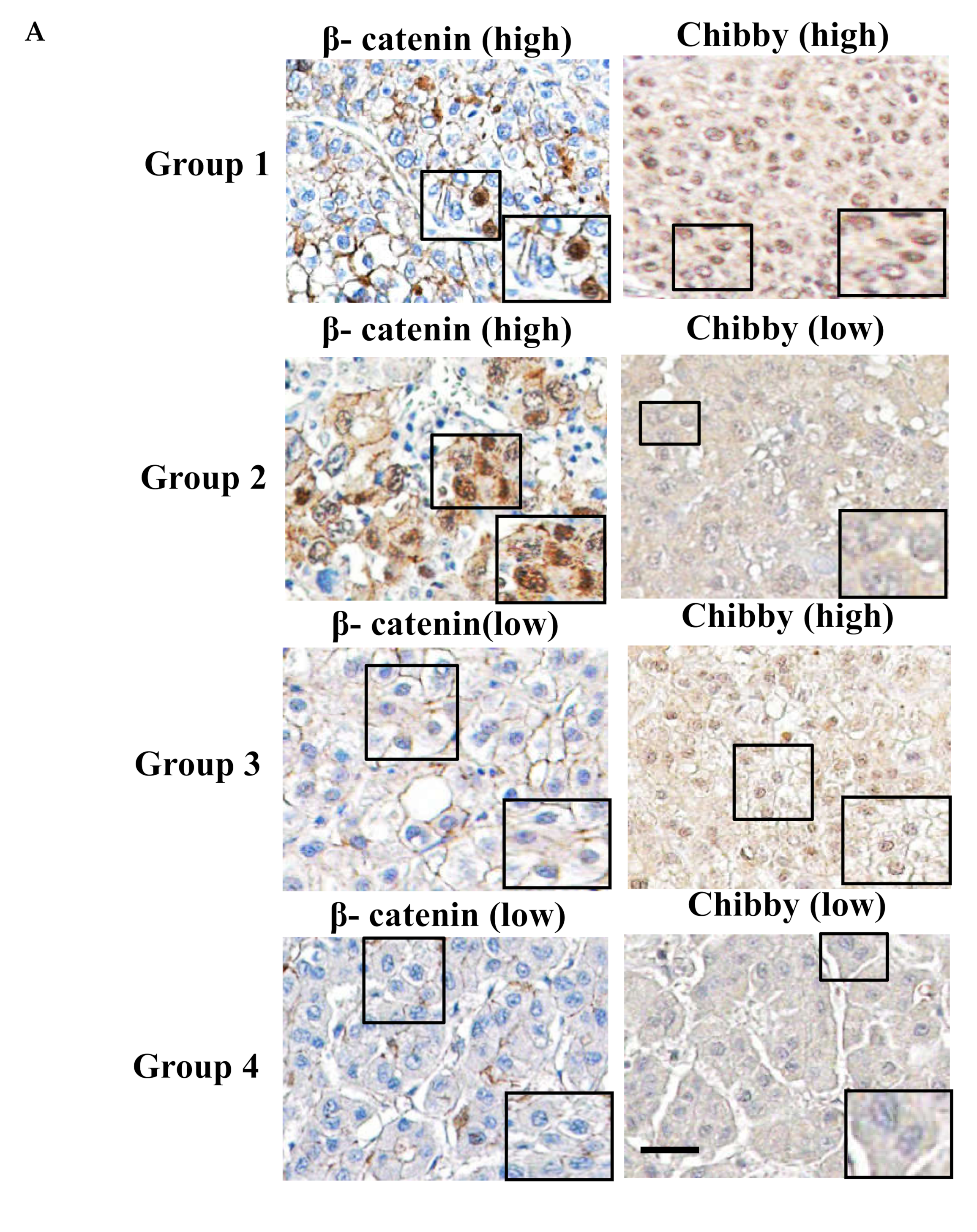
2.2. Correlation between Chibby Expression and β-Catenin Expression in HCC Specimens
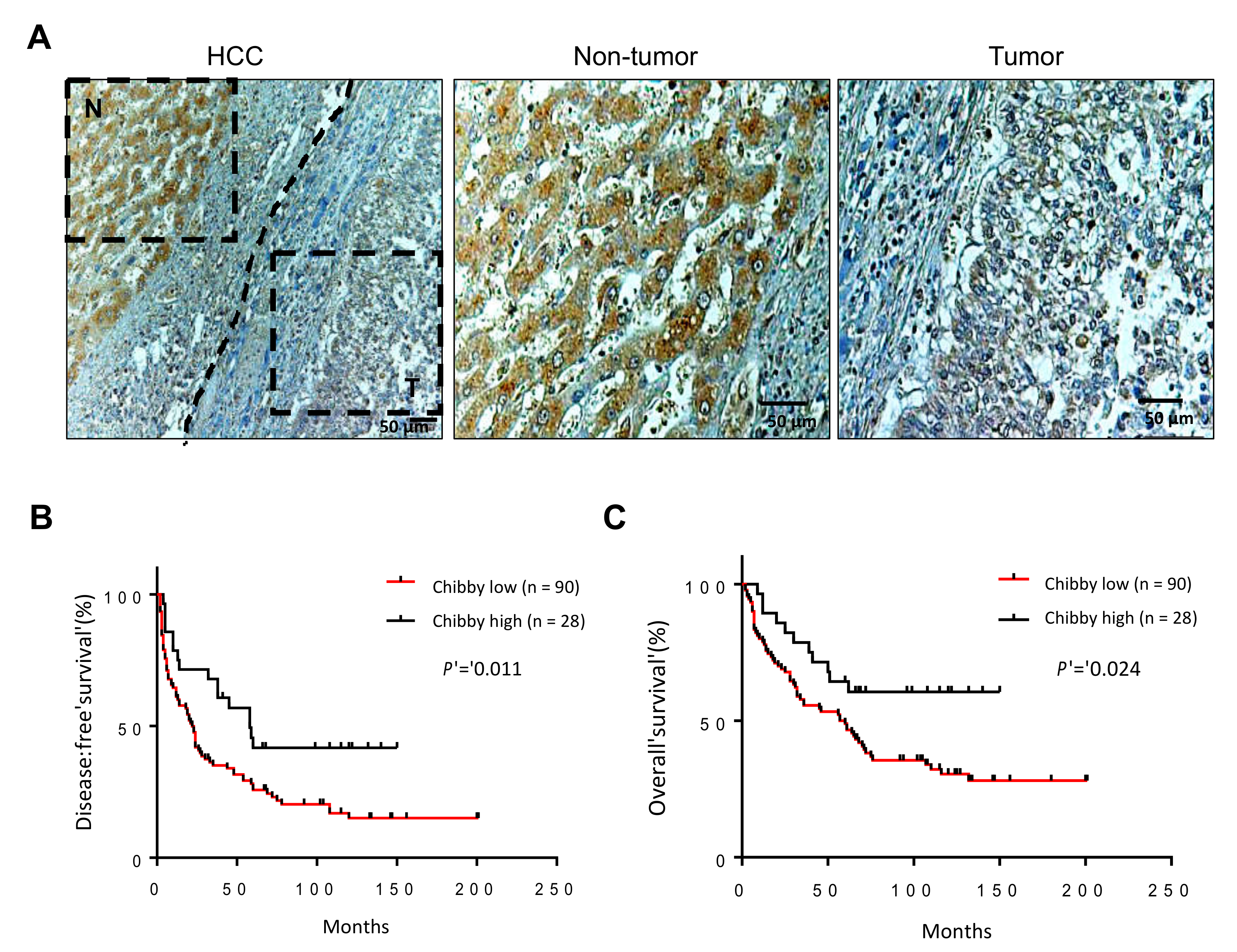
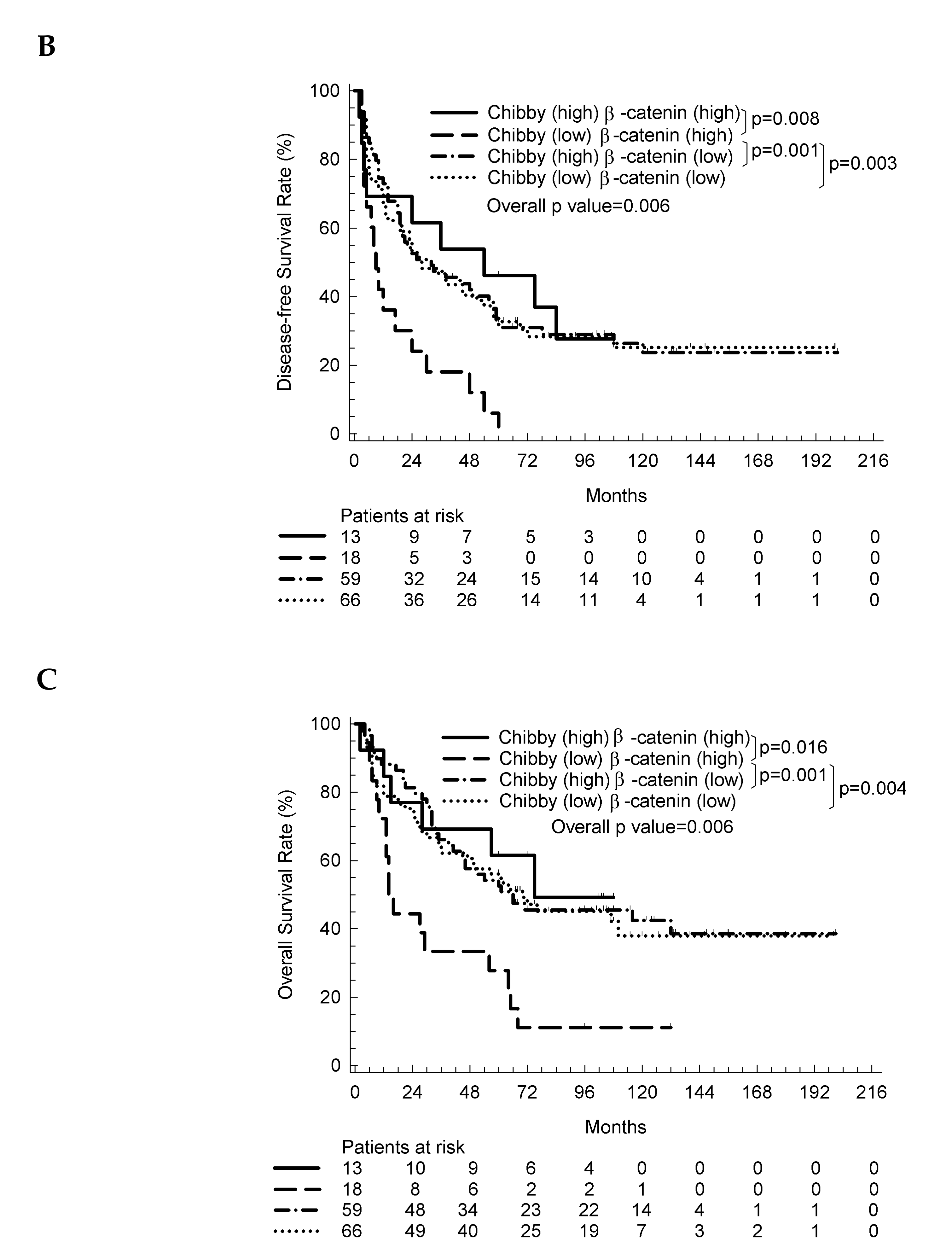
2.3. The Nuclear Expression of Chibby Predict Disease-Free Survival and Overall Survival in HCC Patients with Nuclear Expression of β-Catenin
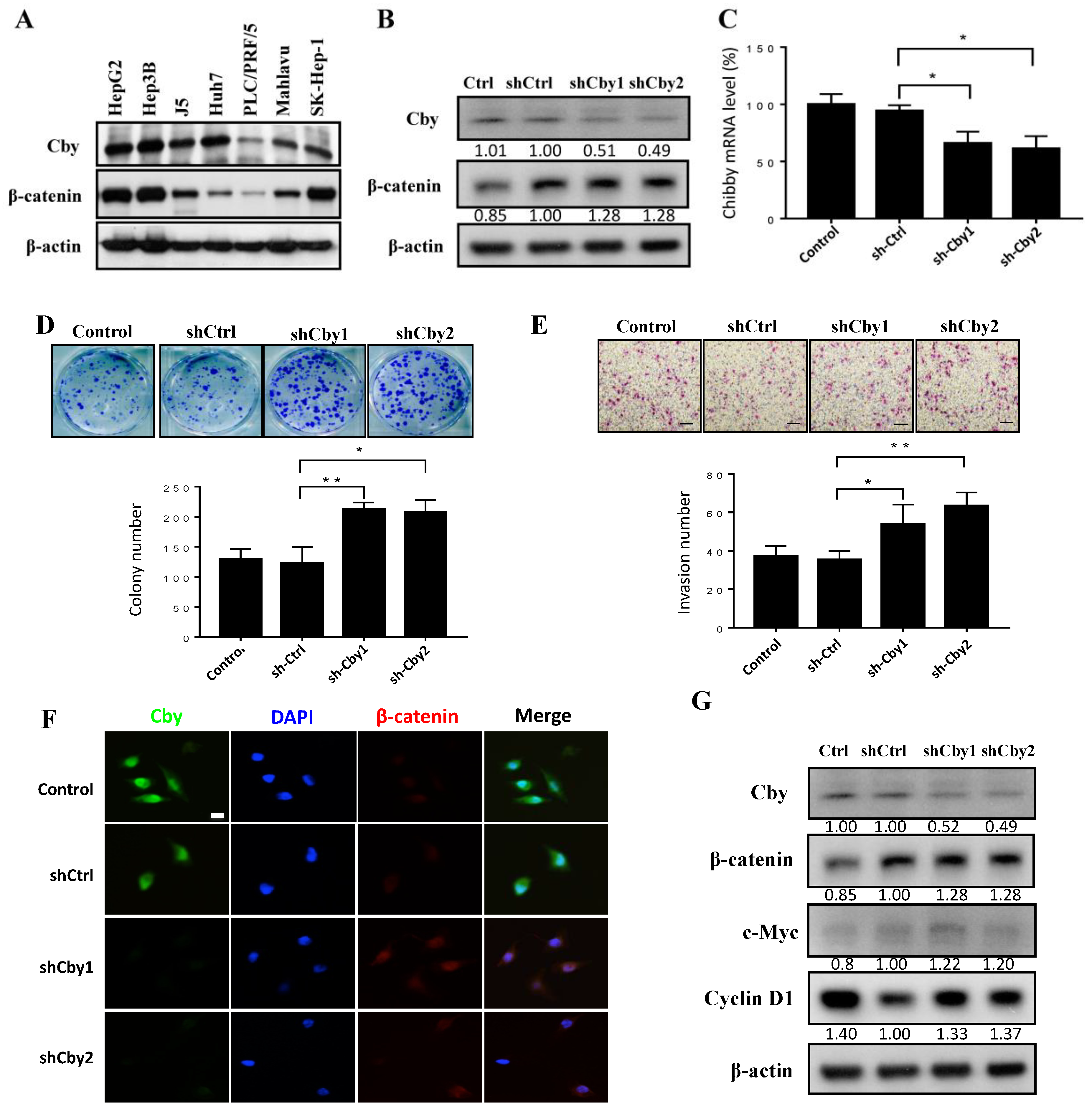
2.4. Knockdown of Chibby Promotes β-Catenin Signaling and HCC Cell Proliferation and Invasion
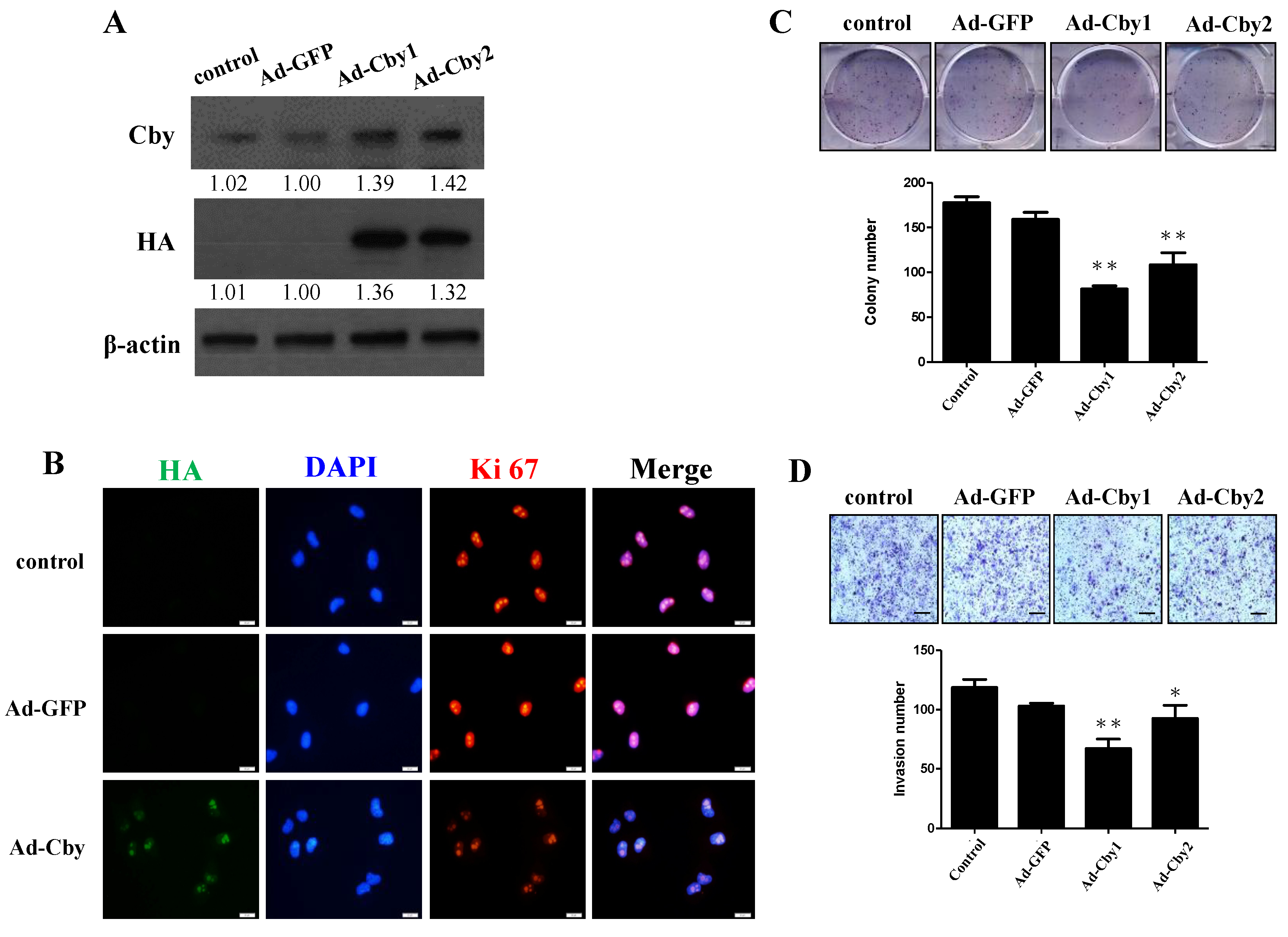
2.5. Expression of Chibby Inhibits β-Catenin Signaling and HCC Cell Proliferation and Invasion
3. Discussion
4. Materials and Methods
4.1. Patients and Tissue Samples
4.2. Immunohistochemical Staining and Scoring
4.3. RNA Isolation and Quantitative Reverse-Transcriptase Polymerase Chain Reaction
4.4. Western Blot Analysis
4.5. Immunofluorescence Staining
4.6. Cell Culture and Transfection Protocols
4.7. Adenoviral Vectors-Mediated Gene Delivery
4.8. Knockdown of Endogenous Chibby
4.9. Colonies Formation Assay
4.10. Invasion Assay
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA A Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Burroughs, A.; Bruix, J. Hepatocellular carcinoma. Lancet 2003, 362, 1907–1917. [Google Scholar] [CrossRef]
