An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity
Abstract
1. Introduction
2. Results
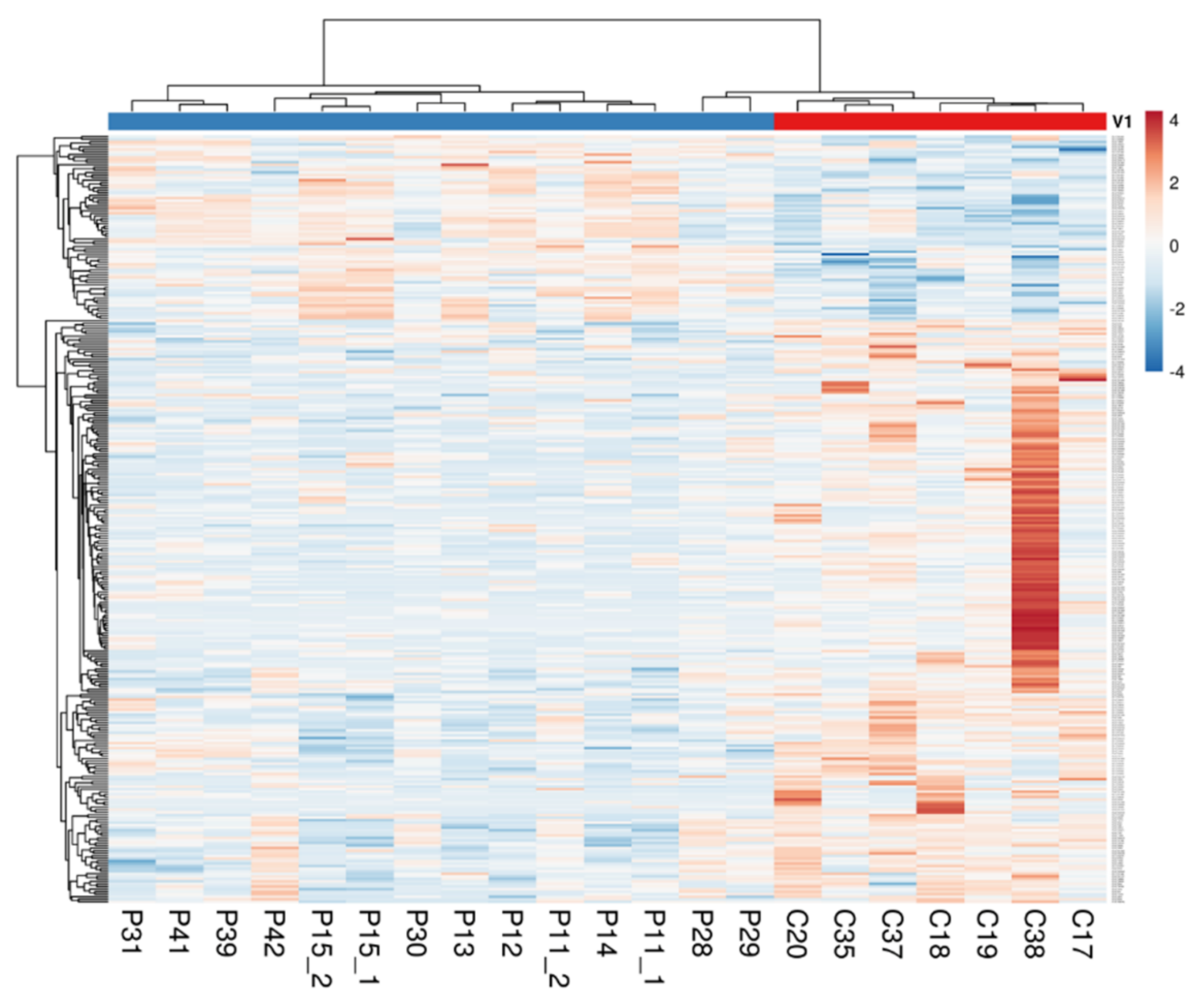
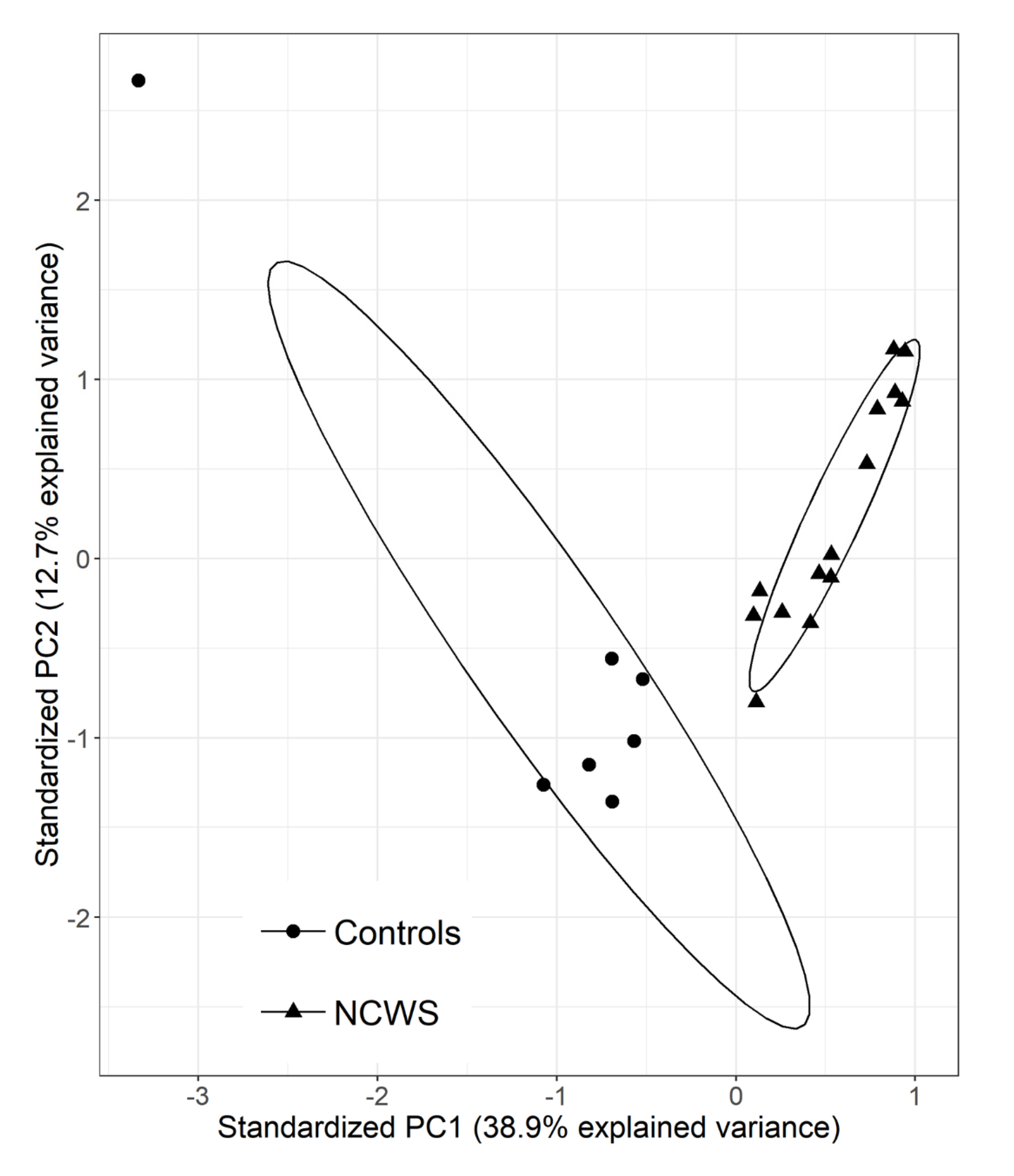
2.1. The Intestinal Mucosa of NCWS Patients Showed a Gene Expression Pattern Different from Controls
2.2. Gene Expression can Contribute to Identify the NCWS Status
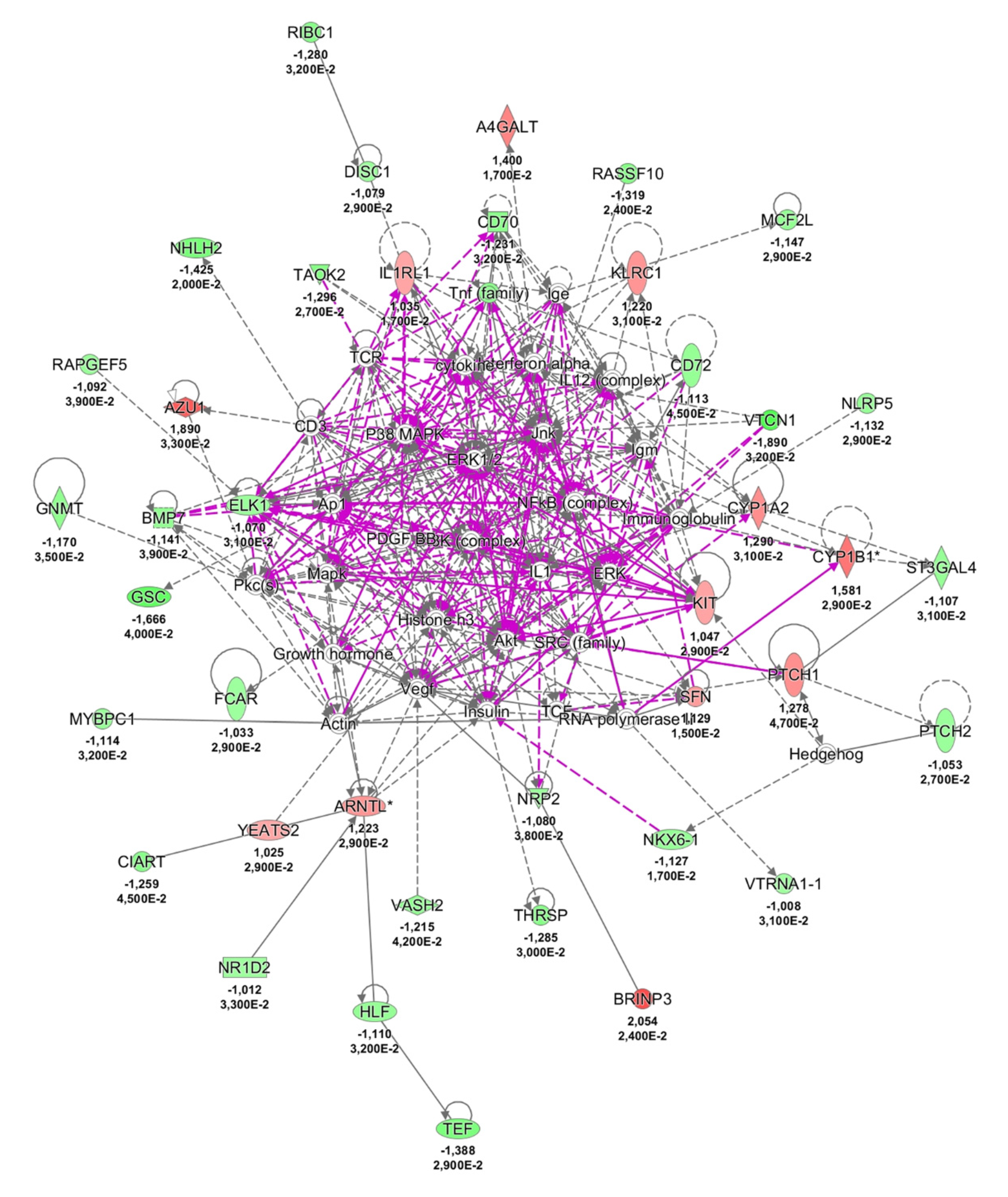
2.3. Possible Pathological Mechanisms Engaged in NCWS
3. Discussion
4. Material and Methods
4.1. Patient’s Recruitment
4.2. RNA Isolation and Microarray Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Ethics Approval and Consent to Participate
Funding
Conflicts of Interest
Abbreviations
| ATI | amylase trypsin inhibitor |
| AUC | area under the curve |
| DE | differentially expressed |
| FDR | false discovery rate |
| FODMAPs | fermentable oligo-, di-, monosaccharides |
| HLA | human leukocyte antigen |
| IBS | irritable bowel syndrome |
| IPA | ingenuity pathway analysis |
| LASSO | least absolute shrinkage and selection operator |
| lncRNA | long non-coding RNA |
| LPS | lipopolysaccharide |
| NCWS | non-celiac wheat sensitivity |
| PCA | principal component analysis |
| SIBO | small intestinal bacterial overgrowth |
References
- Skodje, G.I.; Sarna, V.K.; Minelle, I.H.; Rolfsen, K.L.; Muir, J.G.; Gibson, P.R.; Veierod, M.B.; Henriksen, C.; Lundin, K.E.A. Fructan, rather than gluten, induces symptoms in patients with self-reported non-celiac gluten sensitivity. Gastroenterology 2018, 154, 529.e2–539.e2. [Google Scholar] [CrossRef]
- Caminero, A.; McCarville, J.L.; Zevallos, V.F.; Pigrau, M.; Yu, X.B.; Jury, J.; Galipeau, H.J.; Clarizio, A.V.; Casqueiro, J.; Murray, J.A.; et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat proteins. Gastroenterology 2019, 156, 2266–2280. [Google Scholar] [CrossRef]
