Abstract
Actin plays a critical role in the rhizobium–legume symbiosis. Cytoskeletal rearrangements and changes in actin occur in response to Nod factors secreted by rhizobia during symbiotic interactions with legumes. These cytoskeletal rearrangements are mediated by diverse actin-binding proteins, such as actin depolymerization factors (ADFs). We examined the function of an ADF in the Phaseolus vulgaris–rhizobia symbiotic interaction (PvADFE). PvADFE was preferentially expressed in rhizobia-inoculated roots and nodules. PvADFE promoter activity was associated with root hairs harbouring growing infection threads, cortical cell divisions beneath root hairs, and vascular bundles in mature nodules. Silencing of PvADFE using RNA interference increased the number of infection threads in the transgenic roots, resulting in increased nodule number, nitrogen fixation activity, and average nodule diameter. Conversely, overexpression of PvADFE reduced the nodule number, nitrogen fixation activity, average nodule diameter, as well as NODULE INCEPTION (NIN) and EARLY NODULIN2 (ENOD2) transcript accumulation. Hence, changes in ADFE transcript levels affect rhizobial infection and nodulation, suggesting that ADFE is fine-tuning these processes.
1. Introduction
Legumes have the ability to establish a symbiotic association with Gram-negative soil bacteria belonging to several genera, including Rhizobium, Bradyrhizobium, Sinorhizobium, and Azorhizobium, commonly called rhizobia [1]. This mutualistic interaction is initiated by a molecular dialogue in which plant roots exude flavonoids that activate the expression of bacterial genes (nod genes) encoding proteins involved in the synthesis and secretion of lipochitooligosaccharides, called Nod factors (NFs). These signal molecules are specifically recognized by the root hair, where they activate a signaling pathway. The NFs perceived by legume root hairs trigger a variety of physiological responses, such as calcium fluxes, including perinuclear calcium oscillations, reactive oxygen species production, ion influxes and effluxes, cell-membrane depolarization, cytoplasm alkalinization; cytoskeletal rearrangements, and early nodulin gene expression changes [2,3,4,5,6,7]. These changes allow the bacteria to enter the root hair through a structure called an infection thread (IT) and simultaneously induce cortical cell divisions. When the ITs reach the nodule primordia cells, rhizobia are released into the host cell within a peribacteroid membrane; subsequently, they differentiate into nitrogen-fixing bacteroids [8,9]. After root hair curling, the rhizobia trapped in an infection chamber inside the root hair curl, which is followed by an invagination of the host cell plasma membrane and the subsequent formation of an IT [10]. The growing IT induces changes in the plasmodesmata, cytoskeleton and cell membrane and wall synthesis machineries [11,12,13].
Actin cytoskeleton rearrangements take place at different stages of the symbiotic interaction between rhizobia and their host plants, i.e., during root hair growth and curling, IT formation, bacterial internalization from root hairs to nodule cells, symbiosome organization within the infected cells of root nodules, and cell trafficking [3,14,15,16]. In response to NFs, the actin cytoskeleton changes within 5–10 min; the longitudinal bundles of actin filaments become fragmented and short and fine actin bundles accumulate in the apical and subapical region, and there is a rapid increase in the number of F-actin plus ends at the tips of the responding root hairs [3,17,18].
In nodule cells, the actin cytoskeleton encloses IT and infection droplets, guiding the elongation of IT and the rhizobial release. During later steps of cell colonization, actin microfilaments embraces the developing symbiosomes and, in a mature nodule, actin reorganizes in several short or dot-like actin filaments aligned with mature symbiosomes [19]. Actin cytoskeleton architecture and dynamics are temporally and spatially modulated by diverse actin-binding proteins (ABPs), including profilin, villin, formin, ARPs, and actin depolymerizing factor (ADF/cofilin) [20]. The functions of ABPs include G-actin sequestration, actin nucleation, polymerization, depolymerization, and stabilization of F-actin filaments [20,21]. In general, it is well known that in plant cells upon several stimuli, the actin remodeling and assembly is triggered by small GTPases (ROPs in plants) [22]. These ROPs can activate the nucleation promoting factors (NPFs) such as Wave Regulatory Complex (WRC) consisting of SCAR/WAVE, nap1, pir1, abi and brick, which are a WASP family members that induce actin nucleation and rearrangements via recruitment and activation of the Arp2/3 complex [15]. Then, the Arp2/3 complex (consisting of 7 subunits), induces F-actin formation and the turnover is promoted by ADF/cofilins [23]. In Lotus japonicus, mutants of two components of the SCAR/WAVE complex, nap1 (for Nck-associated protein 1) and pir1 (for 121F-specific p53 inducible RNA), result in disruption of actin rearrangements, short root hairs, impaired formation and progression of ITs into the root cortex, and uncolonized nodule primordia [15]. In fact, silencing of the gene ARP3 causes defects in symbiosome development in Medicago truncatula nodules [24] and L. japonicus mutants in SCARN (encoding suppressor of cAMP receptor defect-nodulation, a protein that binds to the ARP3 complex), present a strong phenotype with reduced root hair growth and aberrant formation and progression of ITs, resulting in uninfected nodules [25]. These results add weight to the idea that actin assembly is an important player during the symbiotic interaction.
ADFs are a family of small proteins of approximately 15–22 kDa found in all eukaryotes [26]. They share a conserved structural motif known as the actin-depolymerizing factor homology (ADF-H) domain consisting of five β-strands surrounded by three or more α-helices [27,28]. These proteins regulate actin filament depolymerization through binding both G-actin and F-actin in a twisted region of the actin filament, which further increases the twisting of the neighboring region [29]. This over-twisting results in the severing of actin filaments into shorter fragments and enhanced dissociation of G-actin from the minus ends of actin filaments [28]. The binding between actin and ADFs is regulated by phosphorylation, pH, phosphoinositides, and Ca2+ signaling [30,31,32,33], demonstrating that ADF activity is highly regulated [20]. Whereas metazoan animals harbour 1–4 ADF/cofilin genes [34], plant ADF genes form a large family in angiosperms; for example, the Arabidopsis thaliana genome contains at least 11 members (AtADF1 to AtADF11) showing organ-specific expression patterns [35]. Although a role for ADFs in defense signaling following pathogen infection has been reported [36,37,38,39,40], information about legume ADFs and their relationship to the symbiotic process are lacking.
We previously demonstrated that actin dynamics and polymerization regulate the early infection process in Phaseolus vulgaris root hairs incubated with purified specific NFs [3,18]. These actin dynamics could be mediated by ADFs, which are involved in F-actin depolymerization. Herein, we examined the function of a P. vulgaris ADF during symbiotic interactions with rhizobia. We found that promoter activity PvADFE was detected in the rhizobially infected root hairs and vascular bundles of mature nodules. The participation of PvADFE in the common bean symbiosis with Rhizobium tropici by down-regulating or overexpressing PvADFE in transgenic composite plants revealed a role of ADFE likely fine-tuning nodulation.
2. Results
2.1. ADF Genes Constitute a Family of Nine Members in P. vulgaris
To investigate the participation of P. vulgaris ADF during symbiosis with R. tropici, first we searched the Phytozome v12 database (http://phytozome.jgi.doe.gov/pz/portal.html#) [41]. Nine ADF genes were identified in the P. vulgaris genome. The deduced protein sequences were arbitrarily denominated PvADFA through PvADFI. The size of the PvADF gene family in P. vulgaris is similar to that of other legumes (Table S1). Based on gene structure analysis, the coding region of each PvADF gene is organized in three exons (Figure S1A), similar to the two- to three-exon organization of Glycine max (Figure S1B), A. thaliana, and Solanum lycopersicum ADF genes [42,43]. In eight of the nine PvADF genes, the first or first few amino acids are encoded by a separate exon, as described for A. thaliana ADF genes [28]. The nucleotide identity among PvADF genes varied from 48.9% to 85.2% (Table S2), whereas the identity between deduced amino acid sequences of PvADF ranged from 40.0% to 89.5% (Table S3). Multiple sequence alignment of the deduced protein sequences of PvADF and Arabidopsis AthADF1 revealed several conserved residues (Figure S2A). Ser-6, identified in other ADFs as a phosphorylation site [44], is followed by a G-actin binding motif (amino acids 6, 7, 125, and 128). This actin-binding motif is accompanied by a second signature specific for F-actin binding, with the amino acids 82, 84, 98, 136, and 137 conserved [45]. In addition, PvADF proteins contain a predicted nuclear localization signal (amino acids 22–28) [46]. The short sequence Trp90 through Met102 is also highly conserved in all PvADF proteins and has been identified as a binding site for both actin and phosphatidylinositol 4,5-bisphosphate (PIP2) [47]. Next, we predicted the three-dimensional structure of PvADFE using the crystal structure of AthADF1, which shares 82% amino acid sequence identity (Table S3), as the template. The predicted tertiary structure of PvADFE is similar to that of Arabidopsis AthADF1 [48], showing three α-helices surrounded by five β-sheets and a putative actin-binding surface (Figure S2B), a feature conserved among ADFs that is critical for binding and/or depolymerization of actin filaments [48]. In addition, there is 82% amino acid sequence identity among AthADF1 and PvADFE, as noted in Table S3.
To study the relationship between PvADF proteins, full-length ADF sequences from legumes (G. max, L. japonicus, M. truncatula, and Vigna unguiculata) and nonlegumes (A. thaliana, Oriza sativa, and Zea mays) were used to reconstruct a maximum likelihood phylogenetic tree. In relation to the Arabidopsis ADF gene family, which is grouped phylogenetically into four ancient subclasses based on differential expression [35], PvADFD and PvADFE protein sequences and other legume ADFs clustered in a subclade (Figure S3) closely related to AthADF1 through AthADF4 (Arabidopsis subclass I), which showed high transcript levels in roots, seedlings, mature leaves, and flowers [35].
2.2. PvADF Genes Are Expressed in Roots and after Rhizobial Inoculation
In A. thaliana, transcript accumulation of certain ADF genes is especially high in particular tissues; for example, AthADF8 and AthADF11 transcripts are elevated in roots compared with other tissues [35,49]. We analyzed transcript abundance of the nine PvADF genes using RT-qPCR in root hairs, apices, and stripped roots from common bean. As shown in Figure S4, accumulation of PvADF transcripts showed different patterns in the tissues examined. Transcripts of PvADFA and F were barely detected in root sections, and PvADFI expression was not detectable. The remaining PvADF genes were expressed in all root sections, with considerably higher expression of PvADFD and H than the other genes. In root hairs, transcripts of PvADFD, E, and H were the most abundant. However, PvADFD transcript abundance was highest in all tissues tested.