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346. [Google Scholar] [CrossRef] [PubMed]
- De La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef]
- Inagawa, S.; Itabashi, M.; Adachi, S.; Kawamoto, T.; Hori, M.; Shimazaki, J.; Yoshimi, F.; Fukao, K. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: Correlation with tumor progression and postoperative survival. Clin. Cancer Res. 2002, 8, 450–456. [Google Scholar]
- Nhieu, J.T.; Renard, C.A.; Wei, Y.; Cherqui, D.; Zafrani, E.S.; Buendia, M.A. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am. J. Pathol. 1999, 155, 703–710. [Google Scholar] [CrossRef]
- Takemaru, K.; Yamaguchi, S.; Lee, Y.S.; Zhang, Y.; Carthew, R.W.; Moon, R.T. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 2003, 422, 905–909. [Google Scholar] [CrossRef]
- Tolwinski, N.S.; Wieschaus, E. A nuclear function for armadillo/beta-catenin. PLoS Biol. 2004, 2, E95. [Google Scholar] [CrossRef] [PubMed]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Takemaru, K.; Fischer, V.; Li, F.Q. Fine-tuning of nuclear-catenin by Chibby and 14-3-3. Cell Cycle 2009, 8, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Fischer, V.; Brown-Grant, D.A.; Li, F.Q. Chibby suppresses growth of human SW480 colon adenocarcinoma