- Sapone, A.; Bai, J.C.; Ciacci, C.; Dolinsek, J.; Green, P.H.; Hadjivassiliou, M.; Kaukinen, K.; Rostami, K.; Sanders, D.S.; Schumann, M.; et al. Spectrum of gluten-related disorders: Consensus on new nomenclature and classification. BMC Med. 2012, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Carroccio, A.; Mansueto, P.; Iacono, G.; Soresi, M.; D’Alcamo, A.; Cavataio, F.; Brusca, I.; Florena, A.M.; Ambrosiano, G.; Seidita, A.; et al. Non-celiac wheat sensitivity diagnosed by double-blind placebo-controlled challenge: Exploring a new clinical entity. Am. J. Gastroenterol. 2012, 107, 1898–1906. [Google Scholar] [CrossRef] [PubMed]
- Makharia, A.; Catassi, C.; Makharia, G.K. The overlap between irritable bowel syndrome and non-celiac gluten sensitivity: a clinical dilemma. Nutrients 2015, 7, 10417–10426. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Elli, L.; Bonaz, B.; Bouma, G.; Carroccio, A.; Castillejo, G.; Cellier, C.; Cristofori, F.; de Magistris, L.; Dolinsek, J.; et al. Diagnosis of Non-Celiac Gluten Sensitivity (NCGS): The salerno experts’ criteria. Nutrients 2015, 7, 4966–4977. [Google Scholar] [CrossRef]
- Molina-Infante, J.; Carroccio, A. Suspected nonceliac gluten sensitivity confirmed in few patients after gluten challenge in double-blind, placebo-controlled trials. Clin. Gastroenterol. Hepatol. 2017, 15, 339–348. [Google Scholar] [CrossRef]
- Schuppan, D.; Zevallos, V. Wheat amylase trypsin inhibitors as nutritional activators of innate immunity. Dig. Dis. 2015, 33, 260–263. [Google Scholar] [CrossRef]
- Sapone, A.; Lammers, K.M.; Casolaro, V.; Cammarota, M.; Giuliano, M.T.; De Rosa, M.; Stefanile, R.; Mazzarella, G.; Tolone, C.; Russo, M.I.; et al. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: Celiac disease and gluten sensitivity. BMC Med 2011, 9, 23. [Google Scholar] [CrossRef]
- Uhde, M.; Ajamian, M.; Caio, G.; De Giorgio, R.; Indart, A.; Green, P.H.; Verna, E.C.; Volta, U.; Alaedini, A. Intestinal cell damage and systemic immune activation in individuals reporting sensitivity to wheat in the absence of coeliac disease. Gut 2016, 65, 1930–1937. [Google Scholar] [CrossRef]