To examine the expression patterns of individual ADF genes after rhizobial inoculation, we performed an in silico analysis based on RNA-sequencing data reported in the Phaseolus vulgaris Gene Expression Atlas (PvGEA, http://plantgrn.noble.org/PvGEA/). According to PvGEA, the nine PvADF genes are expressed in aerial tissues, seeds, roots, and nodules. Interestingly, the PvADFE gene has the highest transcript levels expression among PvADF genes in both inoculated roots and nodules, at 5 and 21 days post-inoculation (dpi); PvADFA, B, C, and F are barely detected in all tissues tested, whereas PvADFG, H, and I are expressed weakly (Figure S5). Besides, ADFE encodes a protein of interest previously identified by our group using a phosphoproteomic approach in which the relative abundance of PvADFE increased after NF treatment in bean roots at 30 min. Then PvADFE was selected for further analysis during symbiosis with R. tropici.
2.3. PvADFE Promoter Activity in Transgenic P. vulgaris after Rhizobial Inoculation
PvADFE promoter activity was monitored by measuring β-glucuronidase (GUS) activity or green fluorescent protein (GFP) fluorescence in hairy roots from composite plants. We analyzed the activity of the PvADFE promoter in infected root hairs of transgenic roots inoculated with R. tropici–DsRed. Promoter activity was detected in root hairs harboring growing ITs and in adjacent cortical cells undergoing division (Figure 1A–D). Furthermore, GUS activity was observed in cortical cells undergoing the initial cell divisions that form the nodule primordium at 5, 7, and 14 dpi (Figure 1E–G), and was restricted to vascular bundles in mature nodules (Figure 1H). Promoter activity was also found at sites of lateral root primordia, and subsequently became confined to the apical region and vascular bundles of fully developed lateral roots (Figure 1I–L). These spatiotemporal expression patterns of the PvADFE promoter compared to control roots (Figure 1M–O) suggest that PvADFE participates in IT progression and nodule organogenesis, as well as lateral root development.
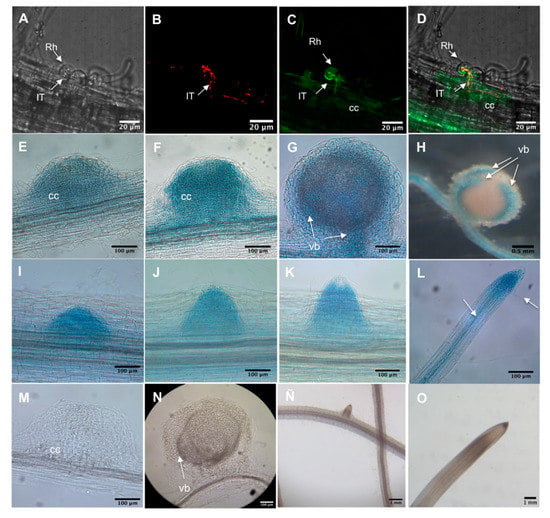
Figure 1.
Spatiotemporal activity of the Phaseolus vulgaris–rhizobia symbiotic interaction (PvADFE) promoter in P. vulgaris transgenic roots during nodulation. Promoter activity was observed in transgenic roots expressing 1.3 kb of the PvADFE promoter region fused to β-glucuronidase (GUS) activity and green fluorescent protein (GFP) (pPvADFE:GUS-GFP). (A–D) Activity of the promoter observed by confocal microscopy in infected root hairs of transgenic roots inoculated with R. tropici–DsRed at 5 dpi. (A) Transmitted light image; (B) red fluorescence emitted by R. tropici CIAT899–DsRed; (C) pPvADFE:GUS-GFP expression; (D) overlay image. (E–H) Promoter activity detected by GUS staining in a nodule primordium, young nodule, and vascular bundles of a mature nodule at (E) 5, (F) 7, (G) 14, and (H) 21 dpi, respectively. (I–L) GUS staining during development of a lateral root: (I–K) root primordium, and (L) apical region and vascular tissue (indicted by arrows) of a fully developed lateral root. (M–O) GUS staining in control roots. (M) Nodule primordium, (N) mature nodule, (O) root vascular tissue, and (P) root apical region. cc, cortical cells; IT, infection thread; Rh, root hair; vb, vascular bundles.
2.4. PvADFE Down-Regulation Increases the Number of Infection Events, Nodule Number, and Nitrogen Fixation in Rhizobium-Inoculated Transgenic Roots
To gain an insight into PvADFE gene function in symbiotic nodulation, its expression was altered by either RNAi knockdown or overexpression. The effectiveness and specificity of RNAi silencing were assessed by RT-qPCR expression analysis of PvADF genes in several independent transgenic roots. The PvADFE transcript level was 60% lower in RNAi transgenic roots than in control transgenic roots (Figure S6). Transcript levels of the other eight PvADF genes indicated that the PvADFE-RNAi construct specifically down-regulated PvADFE transcript in transgenic roots, validating the specificity of the construct designed (Figure S7).
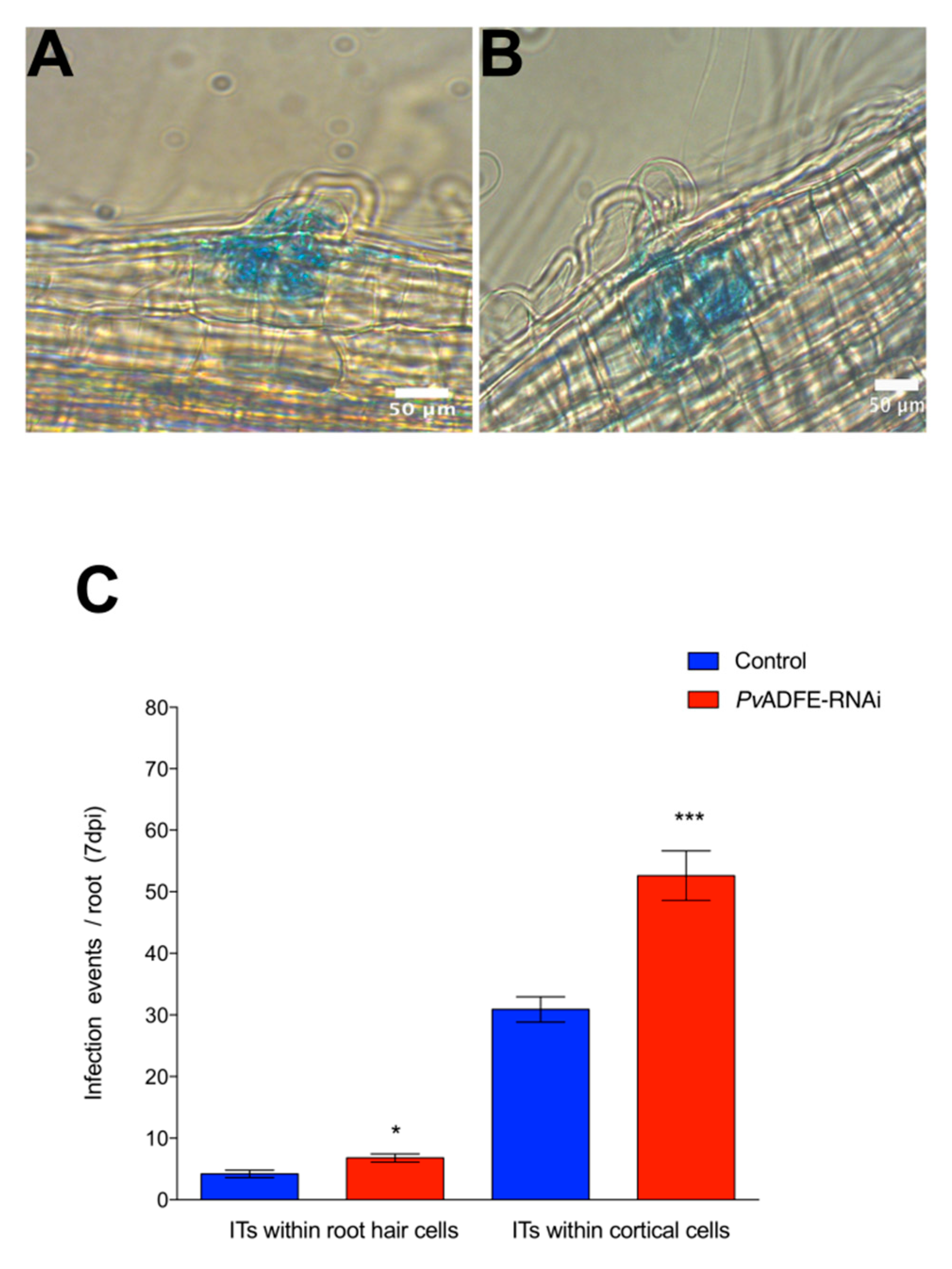
The participation of PvADFE in symbiosis was investigated in silenced inoculated roots and nodules. At 7 dpi, both PvADFE-RNAi and control transgenic roots showed ITs within cortical cells (Figure 2A and B, respectively); however, the number of infection events in PvADFE-RNAi transgenic roots was significantly higher than that in controls within root hair cells and cortical cells (Figure 2C). These results indicate that ITs in silenced roots can progress to characteristic nodule primordia that will develop determinate nodules.
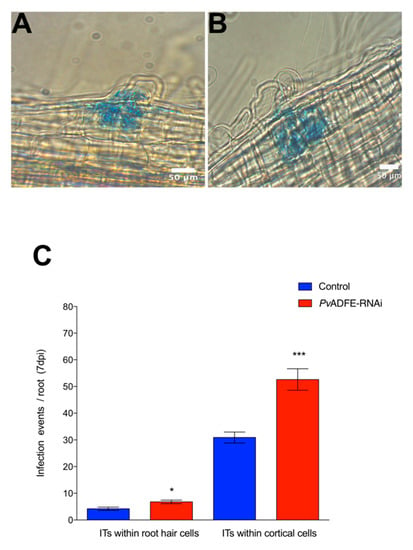
Figure 2.
Analysis of infection events in root hairs of PvADFE-RNAi and control transgenic roots at 7 dpi with R. tropici expressing GUS. Micrographs show (A) branched infection threads (ITs) invading root cell layers in control transgenic roots and (B) ITs within the cortical cells in PvADFE-RNAi transgenic roots. (C) Number of infection events per root in PvADFE-RNAi and control transgenic roots at 7 dpi. Values are mean ± SEM with n > 9 roots per condition. * p < 0.05 and *** p < 0.005 according to Student’s t-test. Bars (A,B), 50 μm.
