Identification of Novel Genomic Regions and Superior Alleles Associated with Zn Accumulation in Wheat Using a Genome-Wide Association Analysis Method
Abstract
1. Introduction
2. Results
2.1. Variation in Zn Contents among the Wheat Lines
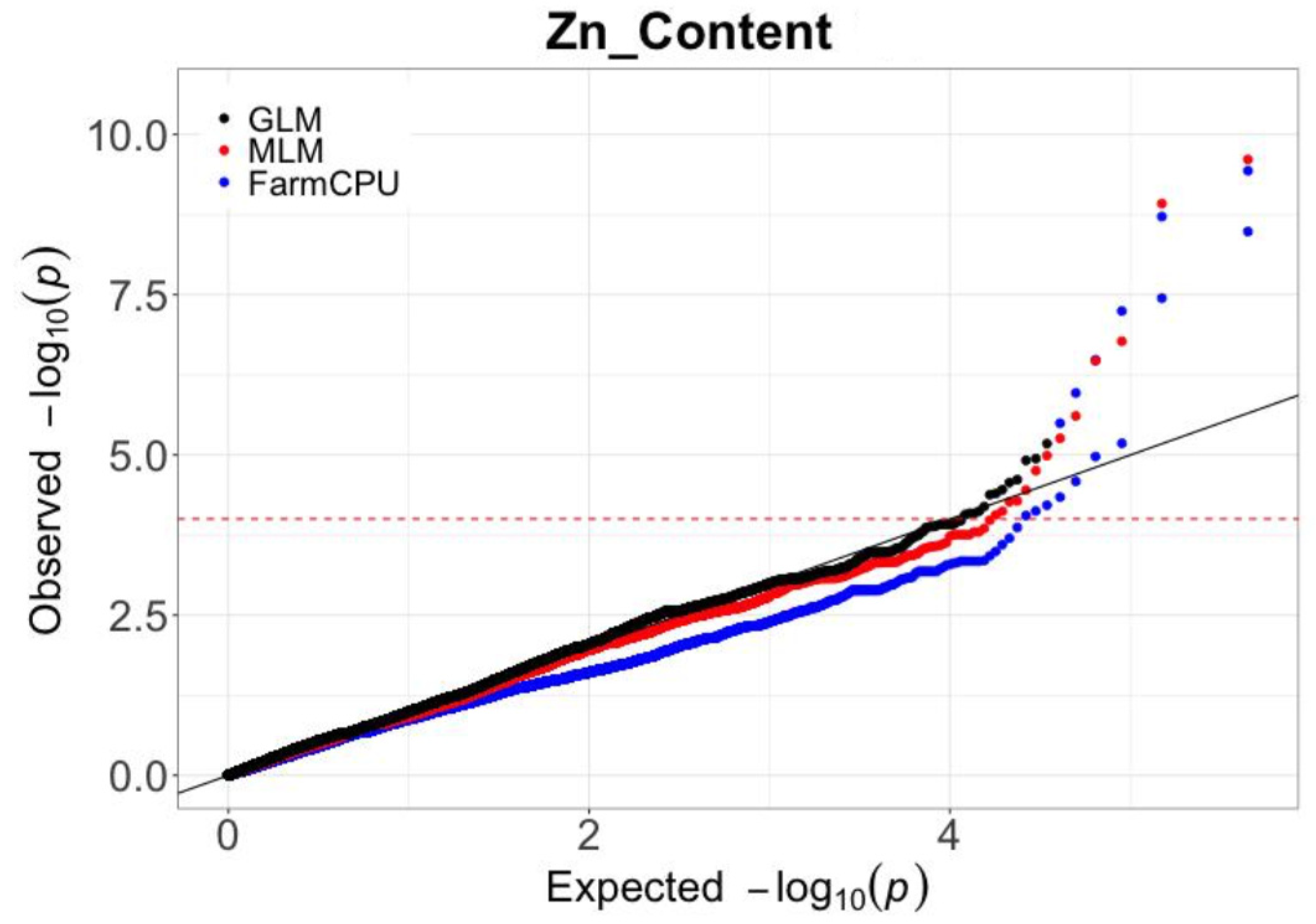
2.2. Screening of SNPs and Statistical Model Selection
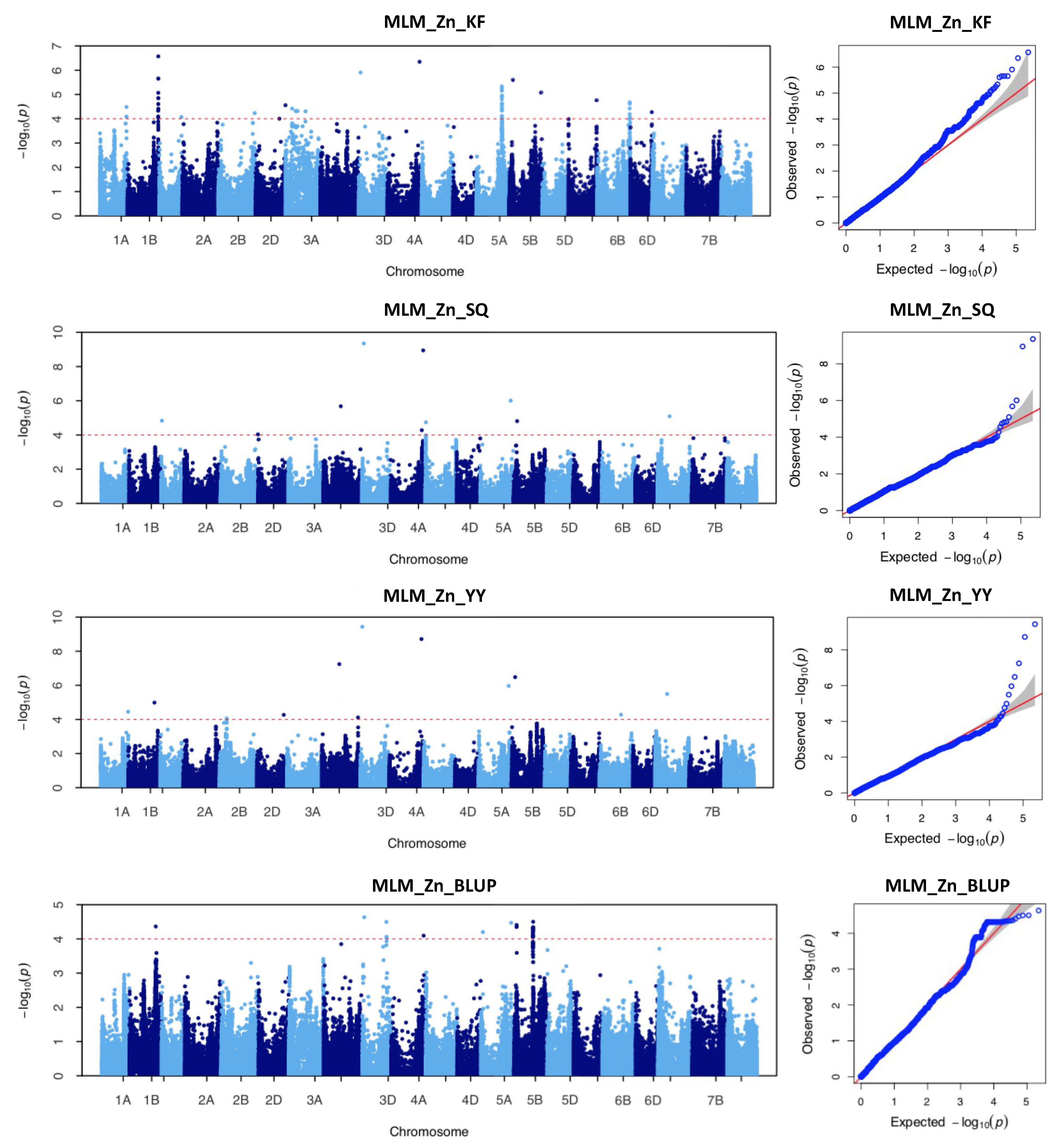
2.3. Genome-Wide Association Study with Individual SNP Markers