cells through inhibition of beta-catenin signaling. J. Mol. Signal 2012, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ren, G.; Zhao, D.A.; Li, B.A.; Cai, C.F.; Zhou, Y.; Luo, X.Y. Downregulated chibby in laryngeal squamous cell carcinoma with increased expression in laryngeal carcinoma Hep-2 cells. Oncol. Rep. 2014, 32, 1947–1956. [Google Scholar] [CrossRef][Green Version]
- Xu, H.T.; Li, Q.C.; Dai, S.D.; Xie, X.M.; Liu, D.I.; Wang, E.H. The expression patterns and correlations of chibby, beta-catenin, and DNA methyltransferase-1 and their clinicopathological significance in lung cancers. APMIS 2011, 119, 750–758. [Google Scholar] [CrossRef]
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma. Hepatology 2005, 42, 1208–1236. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M.; Llovet, J.M.; Beaugrand, M.; Lencioni, R.; Burroughs, A.K.; Christensen, E.; Pagliaro, L.; Colombo, M.; Rodes, J.; et al. Clinical management of hepatocellular carcinoma. Conclusions of the barcelona-2000 easl conference. european association for the study of the liver. J. Hepatol. 2001, 35, 421–430. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, H.A.; Steiner, P.E. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer 1954, 7, 462–503. [Google Scholar] [CrossRef]