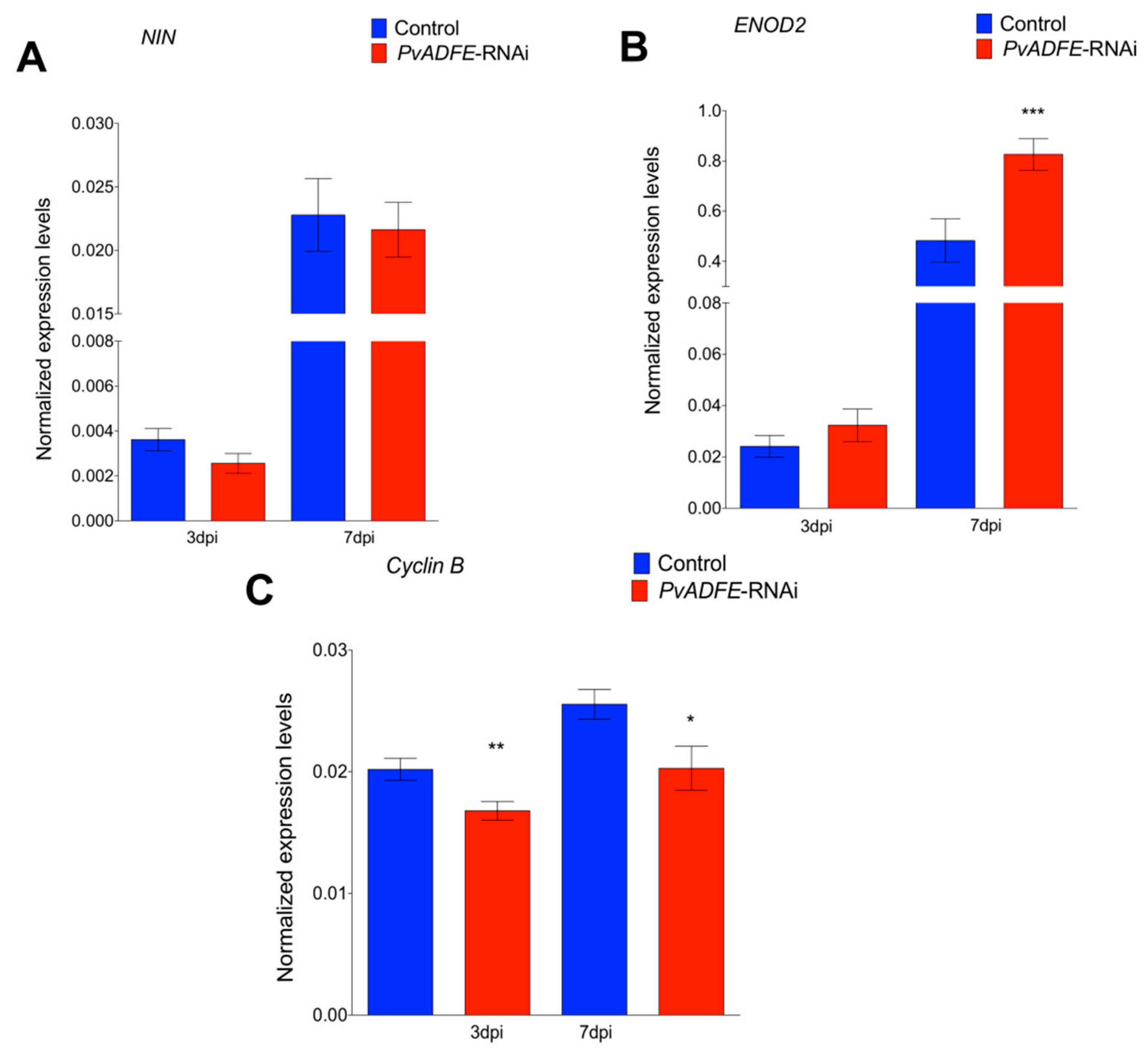
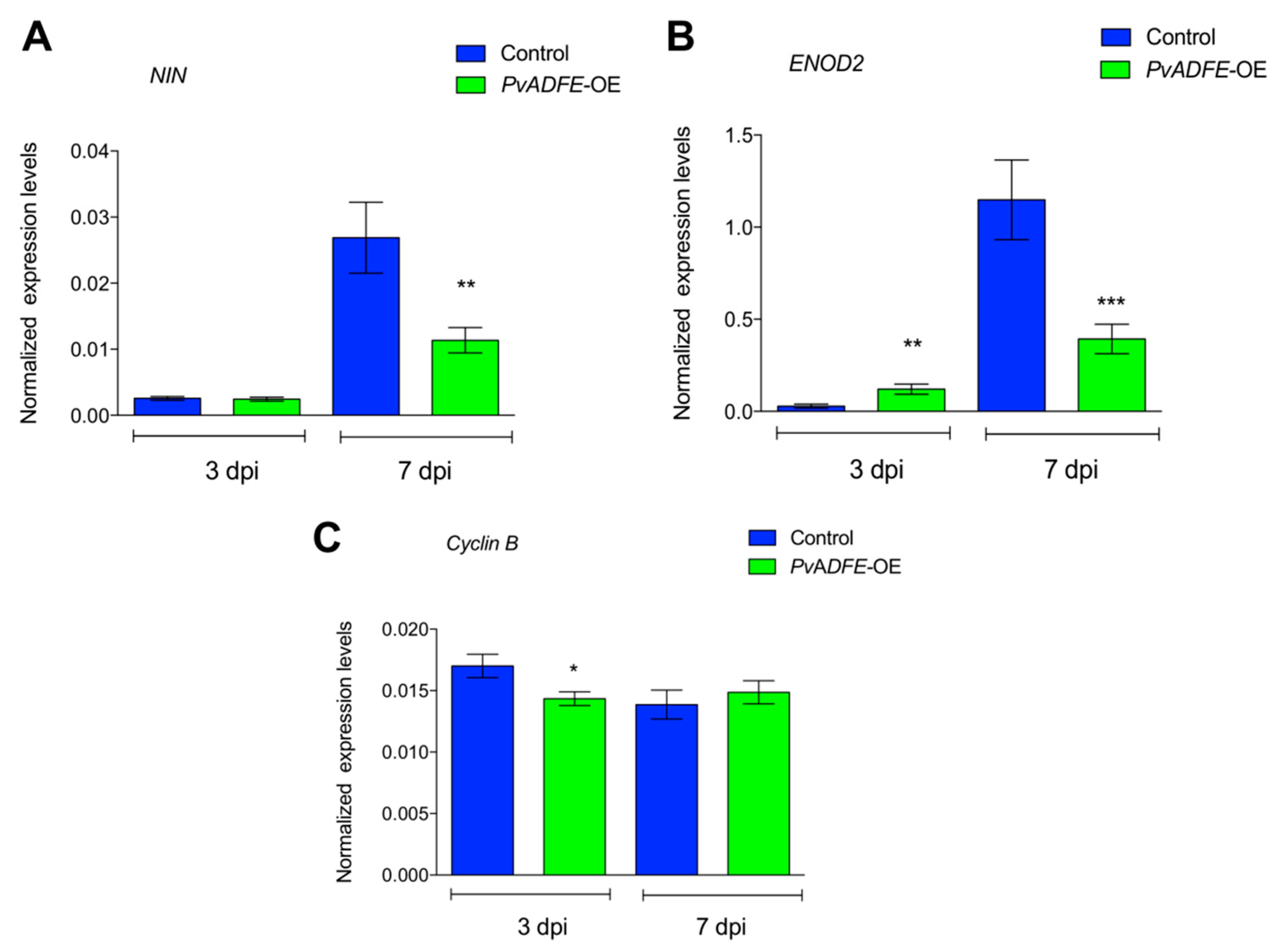
We next monitored the transcript accumulation profiles of NIN and ENOD2, which are associated with changes in nodulation signaling. Transcriptional activation of NIN [50] regulates the early steps of nodulation, such as NF-induced gene expression, IT formation, and initiation of nodule primordia [51]; hence, NIN is a specific marker for IT formation. ENOD2 is induced during cortical cell division in the early phases of nodule development [52]. Even though we did not observe increased expression of NIN at 7 dpi (Figure 3A), a significantly increased transcript abundance of ENOD2 in PvADFE-RNAi transgenic roots relative to controls was found (Figure 3B). Cyclin genes are suitable markers of dividing cells and are used in studies of plant developmental processes, such as nodule formation [53]. The B-type cyclins are highly expressed in meristem [54]. For this reason, we also analyzed cyclin B transcript levels at the same time points, and observed that the down-regulation of PvADFE significantly reduced cyclin B gene expression at 3 and 7 dpi (Figure 3C).
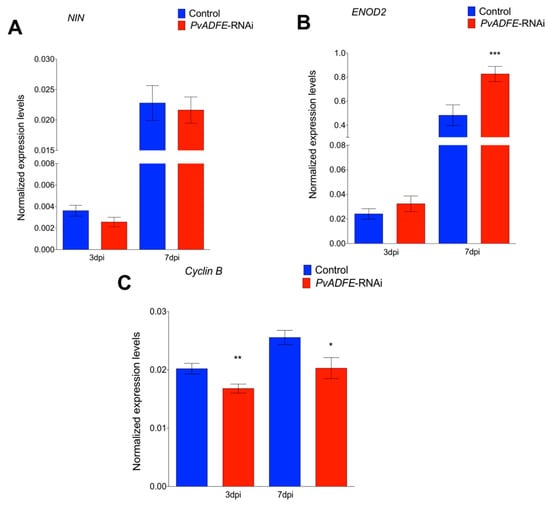
Figure 3.
Reverse-transcription quantitative PCR analysis of early nodulins in PvADFE-RNAi and control transgenic roots after inoculation with R. tropici CIAT-899. (A) NIN, (B) ENOD2, and (C) Cyclin B transcript accumulation profiles determined by RT-qPCR of total RNA isolated from PvADFE-RNAi and control transgenic roots at 3 and 7 days after inoculation with rhizobia. Elongation factor EF1α was used as an endogenous reference gene for normalizing expression levels. Each bar represents mean ± SEM of two independent biological replicates with three technical repeats. * p < 0.05, ** p < 0.005, and *** p < 0.0001 based on Student’s t-test.