- Brottveit, M.; Beitnes, A.C.; Tollefsen, S.; Bratlie, J.E.; Jahnsen, F.L.; Johansen, F.E.; Sollid, L.M.; Lundin, K.E. Mucosal cytokine response after short-term gluten challenge in celiac disease and non-celiac gluten sensitivity. Am. J. Gastroenterol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Zanini, B.; Villanacci, V.; Marullo, M.; Cadei, M.; Lanzarotto, F.; Bozzola, A.; Ricci, C. Duodenal histological features in suspected non-celiac gluten sensitivity: New insights into a still undefined condition. Virchows Arch. 2018. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Samant, H. Wheat allergy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Lebwohl, B.; Sanders, D.S.; Green, P.H.R. Coeliac disease. Lancet 2018, 391, 70–81. [Google Scholar] [CrossRef]
- Bragde, H.; Jansson, U.; Jarlsfelt, I.; Soderman, J. Gene expression profiling of duodenal biopsies discriminates celiac disease mucosa from normal mucosa. Pediatr. Res. 2011, 69, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Croce, C.M. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 7413–7417. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Li, K.; Ramchandran, R. Natural antisense transcript: A concomitant engagement with protein-coding transcript. Oncotarget 2010, 1, 447–452. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- NE, I.I.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014, 5, 3979. [Google Scholar] [CrossRef]
- Ma, S.; Ming, Z.; Gong, A.Y.; Wang, Y.; Chen, X.; Hu, G.; Zhou, R.; Shibata, A.; Swanson, P.C.; Chen, X.M. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-kappaB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017, 31, 1215–1225. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013, 2, e00762. [Google Scholar] [CrossRef]
- Krawczyk, M.; Emerson, B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife 2014, 3, e01776. [Google Scholar] [CrossRef]
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef]
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef]
- Kim, Y.K.; Shin, J.S.; Nahm, M.H. NOD-like receptors in infection, immunity, and diseases. Yonsei Med. J. 2016, 57, 5–14. [Google Scholar] [CrossRef]
- De Tymowski, C.; Heming, N.; Correia, M.D.T.; Abbad, L.; Chavarot, N.; Le Stang, M.B.; Flament, H.; Bex, J.; Boedec, E.; Bounaix, C.; et al. CD89 is a potent innate receptor for bacteria and mediates host protection from sepsis. Cell Rep. 2019, 27, 762–775. [Google Scholar] [CrossRef]