| Clinicopathologic | β-Catenin (high)/Chibby (high) (n = 13) | β-Catenin (high)/Chibby (low) (n = 18) | β-Catenin (low)/Chibby (high) (n = 59) | β-Catenin (low)/Chibby (low) (n = 66) | p value |
|---|---|---|---|---|---|
| Age (>60 vs. ≤60 years) | 4/9 | 9/9 | 15/44 | 28/38 | 0.125 |
| Sex (Male vs. Female) | 11/2 | 13/5 | 46/13 | 55/11 | 0.689 |
| AFP (>200 vs. ≤200 ng/L) | 6/7 | 8/10 | 26/33 | 25/41 | 0.875 |
| HBsAg (positive vs. negative) | 9/4 | 12/6 | 40/16 | 47/16 | 0.914 |
| HCV Ab (positive vs. negative) | 3/10 | 3/15 | 14/42 | 15/48 | 0.909 |
| Liver cirrhosis (yes vs. no) | 7/6 | 11/7 | 38/21 | 78/28 | 0.837 |
| Tumor size (>5 vs. ≤5 cm) | 10/3 | 11/7 | 30/29 | 32/34 | 0.250 |
| Tumor number (multiple vs. single) * | 2/11 | 9/9 | 16/43 | 11/55 | 0.025 |
| TNM Stage (III + IV vs. I + II) | 9/4 | 13/5 | 28/31 | 30/36 | 0.108 |
| Histology grade (Poor vs. Well + Moderate) | 6/7 | 6/12 | 17/42 | 15/51 | 0.349 |
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Variable | Comparison | HR (95%CI) | p value | HR (95%CI) | p value |
| Age (years) | >60 vs. ≤60 | 0.755 (0.513–1.110) | 0.153 | NS | |
| Sex | Male vs. Female | 1.362 (0.856–2.167) | 0.193 | NS | |
| AFP (ng/mL) | >200 vs. ≤200 | 1.676 (1.160–2.421) | 0.006 | 1.907 (1.297–2.803) | 0.001 |