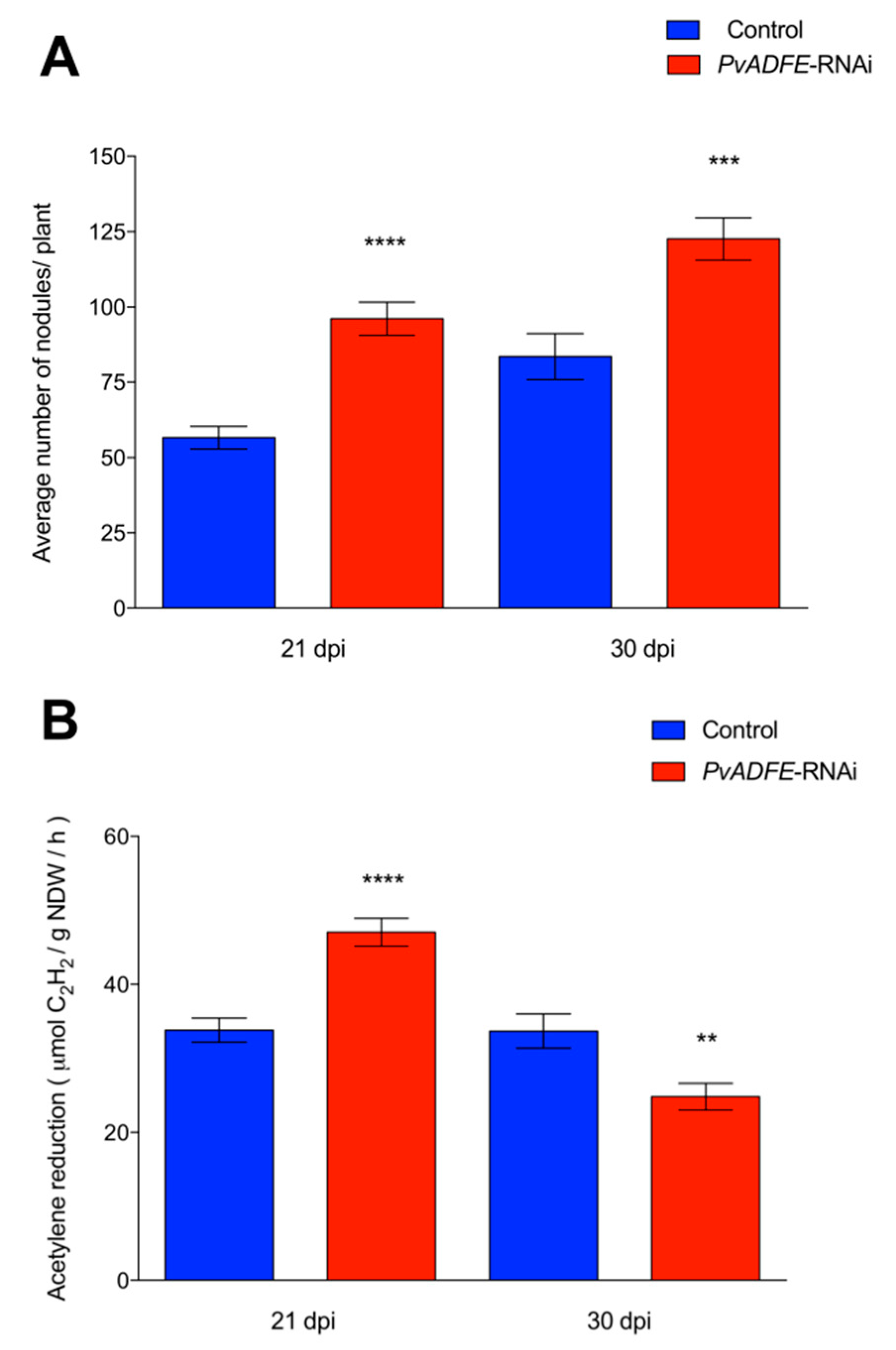
PvADFE-RNAi transgenic roots had 55 and 33% more nodules at 21 and 30 dpi than was observed in the control, respectively (Figure 4A). Interestingly, the overall distribution of nodule size and average nodule diameter on silenced roots was similar to that on control roots at all time points examined (Figure S8A,B). Furthermore, PvADFE-RNAi nodules at 21 dpi were 55% more effective in nitrogen fixation than control nodules, whereas the nitrogen-fixing ability of the PvADFE-RNAi nodules was 28% lower than that of controls at 30 dpi (Figure 4B). Altogether, our data point toward a fine-tuning role of the nodulation process mediated by PvADFE.
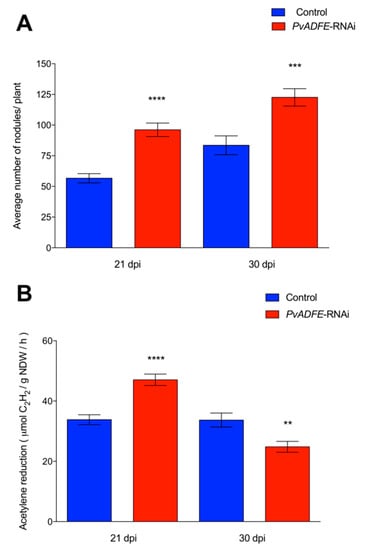
Figure 4.
Nodulation and nitrogen fixation capacity of PvADFE-RNAi and control transgenic roots. (A) Nodules were collected and counted from control and PvADFE-RNAi transgenic roots inoculated with R. tropici CIAT-899 at 21 and 30 dpi. (B) Nitrogenase activity determined by acetylene reduction in control and PvADFE-RNAi nodules of transgenic roots at 21 and 30 dpi. NDW: Nodule dry weight. Bars represent mean ± SEM for three biological replicates with n > 7. ** p < 0.005, *** p < 0.0005, and **** p < 0.0001 based on Student’s t-test.
2.5. IT Progression, Nodule Number, and Nitrogen Fixation Are Impaired in PvADFE-Overexpressing Hairy Roots
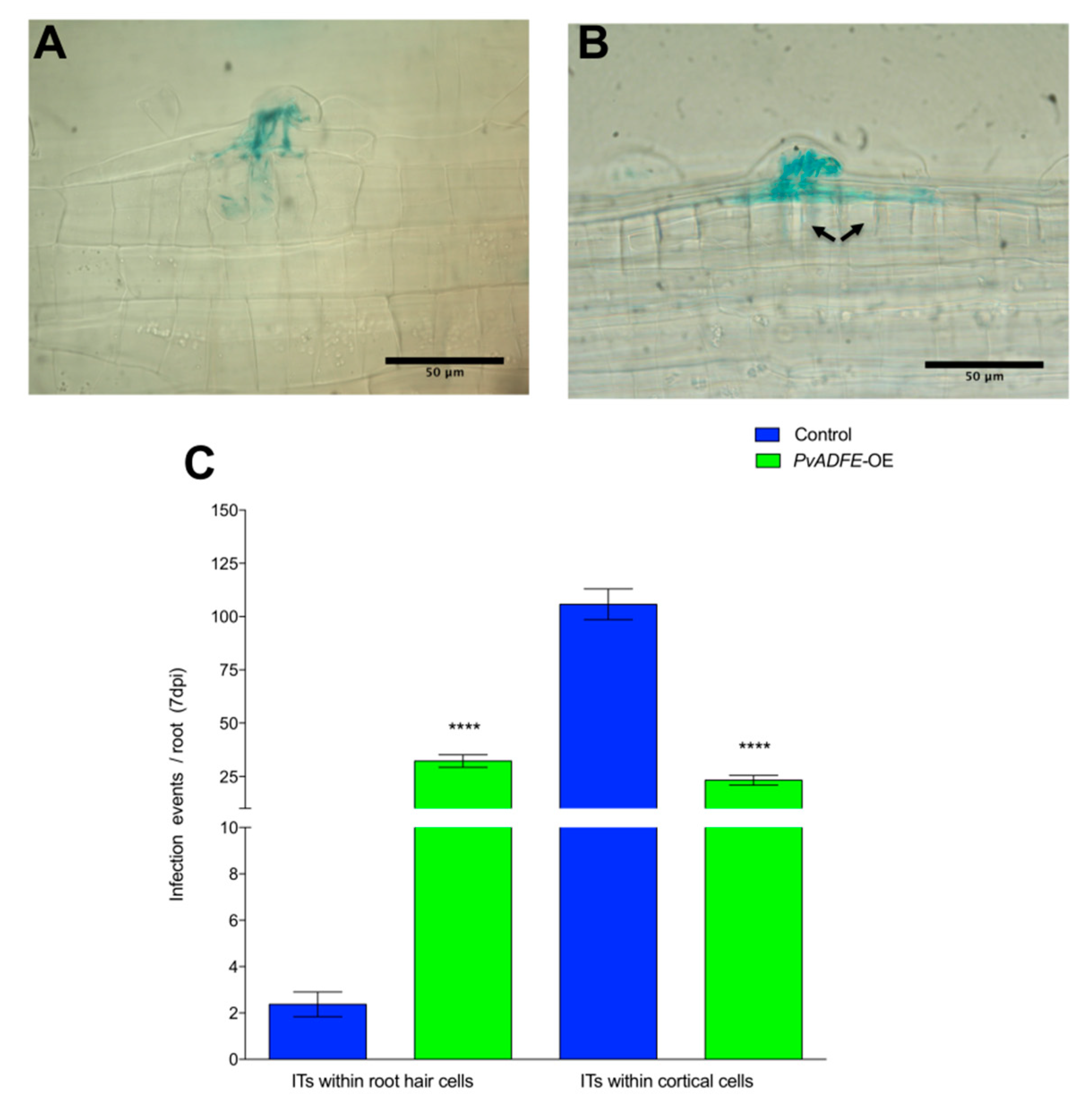
To further evaluate the function of ADF in inoculated roots and nodules, we overexpressed PvADFE (using the 35S promoter) in P. vulgaris transgenic roots. The PvADFE transcript level was 32-fold higher (p < 0.001, Student’s t-test) than in control transgenic roots, which contained an empty pH7WG2tdT vector (Figure S9). Contrary to the PvADFE-RNAi results, ITs were arrested at the base of the root hairs of PvADFE-overexpressing (PvADFE-OE) transgenic roots, although cell division was still observed in the outer cortex (Figure 5A,B, respectively), which can eventually develop into nodule primordia. The number of infection events in PvADFE-OE transgenic roots was significantly smaller than that in control roots. The 58% of the ITs were confined within the root hairs whereas 97% of ITs reached the cortical cells in control roots (Figure 5C). These results reinforce those obtained by PvADFE-RNAi, suggesting a role of PvADFE in fine-tuning nodulation.
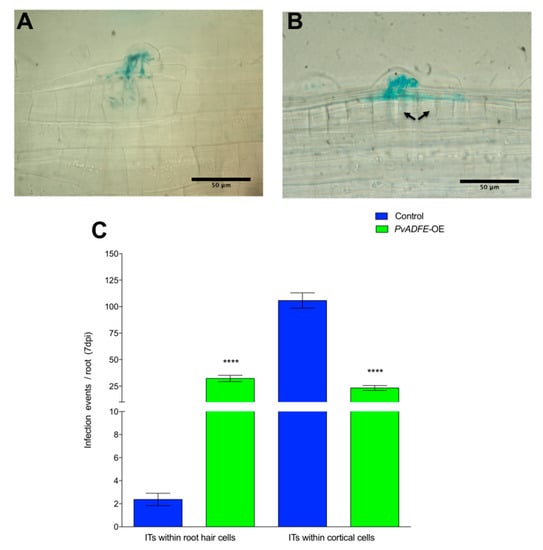
Figure 5.
Infection events in root hairs of PvADFE-OE and control transgenic roots at 7 dpi with R. tropici expressing GUS. Micrographs show (A) branched ITs invading root cell layers in control transgenic roots and (B) ITs arrested within the epidermal cells in PvADFE-OE transgenic roots. Cortical cell division is indicated by black arrows. (C) Number of infection events per root in PvADFE-OE and control transgenic roots at 7 dpi. Values are mean ± SEM with n > 9 roots per each condition. **** p < 0.0001 according to Student’s t-test. Bars (A,B), 50 μm.