- Pasquier, B.; Launay, P.; Kanamaru, Y.; Moura, I.C.; Pfirsch, S.; Ruffie, C.; Henin, D.; Benhamou, M.; Pretolani, M.; Blank, U.; et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: Dual role of FcRgamma ITAM. Immunity 2005, 22, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, F.M.; Monteiro, R.C.; Volanakis, J.E.; Cooper, M.D. IgA deficiency. Immunodefic Rev. 1991, 3, 15–44. [Google Scholar] [PubMed]
- Kasus-Jacobi, A.; Griffith, G.L.; Lerner, M.; Pereira, H.A. Effect of cationic antimicrobial protein CAP37 on cytokine profile during corneal wound healing. J. Ocul. Dis. Ther. 2017, 5, 19–27. [Google Scholar]
- Walsh, C.E.; Ryan, E.J.; O’Farrelly, C.; Golden-Mason, L.; FitzGerald, O.; Veale, D.J.; Bresnihan, B.; Fearon, U. Differential expression of NK receptors CD94 and NKG2A by T cells in rheumatoid arthritis patients in remission compared to active disease. PLoS ONE 2011, 6, e27182. [Google Scholar] [CrossRef] [PubMed]
- Cauli, A.; Dessole, G.; Piga, M.; Angioni, M.M.; Pinna, S.; Floris, A.; Congia, M.; Mascia, E.; Paladini, F.; Tedeschi, V.; et al. Expression analysis of HLA-E and NKG2A and NKG2C receptors points at a role for natural killer function in ankylosing spondylitis. RMD Open 2018, 4, e000597. [Google Scholar] [CrossRef] [PubMed]
- Son, S.W.; Kim, E.O.; Ryu, E.S.; Kim, T.J.; Kim, J.N.; Choi, J.E.; Kye, Y.C.; Lee, K.M. Upregulation of Fas and downregulation of CD94/NKG2A inhibitory receptors on circulating natural killer cells in patients with new-onset psoriasis. Br. J. Dermatol. 2009, 161, 281–288. [Google Scholar] [CrossRef]
- Rocher, C.; Singla, D.K. SMAD-PI3K-Akt-mTOR pathway mediates BMP-7 polarization of monocytes into M2 macrophages. PLoS ONE 2013, 8, e84009. [Google Scholar] [CrossRef] [PubMed]
- Maric, I.; Poljak, L.; Zoricic, S.; Bobinac, D.; Bosukonda, D.; Sampath, K.T.; Vukicevic, S. Bone morphogenetic protein-7 reduces the severity of colon tissue damage and accelerates the healing of inflammatory bowel disease in rats. J. Cell Physiol. 2003, 196, 258–264. [Google Scholar] [CrossRef]