| HBsAg | Positive vs. Negative | 1.306 (0.855–1.996) | 0.217 | NS | |
| HCV Ab | Positive vs. Negative | 0.811 (0.519–1.267) | 0.358 | NA | |
| Liver cirrhosis | Yes vs. No | 1.494 (1.001–2.169) | 0.049 | 1.795 (1.201–2.683) | 0.004 |
| Tumor size (cm) | >5 vs. ≤5 | 1.904 (1.313–2.761) | 0.001 | NS | |
| Tumor number | Multiple vs. Single | 1.534 (1.019–2.310) | 0.041 | NS | |
| TNM stages | III + IV vs. I + II | 2.730 (1.870–3.983) | <0.001 | 2.808 (1.898–4.156) | <0.001 |
| Histology grade | Poor vs. Well + Moderate | 1.557 (1.052–2.304) | 0.027 | NS | |
| β-catenin | High vs. Low | 1.505 (0.971–2.332) | 0.068 | NS | |
| Chibby | High vs. Low | 0.802 (0.556–1.157) | 0.239 | NS | |
| β-catenin/Chibby | High/Low vs. others | 2.450 (1.448–4.144) | 0.001 | 1.986 (1.160–3.400) | 0.012 |
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Variable | Comparison | HR (95%CI) | p value | HR (95%CI) | p value |
| Age (years) | >60 vs. ≤60 | 0.966 (0.633–1.474) | 0.892 | NA | |
| Sex | Male vs. Female | 1.204 (0.719–2.014) | 0.480 | NA | |
| AFP (ng/mL) | >200 vs. ≤200 | 1.580 (1.050–2.377) | 0.028 | 1.638 (1.076–2.493) | 0.021 |
| HBsAg | Positive vs. Negative | 1.264 (0.787–2.031) | 0.333 | NA | |
| HCV Ab | Positive vs. Negative | 0.804 (0.489–1.322) | 0.389 | NA | |