Next, the transcript accumulation profiles of NIN and ENOD2 genes were evaluated. Levels of NIN in PvADFE-OE transgenic roots were significantly lower (30-fold) than those in control roots at 7 dpi (Figure 6A). This down-regulated expression of NIN in PvADFE-OE transgenic roots was associated with a decreased number of ITs in the cortical cells at 7 dpi (Figure 5C). ENOD2 transcript level was significantly higher in PvADFE-OE transgenic roots at 3 dpi compared with control roots; however, ENOD2 transcript accumulation was significantly lower in the PvADFE-OE roots compared with the control transgenic roots at 7 dpi (Figure 6B). At 3 dpi, cyclin B transcript levels were slightly lower in PvADFE-OE transgenic roots compared with control roots, while only a slight difference was observed at 7 dpi in comparison with control transgenic roots (Figure 6C). The low expression levels of NIN and ENOD2 at 7 dpi could explain the inefficiency of IT formation and progression, as well as the reduced number of nodules on PvADFE-OE roots; however, the levels of cyclin B transcript in PvADFE-OE roots were only affected at 3 dpi. We therefore next addressed whether this also affected cell division, primordia, and nodule development.
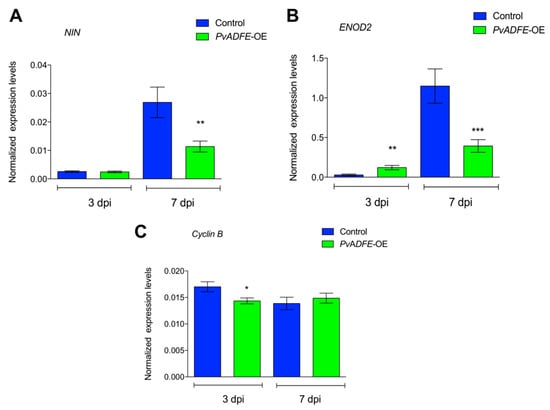
Figure 6.
Reverse-transcription quantitative PCR analysis of early nodulins in PvADFE-OE and control transgenic roots after inoculation with R. tropici CIAT-899. (A) NIN, (B) ENOD2, and (C) Cyclin B transcript accumulation profiles determined by qRT-PCR of total RNA isolated from PvADFE-OE and control transgenic roots at 3 and 7 days after inoculation with rhizobia. Elongation factor EF1α was used as an endogenous reference gene for normalizing expression levels. Each bar represents mean ± SEM for two independent biological replicates with three technical repeats. * p < 0.05, ** p < 0.005, and *** p < 0.001 according to Student’s t-test.
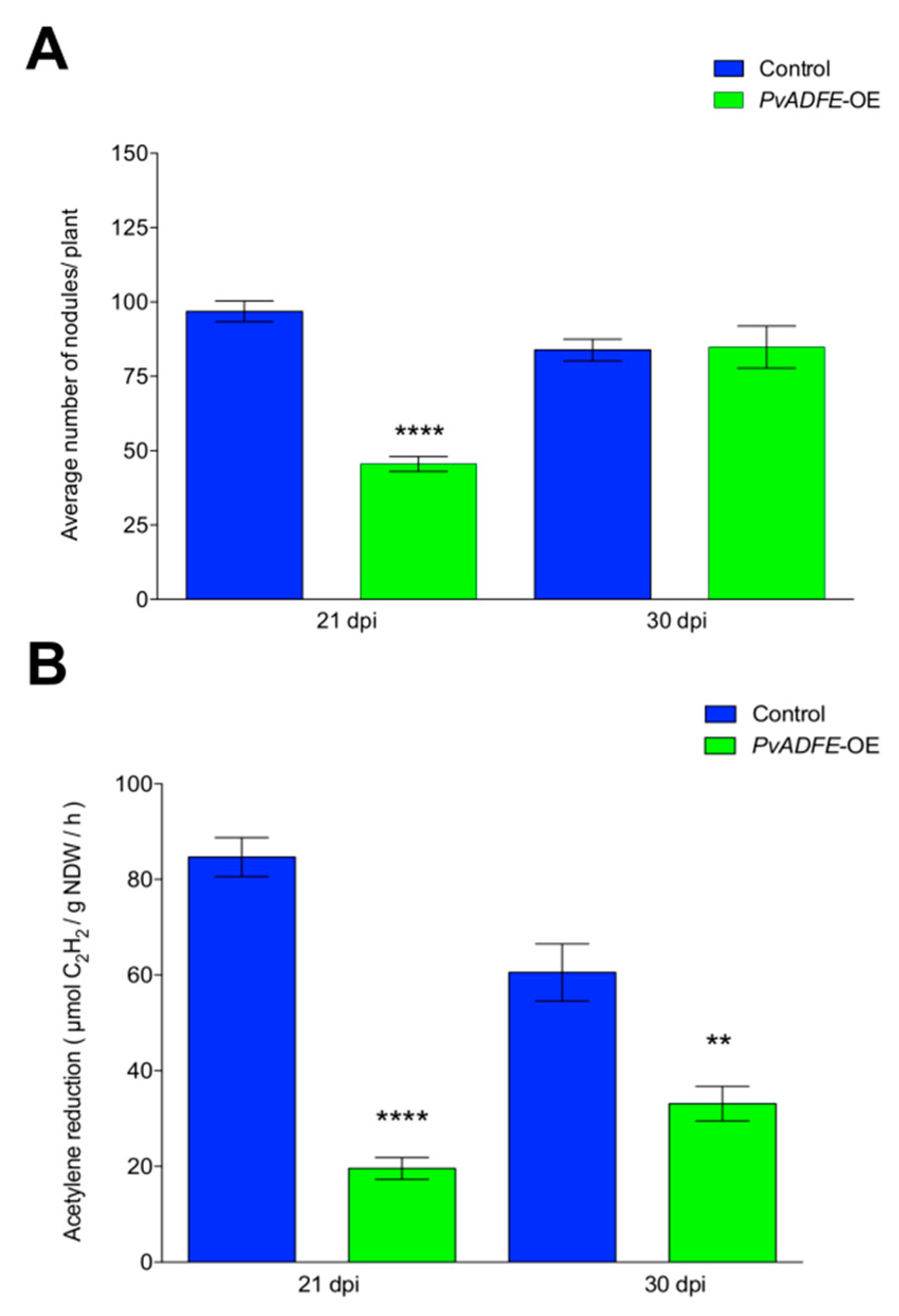
Overexpression of PvADFE resulted in a 45% reduction in nodule number at 21 dpi in comparison to control roots. However, no changes in nodule number were observed at 30 dpi (Figure 7A). Although 50% less of ITs reach the cortical cells in PvADFE-OE transgenic roots (Figure 5C); roots are continuously infected by rhizobia, which may explain the similar number of nodules in control and PvADFE-OE transgenic roots at 30 dpi (Figure 7A). Based on an acetylene reduction assay, nodules from PvADFE-OE transgenic roots had 75% less nitrogen-fixing ability than control roots at 21 dpi and 50% less at 30 dpi (Figure 7B). We also found that the diameter and distribution of nodule size differed in PvADFE-OE and control transgenic roots at 7, 14, 21, and 30 dpi (Figure S10). Specifically, at all time points, PvADFE-OE had a higher proportion of small nodules (Group I, see Materials and Methods Section) compared to control transgenic roots (Figure S10A). In PvADFE-OE transgenic roots, the proportion of large nodules (Group III) at 30 dpi was low (24%), whereas this proportion was 39% in control roots. In addition, the average diameter of nodules in PvADFE-OE was smaller than that of nodules in control roots at the same stages (Figure S10B). These results indicate that overexpression or the general presence of PvADFE transcripts affects nodule development and function, confirming a role of PvADFE under silencing conditions.
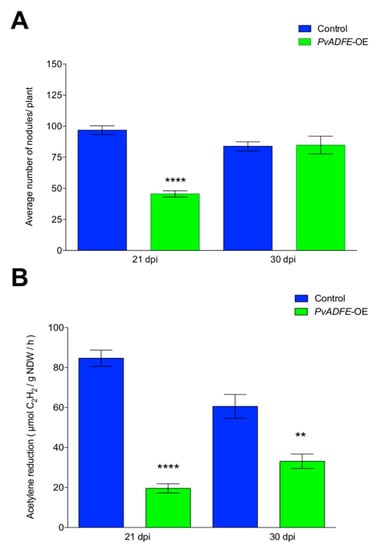
Figure 7.
Nodulation and nitrogen fixation capacity of PvADFE-OE and control transgenic roots. (A) Nodules were collected and counted from control and PvADFE-OE transgenic roots inoculated with R. tropici CIAT-899 at 21 and 30 dpi. (B) Nitrogenase activity determined by the acetylene reduction assay in control and PvADFE-OE nodules of transgenic roots at 21 and 30 dpi. NDW: Nodule dry weight. Bars represent mean ± SEM for three biological replicates with n > 7. ** p < 0.005 and **** p < 0.0001 according to Student’s t-test.
3. Discussion
Plant ADF genes in angiosperms comprise a large family, which can be classified into four subclasses according to their tissue-specific expression and phylogeny. For example, Arabidopsis subclass I ADFs, ADF1 to ADF4, are expressed strongly and constitutively in all vegetative tissues [35]. This ADF subclass has been shown to function in plant resistance to pathogenic microbes, fungi, and pests. Arabidopsis ADF4 functions as a susceptibility factor between the host plant and the powdery mildew fungus [55]. Control of Myzus persicae Sülzer infestation in adf3 was restored by overexpression of the related ADF4 or treatment with the actin cytoskeleton destabilizers cytochalasin D and latrunculin B [40]. Despite symbiotic and pathogenic interactions being different manifestations of the bacteria–host interaction, similar mechanisms exist in both processes to facilitate successful colonization. Here, we suggest that PvADFE participates fine-tuning the signaling pathway leading to the infection process and nodule organogenesis in P. vulgaris symbiosis.
In previous studies, we described early actin cytoskeleton rearrangements in bean root hair cells in response to specific NFs [3,18]. Actin dynamics are the result of highly regulated polymerization and depolymerization processes. A proteomics analysis conducted by our group showed that one of the P. vulgaris ADFs, PvADFE, increases 11.7-fold in relative abundance 30 min after rhizobial NF treatment. Herein, we establish that ADFE is required for both early and late symbiotic processes in bean.