- Smith, P.J.; Levine, A.P.; Dunne, J.; Guilhamon, P.; Turmaine, M.; Sewell, G.W.; O’Shea, N.R.; Vega, R.; Paterson, J.C.; Oukrif, D.; et al. Mucosal transcriptomics implicates under expression of BRINP3 in the pathogenesis of ulcerative colitis. Inflamm. Bowel. Dis. 2014, 20, 1802–1812. [Google Scholar] [CrossRef]
- Han, B.K.; Olsen, N.J.; Bottaro, A. The CD27-CD70 pathway and pathogenesis of autoimmune disease. Semin. Arthritis Rheum. 2016, 45, 496–501. [Google Scholar] [CrossRef]
- Tsubata, T. Negative regulation of B cell responses and self-tolerance to RNA-related lupus self-antigen. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Lin, M.H.; Chu, S.H.; Tu, P.H.; Fang, C.C.; Yen, C.H.; Liang, P.I.; Huang, J.C.; Su, Y.C.; Sytwu, H.K.; et al. Role of glycine N-methyltransferase in the regulation of T-cell responses in experimental autoimmune encephalomyelitis. Mol. Med. 2015, 20, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Steen, V.M.; Nepal, C.; Ersland, K.M.; Holdhus, R.; Naevdal, M.; Ratvik, S.M.; Skrede, S.; Havik, B. Neuropsychological deficits in mice depleted of the schizophrenia susceptibility gene CSMD1. PLoS ONE 2013, 8, e79501. [Google Scholar] [CrossRef] [PubMed]
- Hanna, A.; Metge, B.J.; Bailey, S.K.; Chen, D.; Chandrashekar, D.S.; Varambally, S.; Samant, R.S.; Shevde, L.A. Inhibition of Hedgehog signaling reprograms the dysfunctional immune microenvironment in breast cancer. Oncoimmunology 2019, 8, 1548241. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, A.; Dreier, J.; Cheng, P.F.; Nageli, M.; Lehmann, H.; Felderer, L.; Frew, I.J.; Matsushita, S.; Levesque, M.P.; Dummer, R. Hedgehog pathway inhibitors promote adaptive immune responses in basal cell carcinoma. Clin. Cancer Res. 2015, 21, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Senger, S.; Sapone, A.; Fiorentino, M.R.; Mazzarella, G.; Lauwers, G.Y.; Fasano, A. Celiac disease histopathology recapitulates hedgehog downregulation, consistent with wound healing processes activation. PLoS ONE 2015, 10, e0144634. [Google Scholar] [CrossRef]