| Liver cirrhosis | Yes vs. No | 1.307 (0.854–2.001) | 0.217 | NS | |
| Tumor size (cm) | >5 vs. ≤5 | 2.407 (1.343–3.119) | 0.001 | NS | |
| Tumor number | Multiple vs. Single | 1.221 (0.762–1.956) | 0.407 | NA | |
| TNM stages | III + IV vs. I + II | 3.351 (2.165–5.188) | <0.001 | 3.135 (1.979–4.968) | <0.001 |
| Histology grade | Poor vs. well + moderate | 1.858 (1.212–2.850) | 0.004 | NS | |
| β-catenin | High vs. Low | 1.612 (0.996–2.609) | 0.052 | NS | |
| Chibby | High vs. Low | 0.756 (0.501–1.140) | 0.182 | NS | |
| β-catenin/Chibby | High/Low vs. others | 2.534 (1.471–4.363) | 0.001 | 2.207 (1.270–3.835) | 0.005 |
| Patient Demographics | |
|---|---|
| Age [years; median (range)] | 56 (5–82) |
| Sex (M:F) | 125:31 |
| AFP [ng/mL; median (range)] | 87 (2–80000) |
| Tumor size [cm; median (range)] a | 5 (1–20) |
| Tumor number (single:multiple) | 37:119 |
| Liver cirrhosis, n (%) | 94 (59.9) |
| Hepatitis (B:C:B+C:NBNC) | 101:28:7:20 |
| TNM stage (I:II:III:IV) | 22:54:46:34 |
| Pathological Features | |
| Capsule (Yes:No) | 69:86 |
| Microvascular invasion (Yes:No) | 76:77 |