Some reports have described actin cytoskeleton remodeling at the site of plant cell contact with pathogens [56,57,58,59]. Similar remodeling has been observed in symbioses; for instance, IT development requires the rearrangement of the actin cytoskeleton [60]. NF perception leads to fragmentation of longitudinal thick actin bundles, causing accumulation of finer and more diffuse actin at the root-hair tips [3,17]. The number of F-actin plus ends also increases, and F-actin is relocalized to the IT initiation sites [18]. ADFs are the first regulators of actin cytoskeleton organization and function in the disassembly of actin filaments [61]. We showed that the PvADFE promoter is activated in infected root hairs and in cortical cells during division (Figure 1A–D). Knockdown and overexpression of PvADFE had opposing effects on the number of infection events, nodule number, and nitrogen fixation. PvADFE silencing increased the number of infection events (Figure 2C), whereas PvADFE overexpression resulted in 58% of the ITs being confined within the root hairs (Figure 5C). Although the link between ADF and actin during symbiosis is unknown, cytoskeletal rearrangements have been linked to ADF after pathogen perception [37,55,62]. In A. thaliana, the adf4 mutant fails to undergo actin rearrangements during innate immune signaling in response to treatment with a bacterial microbe-associated molecular pattern, elf26, but responds normally to a fungal microbe-associated molecular pattern, chitin [62]. Also, AtADF4 is specifically required for the activation of RPS5-mediated resistance as well as MAPK signaling via the coordinated regulation of actin cytoskeletal dynamics and R-gene transcription [37]. The knockdown of AtADF1 to AtADF4 genes enhances resistance to the powdery mildew fungus Golovinomyces orontii [55], which is contrary to our results, suggesting a different role of ADF in symbiosis and fungal pathogen response. Then, we propose that cytoskeletal changes triggered by the depolymerization of F-actin during rhizobial invasion might be mediated by the participation of PvADFE, which may decrease the viscosity of the cytoplasm, and the relaxation of the cytoskeleton may facilitate successful infection, as previously suggested for nematode-infected roots [63]. However, in excess of ADF, this leads to actin filament stabilization and interferes with actin severing [64,65]. For instance, NtADF1 inhibits pollen tube growth in a dose-dependent manner, and very high levels of GFP-NtADF1 expression result in bundled or patchy regions of actin [44]. A similar scenario may occur in root hairs infected with rhizobia, in which PvADFE excess stabilizes actin filaments, blocking the required reorganization of actin in root hairs during IT initiation and IT progression, which has been widely reported [3,17]. It is possible that during rhizobial infection, ADF protein is accumulated; however, it might remain inactivated (e.g., by phosphorylation or pH) in order to mediate a successful rhizobial infection process.
The initiation of IT growth following NF perception in root hairs includes early changes in nodulin gene expression [9]. In the current study, we observed down-regulation of NIN and ENOD2 in PvADFE-overexpressing transgenic roots at 7 dpi (Figure 6A,B), supporting the idea that PvADFE participates in nodulin signaling, in a similar manner to AtADF4, which is required for MAPK signaling in the presence of the bacterial effector AvrPphB [37]. Surprisingly, overexpression of PvADFE slightly affects the cyclin B expression at 3 dpi, suggesting that, at this stage, PvADF is involved in IT growth rather than primordia development. This role has also been suggested for L. japonicus Nap and Pir1, components of the Suppressor of cAMP receptor defect/WASP family verpolin homologous protein (SCAR/WAVE) complex, which induces actin nucleation and rearrangements via recruitment and activation of the Arp2/3 complex [15].
The silencing and overexpression of PvADFE also cause defects in nodule development. At 21 dpi, PvADFE-RNAi transgenic roots had 41% more nodules and these were 55% more effective in nitrogen fixation compared to the control (Figure 4), whereas 63% fewer nodules and a low level of nitrogen fixation were observed in transgenic roots overexpressing PvADFE (Figure 7). We revealed that the PvADFE promoter is also activated in the vascular bundles of mature nodules (Figure 1H). In this direction, gene expression was reported in vascular bundle tissue of petunia (PhADF1) and rice (OsADF1 and OsADF3) [66,67]. Thus, overexpression of PvADFE might alter the actin filaments in vascular bundles of nodules, compromising the supply of nutrients and water transport to the nodule cells. In addition, nitrogen-fixing symbiosomes develop highly dynamic actin networks to support vesicle transport and promote growth [19,24]. Actin organization and remodeling associated with rhizobium release and symbiosome development can be described in three steps: (a) F-actin arrays channel elongating ITs and mediate the release of infection droplets, (b) a network of actin microfilaments embraces the developing symbiosomes, and (c) short F-actin fragments and actin dots align with the mature symbiosomes [19]. Therefore, the small nodules and low level of nitrogen fixation observed in transgenic roots overexpressing PvADFE suggest that an altered actin rearrangement within infected nodule cells impairs bacteroid functional maturation within symbiosomes. By contrast, as reported [19], actin cytoskeleton channel formation that facilitates the infection droplet release, and symbiosome development and maturation are processes that might be facilitated by ADF silencing.
This work presents evidence that ADF is fine-tuning the symbiotic interaction between legumes and rhizobia. ADFE affects root hair infection, nodule development and nodule fixation. We are currently analyzing adfe mutations affecting Ser-6 (phosphomimetic PvADFE), and colocalization with actin cytoskeleton to better understand the function of this ADF and the actin dynamics in the rhizobia–P. vulgaris symbiosis.
4. Materials and Methods
4.1. Plant Growth Conditions and Rhizobia Inoculation
Seed of P. vulgaris cv. Negro Jamapa (obtained from the local farmers’ markets, Cuernavaca, Mexico) were surface-sterilized and germinated at 28 °C for 2 days in the dark. At 2 days post-germination, the apex of root was sectioned from seedlings and frozen in liquid nitrogen. Separately, the root hairs were gently broken off from roots without apex using a magnetic stir bar in a stainless-steel tank with liquid nitrogen. Consequently, the tripped roots were separated by filtering through a colander, and the root hairs were collected in the liquid nitrogen remained. All the material was stored at −70 °C until subsequent RNA extraction and reverse-transcription quantitative PCR (RT-qPCR) assays.
Composite common bean plants were generated according to the protocol developed by Estrada-Navarrete et al. [68]. Hairy roots (10–13 days post-emergence) were generated using Rhizobium rhizogenes strain K599 [69]. Transgenic composite plants carrying the corresponding construct were observed under epifluorescence microscopy to confirm the presence of the reporter gene (GFP or DsRed), and untransformed roots were removed. Composite common bean plants were planted in pots with vermiculite and inoculated with 20 mL of R. tropici–GUS or R. tropici–DsRed suspension at an optical density at 600 nm (OD600) of 0.05 (undiluted suspension had OD600 = 0.8–1.0). Plants were grown under greenhouse conditions with a controlled environment (26–28 °C, 16 h light:8 h dark) and were watered with B&D medium [70].
4.2. Identification of PvADF Sequences, Three-Dimensional Structure Analysis, and Relative Transcript Accumulation in Different Organs and Tissues
Using the Phvul006G132700.1 sequence as template, searches for ADFs in the Phytozome v12 database (http://phytozome.jgi.doe.gov/pz/portal.html) were performed for P. vulgaris, G. max, M. truncatula, A. thaliana, O. sativa, and Z. mays. L. japonicus, and V. unguiculata protein sequences homologous to ADF were identified and downloaded from the LIS (http://legumeinfo.org) and CGKB (http://cowpeagenomics.med.virginia.edu/CGKB/) databases, respectively (Table S4). PvADF sequences were arbitrarily named from PvADFA to PvADFI based on homology with Arabidopsis ADFs. A phylogenetic tree was generated by the maximum likelihood method based on the JTT matrix and FreeRate with 3 categories using IQ-TREE software version 1.3.11.1 for Linux [71] from 10,000 bootstrap replicates [72]. Multiple sequence alignment of the ADF amino acid sequences was performed using MUSCLE software for Linux [73] and edited using the web service Gblocks 0.91b (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=gblocks). The generated data in Newick tree format were visualized and edited in MEGA X [74].
PvADFE was selected for further analysis. A three-dimensional model of PvADFE was constructed using the I-TASSER server, http://zhanglab.ccmb.med.umich.edu/I-TASSER/ [75], based on homology with the Arabidopsis ADF1 crystal structure, PDB accession number 1FS7 [48]. The model was analyzed using Protean 3D software (DNAStar).
The P. vulgaris Genome Expression Atlas (PvGEA, http://plantgrn.noble.org/PvGEA/) was used in order to estimate the PvADF transcript accumulation profile from different organs and tissues of P. vulgaris analyzed by RNA sequencing and expressed as reads per kilobase of transcript per million mapped reads.
4.3. RT-qPCR Assays
High quality RNA was isolated from frozen tissues using TRIzol® Reagent (Invitrogen, Life Technologies, USA) following the manufacturer’s recommendations. RNA integrity and concentrations were determined by electrophoresis and NanoDrop (2000c, Thermo Scientific) spectrophotometry, respectively. Genomic DNA contamination from RNA samples was removed by incubating the samples with RNase-free DNase (1 U·μL−1) at 37 °C for 15 min and then at 65 °C for 10 min. RT-qPCR assays were performed using an iScript™ One-step RT-PCR Kit with SYBR® Green (Bio-Rad) from DNA-free RNA samples, according the manufacturer’s recommendations, in a LightCycler® Nano cycler (Roche). RNA template concentration was 40 ng (10 ng·μL−1) in each reaction. DNA-free RNA samples were used as a control to confirm the absence of DNA contamination. Relative expression values were calculated using the formula 2−ΔCT, where cycle threshold value (ΔCt) is the cycle threshold (Ct) of the gene of interest minus the Ct of the reference gene [76]. RT-qPCR data were generated from at least two biological replicates with three independent plants each one, together with three technical replicates. The gene encoding elongation factor 1-alpha (EF1α) was used as a reference gene to normalize the experimental data. The genes analyzed are listed in Table S5, together with their specific oligonucleotides used.
4.4. Plasmid Construction
To generate the RNAi construct, a fragment corresponding to the 3′-untranslated region of PvADFE was amplified from cDNA isolated from common bean roots at 2 days post germination using the following primers: PvADFE-RNAi-Up (5′-GTACGCTTTCTGGTGGGAGCAC-3′) and PvADFE-RNAi-Lw (5′-ACAAAAGAAAGCATATATCGTCCAAA-3′). The resulting PCR product was cloned into pENTR/D-TOPO (Invitrogen) and transformed by heat-shock into Escherichia coli TOP10 chemically competent cells. The recombination into the destination vector pTdT-DC-RNAi [77] was performed with the LR clonase, using the Gateway System (Invitrogen). The appropriated orientation of the insert was confirmed by PCR and sequencing using the PvADFE-RNAi-Up primer for the pTdT-PvADFE-RNAi plasmid together with WRKY-5-Rev (5′-GCAGAGGAGGAGAAGCTTCTAG-3′) or WRKY-3-Fwd (5′-CTTCTCCAACCACAGGAATTCATC-3′) primer. As a control, a truncated and irrelevant sequence from A. thaliana pre-mir159 (kindly provided by Dr. José Luis Reyes), lacking the target sequence of miR159 (ACAGTTTGCTTATGTCGGATCCATAATATATTTGACAAGATACTTTGTTTTTCGATAGATCTTGATCTGACGATGGAAGTAGAGCTCTACATCCCGGGTCA), was cloned into the pTdT-DC-RNAi vector. The correct orientation of the sequence in the construct was confirmed by DNA sequencing.