- Catassi, C.; Bai, J.C.; Bonaz, B.; Bouma, G.; Calabro, A.; Carroccio, A.; Castillejo, G.; Ciacci, C.; Cristofori, F.; Dolinsek, J.; et al. Non-Celiac Gluten sensitivity: The new frontier of gluten related disorders. Nutrients 2013, 5, 3839–3853. [Google Scholar] [CrossRef]
- Benjamini, Y.; Krieger, A.M.; Yekutieli, D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 2006, 93, 491–507. [Google Scholar] [CrossRef]

| Variables | CTRL | NCWS | p-Value a |
|---|---|---|---|
| Number of subjects | 7 | 12 | |
| Age (years), mean ± SD | 56.3 ± 22.8 | 46.5 ± 11.7 | 0.228 |
| Gender (M/F) | 1/6 | 0/12 | 0.779 b |
| Marsh classification, n (%) | - | ||
| 0 | 6 (50.0) | ||
| 1 | 6 (50.0) | ||
| 3a | - | ||
| 3b | - | ||
| 3c | - | ||
| TGA-IgA, (U/mL), mean ± SD | 1.8 ± 1.2 | 2.5 ± 1.6 | 0.331 |
| Hp positive, n (%) | 4 (57.1) | 2 (16.7) | 0.187 b |
| HLA-DQ, n (%) | 0.767 b | ||
| Negative | 5 (71.4) | 8 (66.7) | |
| DQ2 | 2 (28.6) | 4 (33.3) | |
| EMA positive | - | - | - |
| ESR (mm/h), mean ± SD | 12.6 ± 4.3 | 14.3 ± 4.7 | 0.444 |
| CRP (mg/L), mean ± SD | 0.4 ± 0.2 | 0.4 ± 0.3 | 0.998 |
| Calprotectin (µg/g), mean ± SD | 68.0 ± 35.5 | 85.3 ± 79.9 | 0.598 |
| Hb (g/dL), mean ± SD | 12.7 ± 0.8 | 13.0 ± 0.7 | 0.404 |
| Ferritin (ng/mL), mean ± SD | 24.2 ± 20.2 | 22.3 ± 16.7 | 0.827 |
| BMI (kg/m2), mean ± SD | 25.8 ± 4.0 | 27.6 ± 4.1 | 0.365 |
| Number of Transcripts Analysed | % | Absolute Mean Difference > 1 | % | Benjamini–Hochberg Correction p-Value < 0.05 | % | |
|---|---|---|---|---|---|---|
| Total | 53,217 | 100 | 429 | 100 | 300 | 100 |
| Protein-coding | 27,883 | 52.4 | 188 | 43.8 | 110 | 36.7 |
| Non-coding | 25,334 | 47.6 | 241 | 56.2 | 190 | 63.3 |
| Xloc or Loc | 4769 | 18.8 | 42 | 17.4 | 30 | 15. 8 |
| LNC | 9200 | 36.3 | 84 | 34.9 | 65 | 34.2 |
| Linc | 1419 | 5.6 | 17 | 7.1 | 17 | 8.9 |
| -AS | 1465 | 5.8 | 13 | 5.4 | 11 | 5.8 |
| SNORA- | 118 | 0.46 | 1 | 0.4 | 1 | 0.5 |
| SNORD- | 185 | 0.73 | 2 | 0.8 | 2 | 1.1 |
| -IT | 63 | 0.24 | 1 | 0.4 | 1 | 0.5 |
| No name | 8115 | 32.0 | 81 | 33.6 | 63 | 33.2 |