| Histological grade (well:moderate:poor) | 38:74:44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, M.-C.; Huang, C.-C.; Wei, Y.-C.; Liu, T.-T.; Lin, M.-T.; Yi, L.-N.; Lin, P.-R.; Wang, C.-C.; Chu, T.-H.; Hsiao, C.-C.; et al. Combined Chibby and β-Catenin Predicts Clinical Outcomes in Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 2060. https://doi.org/10.3390/ijms21062060
Tsai M-C, Huang C-C, Wei Y-C, Liu T-T, Lin M-T, Yi L-N, Lin P-R, Wang C-C, Chu T-H, Hsiao C-C, et al. Combined Chibby and β-Catenin Predicts Clinical Outcomes in Patients with Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2020; 21(6):2060. https://doi.org/10.3390/ijms21062060
Chicago/Turabian StyleTsai, Ming-Chao, Chao-Cheng Huang, Yu-Ching Wei, Ting-Ting Liu, Ming-Tsung Lin, Li-Na Yi, Pey-Ru Lin, Chih-Chi Wang, Tian-Huei Chu, Chang-Chun Hsiao, and et al. 2020. "Combined Chibby and β-Catenin Predicts Clinical Outcomes in Patients with Hepatocellular Carcinoma" International Journal of Molecular Sciences 21, no. 6: 2060. https://doi.org/10.3390/ijms21062060
APA StyleTsai, M.-C., Huang, C.-C., Wei, Y.-C., Liu, T.-T., Lin, M.-T., Yi, L.-N., Lin, P.-R., Wang, C.-C., Chu, T.-H., Hsiao, C.-C., Hu, T.-H., & Tai, M.-H. (2020). Combined Chibby and β-Catenin Predicts Clinical Outcomes in Patients with Hepatocellular Carcinoma. International Journal of Molecular Sciences, 21(6), 2060. https://doi.org/10.3390/ijms21062060