To construct an overexpression vector for PvADFE, the 833-bp ORF of PvADFE (Phvul006G132700.1) including the 5′-untranslated region (154 bp) and a 3’-untranslated (169 bp) fragment was isolated from P. vulgaris cDNA. This region was amplified from P. vulgaris root cDNA at 2 days post-germination using PvADFE-OE-Up and PvADFE-OE-Lw primers (Table S5). The fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) and sequenced. The resulting pENTR-PvADFE plasmid was recombined into the binary vector pH7WG2tdT (constructed by Marco A. Juárez-Verdayes) under the control of the constitutive 35S promoter (Figure S11). Briefly, this vector was derived from vector pH7WG2D [78]; the cassette pEgfpER and the 35S terminator were respectively replaced by a TdTomato (red fluorescent protein) reporter obtained from the pTd-DC-RNAi vector [77] and an E9 terminator. Empty pH7WG2tdT vector was used as the control.
To develop a pPvADFE:GUS-GFP construct, a 1383-bp fragment upstream of the PvADFE translation start site was amplified using bean genomic DNA and primers pPvADFE-Up and pPvADFE-Lw (Table S5) and cloned into vector pENTR/SD/D-TOPO (Invitrogen). The Gateway LR reaction was performed between the entry vector pENTR/SD/D-TOPO-PvADFE and the destination vector pBGWFS7.0 [78], according to the manufacturers’ instructions (Invitrogen). Control transgenic roots harbored a cassette with no promoter sequences upstream of the GFP-GUS sequence.
4.5. Promoter Activity Analysis
Composite P. vulgaris plants harboring pPvADFE:GUS-GFP were transferred into pots of vermiculite and each plant was inoculated with 20 mL of R. tropici–DsRed suspension (with an OD600 of 0.05). Roots and nodules were collected at 5, 7, 14, and 21 dpi (days post-inoculation). Samples were histochemically analyzed for GUS activity according to the method of Jefferson [79] and images were acquired with a Retiga 4000R CCD camera coupled to a Nikon TE300 inverted microscope. Promoter activity during early infection events was detected using an inverted confocal microscope (Nikon eclipse Ti in combination with a Yokogawa CSU-W1 spinning disk confocal system) and images were processed using ImageJ version 1.48 (US National Institutes of Health). GFP fluorescence was excited at 488 nm, while DsRed fluorescence was excited at 543 nm.
4.6. Analysis of Infection Events, Nodule Number, and Nodule Diameter
Transgenic roots expressing red fluorescent protein (TdTomato from vector pH7WG2tdT) were selected as described above. These roots were transferred to pots with vermiculite and inoculated with R. tropici–GUS to analyze IT progression, nodulation, and nitrogen fixation. Infection events were analyzed in the control and PvADFE transgenic roots under a Zeiss Axioskop light microscope (Carl Zeiss, Jena, Germany) equipped with a ×63 objective. Images were captured by a Nikon Coolpix 5000 camera with a UR-E6 adapter and an MDC lens attached to the microscope. GUS activity was analyzed according to the method of Jefferson [79]. Images of nodulated transgenic roots at 7, 14, 21, and 30 dpi stained with GUS were taken using a Perfection 4490 scanner (Epson) and captured in TIFF format at a resolution of 6108 × 6108 pixels. Nodule diameter was measured using ImageJ 1.48 (US National Institutes of Health) and classified according to their diameter (d) into four groups: Group I (d < 0.5 mm), Group II (0.5 < d ≤ 1.0 mm), Group III (1.0 < d ≤ 1.5 mm), and Group IV (1.5 < d < 2.0 mm). The number of nodules was counted manually at 21 and 30 dpi.
4.7. Acetylene Reduction Analysis
Acetylene reduction [80] was used to quantify the nitrogenase activity in transgenic nodules at 21 and 30 dpi. Nodulated plant roots were transferred to bottles with rubber seal stoppers by injecting acetylene to a final concentration of 10% of the gas phase. Each sample was incubated for 120 min at room temperature, and ethylene production was determined by gas chromatography in a Variant model 3300 chromatograph. Specific activity is expressed as μmol ethylene−1·(g nodule dry weight)−1·h−1.
4.8. Statistical Analysis
Statistical analyses were computed using GraphPad Prism version 6.00 for Windows, (GraphPad Software, San Diego, CA, USA). Significance tests were performed using an unpaired Student’s t-test. Differences were considered significant if p < 0.05. Results are presented as means ± standard error of the mean.
5. Conclusions
In this study, we demonstrated that ADFE is preferentially expressed in rhizobia-inoculated roots and nodules. Functional characterization showed that ADF overexpression and silencing affect infection threads number, nodule number, average nodule diameter, and nitrogen fixation. Altogether, our results revealed that ADFE is fine-tuning the symbiotic interaction between common bean and rhizobia.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/6/1970/s1. Figure S1. Exon–intron organization of ADF genes; Figure S2. Protein structure of P. vulgaris actin depolymerizing factors (PvADFs); Figure S3. Phylogenetic tree of ADF family proteins; Figure S4. Expression profile of P. vulgaris ADF genes in root hairs, apices, and stripped roots from seedlings harvested at 2 days post-germination; Figure S5. Transcript abundance of ADF genes in different organs and tissues of P. vulgaris; Figure S6. Reverse-transcription quantitative PCR analysis of PvADFE silencing in composite common beans roots; Figure S7. Expression of PvADF genes in control and PvADFE-RNAi transgenic roots at 10 days post emergence; Figure S8. Nodule diameters on PvADFE-RNAi and control transgenic roots after inoculation with R. tropici expressing GUS; Figure S9. Reverse-transcription quantitative PCR analysis of PvADFE overexpression in composite common bean roots; Figure S10. Nodule diameters on PvADFE-OE and control transgenic roots after inoculation with R. tropici expressing GUS; Figure S11. In silico map of the pH7WG2tdT vector; Table S1. Size of the ADF gene family in various plants; Table S2. Percentage of nucleotide sequence identity among P. vulgaris ADF genes; Table S3. Percentage of amino acid sequence identity between PvADF and AthADF proteins; Table S4. Annotation of ADFs aminoacid sequences used for the phylogenetic analysis; Table S5. Gene specific oligonucleotides used.
Author Contributions
Y.O.-O. performed the experiments, interpreted the data and wrote the manuscript; J.C.-C. interpreted the data and wrote the manuscript; M.A.J.-V. gave bioinformatic advices; R.T.-M. contributed to the draft manuscript; C.F.-G. constructed the phylogenetic tree and assisted also to the draft manuscript; N.N. helped with RT-qPCR and inoculation experiments with rhizobia; L.C. gave experimental advice and contributed to the draft manuscript; C.Q. conceived and designed the whole project, contributed to the drafting of the manuscript and gave final approval of the version to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported partially by Consejo Nacional de Ciencia y Tecnología with a postdoctoral fellowship (174806) to Yolanda Ortega-Ortega.
Acknowledgments
We acknowledge Rosana Sánchez for technical assistance with promoter experiments. The authors are grateful to Biol. Olivia Santana for technical assistance with inoculation experiments with rhizobia, and QFB Xochitl Alvarado-Affantranger (IBT-UNAM) and Alfonso Leija (Centro de Ciencias Genómicas, UNAM), for technical assistance in optical microscopy, epifluorescence microscopy, and acetylene reduction assays.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| ADFs | Actin Depolymerization Factors |
| NFs | Nod factors |
| IT | Infection Thread |
| ARP3 | Actin Related Protein 3 |
| SCARN | Suppressor of cAMP Receptor Refect-Nodulation |
| ABP | Actin-Binding |
| ADF-H | Proteins Actin-Depolymerizing Factor Homology |
| PIP2 | Phosphatidylinositol 4,5-bisphosphate |
| RPS5 | Disease Resistance Protein 5 |
| dpi | days post-inoculation |
| GFP | Green Fluorescent Protein |
| NDW | Nodule Dry Weight |
| GUS | β-glucuronidase |
| NIN | Nodule Inception |
| ENOD2 | Early Nodulin2 |
References
- Laranjo, M.; Alexandre, A.; Oliveira, S. Legume growth-promoting rhizobia: An overview on the Mesorhizobium genus. Microbiol. Res. 2014, 169, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.W.; Wais, R.; Long, S.R. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 1996, 85, 673–681. [Google Scholar] [CrossRef]
- Cárdenas, L.; Vidali, L.; Domínguez, J.; Pérez, H.; Sánchez, F.; Hepler, P.K.; Quinto, C. Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 1998, 116, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Felle, H.H.; Kondorosi, É.; Kondorosi, A.; Schultze, M. Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 1996, 10, 295–301. [Google Scholar] [CrossRef]
- Cárdenas, L.; Martínez, A.; Sánchez, F.; Quinto, C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 2008, 56, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, L.; Holdaway-Clarke, T.L.; Sánchez, F.; Quinto, C.; Feijó, J.A.; Kunkel, J.G.; Hepler, P.K. Ion changes in legume root hairs responding to Nod factors. Plant Physiol. 2000, 123, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.W.; Atkinson, E.; Long, S.R. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 1992, 256, 998–1000. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Niebel, F.; Barker, D.G. Remodeling of the Infection Chamber before Infection Thread Formation Reveals a Two-Step Mechanism for Rhizobial Entry into the Host Legume Root Hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef]
- Long, S.R. SnapShot: Signaling in Symbiosis. Cell 2016, 167, 582. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C. Symbiosis: Plasmodesmata Link Root-Nodule Organogenesis with Infection. Curr. Biol. 2018, 28, R1400–R1403. [Google Scholar] [CrossRef] [PubMed]
- Gaudioso-Pedraza, R.; Beck, M.; Frances, L.; Kirk, P.; Ripodas, C.; Niebel, A.; Oldroyd, G.E.D.; Benitez-Alfonso, Y.; de Carvalho-Niebel, F. Callose-Regulated Symplastic Communication Coordinates Symbiotic Root Nodule Development. Curr. Biol. 2018, 28, 3562–3577. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Newcomb, W. Changes in actin microfilament arrays in developing pea root nodule cells. Can. J. Bot. 2001, 79, 767–776. [Google Scholar]
- Yokota, K.; Fukai, E.; Madsen, L.H.; Jurkiewicz, A.; Rueda, P.; Radutoiu, S.; Held, M.; Hossain, M.S.; Szczyglowski, K.; Morieri, G.; et al. Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 2009, 21, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Timmers, A.C.J. The role of the plant cytoskeleton in the interaction between legumes and rhizobia. J. Microsc. 2008, 231, 247–256. [Google Scholar] [CrossRef]
- De Ruijter, N.C.A.; Bisseling, T.; Emons, A.M.C. Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol. Plant Microbe Interact. 1999, 12, 829–832. [Google Scholar] [CrossRef]