| Probe Name | Gene Symbol | Variable Importance | AUC (95% CI) |
|---|---|---|---|
| A_22_P00006238 | N/A | −2.299 | 0.990 (0.958–1.000) |
| A_21_P0011074 | XLOC_l2_003253 | 1.795 | 0.969 (0.901–1.000) |
| A_21_P0005811 | lnc-DUSP4-2 | 1.413 | 0.939 (0.838–1.000) |
| A_22_P00022828 | lnc-RLBP1-1 | −0.480 | 0.923 (0.806–1.000) |
| A_21_P0011522 | XLOC_l2_005692 | 0.506 | 0.908 (0.784–1.000) |
| A_23_P27207 | SfigureCGB1C1 | −0.028 | 0.908 (0.773–1.000) |
| A_33_P3389286 | SFN | 0.263 | 0.913 (0.779–1.000) |
| A_24_P36229 | DTNA | −0.105 | 0.893 (0.755–1.000) |
| A_21_P0001949 | LOC101927431 | 0.881 | 0.867 (0.643–1.000) |
| A_33_P3266564 | KRTAP5-6 | 0.644 | 0.857 (0.689–1.000) |
| A_21_P0008666 | lnc-SH3GL3-1 | −0.173 | 0.827 (0.568–1.000) |
| A_24_P263284 | CSMD2 | −0.325 | 0.770 (0.543–0.998) |
| A_24_P281395 | N/A | −0.006 | 0.755 (0.462–1.000) |
| A_21_P0007206 | lnc-CTD-2210P24.4.1-3 | 0.226 | 0.745 (0.478–1.000) |
| A_22_P00001191 | lnc-ANGPTL2-3 | −0.121 | 0.684 (0.391–0.977) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efthymakis, K.; Clemente, E.; Marchioni, M.; Di Nicola, M.; Neri, M.; Sallese, M. An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity. Int. J. Mol. Sci. 2020, 21, 1969. https://doi.org/10.3390/ijms21061969
Efthymakis K, Clemente E, Marchioni M, Di Nicola M, Neri M, Sallese M. An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity. International Journal of Molecular Sciences. 2020; 21(6):1969. https://doi.org/10.3390/ijms21061969
Chicago/Turabian StyleEfthymakis, Konstantinos, Emanuela Clemente, Michele Marchioni, Marta Di Nicola, Matteo Neri, and Michele Sallese. 2020. "An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity" International Journal of Molecular Sciences 21, no. 6: 1969. https://doi.org/10.3390/ijms21061969
APA StyleEfthymakis, K., Clemente, E., Marchioni, M., Di Nicola, M., Neri, M., & Sallese, M. (2020). An Exploratory Gene Expression Study of the Intestinal Mucosa of Patients with Non-Celiac Wheat Sensitivity. International Journal of Molecular Sciences, 21(6), 1969. https://doi.org/10.3390/ijms21061969