- Zepeda, I.; Sánchez-López, R.; Kunkel, J.G.; Bañuelos, L.A.; Hernández-Barrera, A.; Sánchez, F.; Quinto, C.; Cárdenas, L. Visualization of highly dynamic F-actin plus ends in growing Phaseolus vulgaris root hair cells and their responses to Rhizobium etli Nod factors. Plant Cell Physiol. 2014, 55, 580–592. [Google Scholar] [CrossRef]
- Zhang, X.; Han, L.; Wang, Q.; Zhang, C.; Yu, Y.; Tian, J.; Kong, Z. The host actin cytoskeleton channels rhizobia release and facilitates symbiosome accommodation during nodulation in Medicago truncatula. New Phytol. 2019, 221, 1049–1059. [Google Scholar] [CrossRef]
- Ketelaar, T. The actin cytoskeleton in root hairs: All is fine at the tip. Curr. Opin. Plant Biol. 2013, 16, 749–756. [Google Scholar] [CrossRef]
- McCurdy, D.W.; Kovar, D.R.; Staiger, C.J. Actin and actin-binding proteins in higher plants. Protoplasma 2001, 215, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Nagawa, S.; Xu, T.; Yang, Z. RHO GTPase in plants: Conservation and invention of regulators and effectors. Small GTPases 2010, 1, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Facette, M.R.; Park, Y.; Sutimantanapi, D.; Luo, A.; Cartwright, H.N.; Yang, B.; Bennett, E.J.; Sylvester, A.W.; Smith, L.G. The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat. Plants 2015, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gavrin, A.; Jansen, V.; Ivanov, S.; Bisseling, T.; Fedorova, E. ARP2/3-mediated actin nucleation associated with symbiosome membrane is essential for the development of symbiosomes in infected cells of Medicago truncatula root nodules. Mol. Plant Microbe Interact. 2015, 28, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Lin, J.-S.; Xu, J.; Sato, S.; Parniske, M.; Wang, T.L.; Downie, J.A.; Xie, F. SCARN a novel class of SCAR protein that is required for root-hair infection during legume Nodulation. PLoS Genet. 2015, 11, e1005623. [Google Scholar] [CrossRef]
- Poukkula, M.; Kremneva, E.; Serlachius, M.; Lappalainen, P. Actin-depolymerizing factor homology domain: A conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton 2011, 68, 471–490. [Google Scholar] [CrossRef]
- Lappalainen, P.; Kessels, M.M.; Cope, M.J.T.V.; Drubin, D.G. The ADF homology (ADF-H) domain: A highly exploited actin-binding module. Mol. Biol. Cell 1998, 9, 1951–1959. [Google Scholar] [CrossRef]
- Maciver, S.K.; Hussey, P.J. The ADF/cofilin family: Actin-remodeling proteins. Genome Biol. 2002, 3, reviews3007-1. [Google Scholar] [CrossRef]
- Hayakawa, K.; Sakakibara, S.; Sokabe, M.; Tatsumi, H. Single-molecule imaging and kinetic analysis of cooperative cofilin–actin filament interactions. Proc. Natl. Acad. Sci. USA 2014, 111, 9810–9815. [Google Scholar] [CrossRef]
- Augustine, R.C.; Vidali, L.; Kleinman, K.P.; Bezanilla, M. Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 2008, 54, 863–875. [Google Scholar] [CrossRef]
- Gungabissoon, R.A.; Jiang, C.-J.; Drøbak, B.K.; Maciver, S.K.; Hussey, P.J. Interaction of maize actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J. 1998, 16, 689–696. [Google Scholar] [CrossRef]
- Allwood, E.G.; Smertenko, A.P.; Hussey, P.J. Phosphorylation of plant actin-depolymerising factor by calmodulin-like domain protein kinase. FEBS Lett. 2001, 499, 97–100. [Google Scholar] [CrossRef]
- Smertenko, A.P.; Jiang, C.-J.; Simmons, N.J.; Weeds, A.G.; Davies, D.R.; Hussey, P.J. Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. 1998, 14, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Roy-Zokan, E.M.; Dyer, K.A.; Meagher, R.B. Phylogenetic patterns of codon evolution in the ACTIN-DEPOLYMERIZING FACTOR/COFILIN (ADF/CFL) gene family. PLoS ONE 2015, 10, e0145917. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, D.R.; Kandasamy, M.K.; McKinney, E.C.; Burgos-Rivera, B.; Meagher, R.B. The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 2007, 52, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Chaudhry, F.; Ruzicka, D.R.; Meagher, R.B.; Staiger, C.J.; Day, B. Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 2009, 150, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.; Shimono, M.; Tian, M.; Day, B. Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog. 2012, 8, e1003006. [Google Scholar] [CrossRef]
- Fu, Y.; Duan, X.; Tang, C.; Li, X.; Voegele, R.T.; Wang, X.; Wei, G.; Kang, Z. TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 2014, 78, 16–30. [Google Scholar] [CrossRef]
- Tang, C.; Deng, L.; Chang, D.; Chen, S.; Wang, X.; Kang, Z. TaADF3, an actin-depolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Mondal, H.A.; Louis, J.; Archer, L.; Patel, M.; Nalam, V.J.; Sarowar, S.; Sivapalan, V.; Root, D.D.; Shah, J. Arabidopsis ACTIN-DEPOLYMERIZING FACTOR3 is required for controlling aphid feeding from the phloem. Plant Physiol. 2018, 176, 879–890. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, Q.; Xue, Q. Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. J. Plant Physiol. 2006, 163, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Khatun, K.; Robin, A.H.K.; Park, J.-I.; Kim, C.K.; Lim, K.-B.; Kim, M.-B.; Lee, D.-J.; Nou, I.S.; Chung, M.-Y. Genome-wide identification, characterization and expression profiling of ADF family genes in Solanum lycopersicum L. Genes 2016, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Wong, E.I.; Vidali, L.; Estavillo, A.; Hepler, P.K.; Wu, H.-M.; Cheung, A.Y. The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 2002, 14, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, P.; Fedorov, E.V.; Fedorov, A.A.; Almo, S.C.; Drubin, D.G. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997, 16, 5520–5530. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-J.; Weeds, A.G.; Hussey, P.J. The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 1997, 12, 1035–1043. [Google Scholar] [CrossRef]
- Yonezawa, N.; Homma, Y.; Yahara, I.; Sakai, H.; Nishida, E. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J. Biol. Chem. 1991, 266, 17218–17221. [Google Scholar]
- Bowman, G.D.; Nodelman, I.M.; Hong, Y.; Chua, N.-H.; Lindberg, U.; Schutt, C.E. A comparative structural analysis of the ADF/cofilin family. Proteins Struct. Funct. Bioinform. 2000, 41, 374–384. [Google Scholar] [CrossRef]
- Bou Daher, F.; van Oostende, C.; Geitmann, A. Spatial and temporal expression of actin depolymerizing factors ADF7 and ADF10 during male gametophyte development in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1177–1192. [Google Scholar] [CrossRef]
- Madsen, L.H.; Tirichine, L.; Jurkiewicz, A.; Sullivan, J.T.; Heckmann, A.B.; Bek, A.S.; Ronson, C.W.; James, E.K.; Stougaard, J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010, 1, 1–12. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Stougaard, J. Regulators and regulation of legume root nodule development. Plant Physiol. 2000, 124, 531–540. [Google Scholar] [CrossRef]
- Jeleńska, J.; Deckert, J.; Kondorosi, E.; Legocki, A.B. Mitotic B-type cyclins are differentially regulated by phytohormones and during yellow lupine nodule development. Plant Sci. 2000, 150, 29–39. [Google Scholar] [CrossRef]
- Murray, J.D. The Cell Cycle in Nodulation: Molecular Cell Biology of the Growth and Differentiation of Plant Cell; Rose, R.J., Ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Inada, N.; Higaki, T.; Hasezawa, S. Nuclear function of subclass I actin-depolymerizing factor contributes to susceptibility in Arabidopsis to an adapted powdery mildew fungus. Plant Physiol. 2016, 170, 1420–1434. [Google Scholar] [CrossRef] [PubMed]
- Henty-Ridilla, J.L.; Shimono, M.; Li, J.; Chang, J.H.; Day, B.; Staiger, C.J. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog. 2013, 9, e1003290. [Google Scholar] [CrossRef]
- Day, B.; Henty, J.L.; Porter, K.J.; Staiger, C.J. The pathogen-actin connection: A platform for defense signaling in plants. Annu. Rev. Phytopathol. 2011, 49, 483–506. [Google Scholar] [CrossRef]
- Porter, K.; Day, B. From filaments to function: The role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J. Integr. Plant Biol. 2015, 58, 299–311. [Google Scholar] [CrossRef]
- Li, P.; Day, B. Battlefield Cytoskeleton: Turning the Tide on Plant Immunity. Mol. Plant Microbe Interact. 2019, 32, 25–34. [Google Scholar] [CrossRef]
- Takemoto, D.; Hardham, A.R. The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol. 2004, 136, 3864–3876. [Google Scholar] [CrossRef]
- Kanellos, G.; Frame, M.C. Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 2016, 129, 3211–3218. [Google Scholar] [CrossRef]
- Henty-Ridilla, J.L.; Li, J.; Day, B.; Staiger, C.J. ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 2014, 26, 340–352. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Engler, J.; Van Poucke, K.; Karimi, M.; De Groodt, R.; Gheysen, G.; Engler, G.; Gheysen, G. Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant J. 2004, 38, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Andrianantoandro, E.; Pollard, T.D. Mechanism of ACTIN filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 2006, 24, 13–23. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, E.M. How cofilin severs an actin filament. Biophys. Rev. 2009, 1, 51–59. [Google Scholar] [CrossRef]
- Mun, J.-H.; Lee, S.-Y.; Yu, H.-J.; Jeong, Y.-M.; Shin, M.-Y.; Kim, H.; Lee, I.; Kim, S.-G. Petunia actin-depolymerizing factor is mainly accumulated in vascular tissue and its gene expression is enhanced by the first intron. Gene 2002, 292, 233–243. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Huang, W.-L.; Hong, C.-Y.; Lur, H.-S.; Chang, M.-C. Comprehensive analysis of differentially expressed rice actin depolymerizing factor gene family and heterologous overexpression of OsADF3 confers Arabidopsis Thaliana drought tolerance. Rice 2012, 5, 33. [Google Scholar] [CrossRef]
- Estrada-Navarrete, G.; Alvarado-Affantranger, X.; Olivares, J.E.; Guillen, G.; Diaz-Camino, C.; Campos, F.; Quinto, C.; Gresshoff, P.M.; Sánchez, F. Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat. Protoc. 2007, 2, 1819–1824. [Google Scholar] [CrossRef]
- Young, J.M.; Kuykendall, L.D.; Martínez-Romero, E.; Kerr, A.; Sawada, H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 2001, 51, 89–103. [Google Scholar]
- Broughton, W.J.; Dilworth, M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Valdés-López, O.; Arenas-Huertero, C.; Ramírez, M.; Girard, L.; Sánchez, F.; Vance, C.P.; Luis Reyes, J.; Hernández, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Ramírez, M.; Valderrama, B.; Arredondo-Peter, R.; Soberón, M.; Mora, J.; Hernández, G. Rhizobium etli genetically engineered for the heterologous expression of Vitreoscilla sp. hemoglobin: Effects on free-living and symbiosis. Mol. Plant Microbe Interact. 1999, 12, 1008–1015. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).