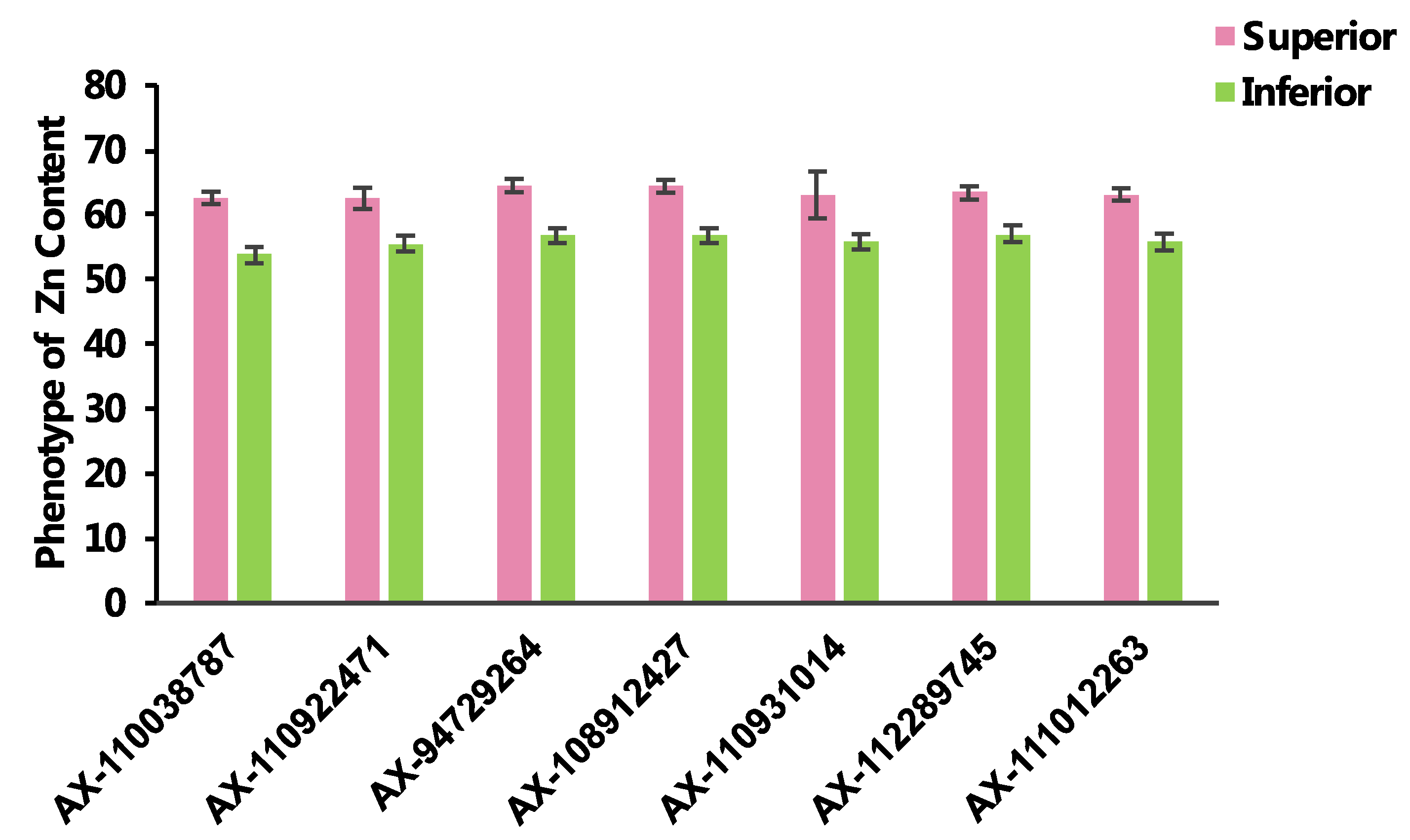
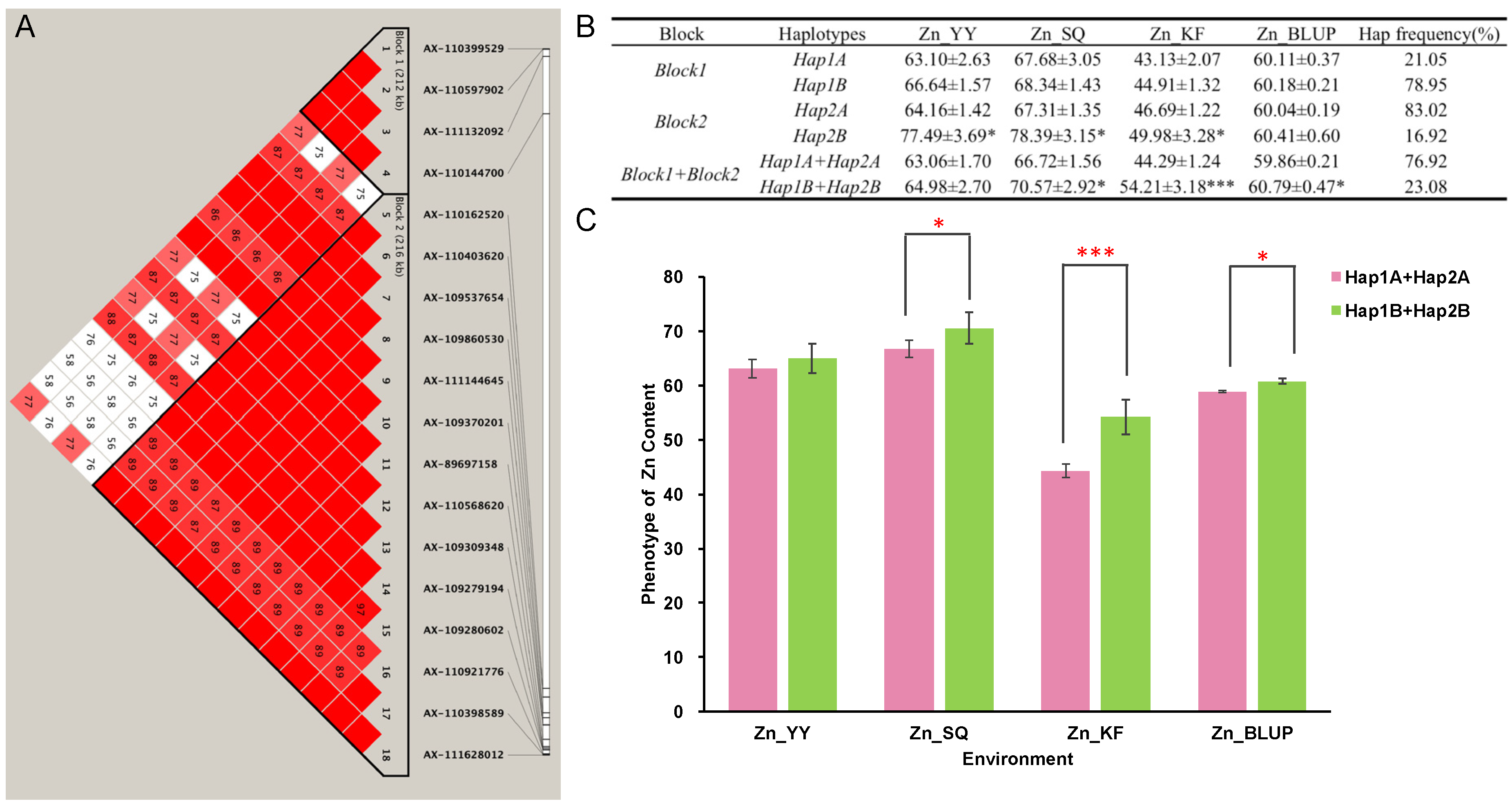
2.4. Haplotypes Associated with Zn Grain Content
2.5. Prediction of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of the Zn Concentration in Wheat Grains
4.3. Statistical Analysis
4.4. SNP Genotyping and Filtering
4.5. GWAS
4.6. Haplotype Block and Superior Allele Estimation
4.7. Candidate Gene Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| BLUP | Best linear unbiased predictors |
| FarmCPU | Fixed and random model Circulating Probability Unification |
| GLM | General linear model |
| GWAS | Genome-wide association studies |
| IWGSC | International Wheat Genome Sequence Consortium |
| MLM | Mixed linear model |
| PVE | Phenotypic variation explained |
| Quantile–quantile | |
| QTL | Quantitative trait loci |
| SNP | Single nucleotide polymorphism |
| Zn | Zinc |
References
- Bouis, H.E.; Hotz, C.; McClafferty, B.; Meenakshi, J.V.; Pfeiffer, W.H. Biofortification: A new tool to reduce micronutrient malnutrition. Food Nutr. Bull. 2011, 32, S31–S40. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.H.; Wuehler, S.E.; Peerson, J.M. The importance of zinc in human nutrition and estimation of the global prevalence of zinc deficiency. Food Nutr. Bull. 2001, 22, 113–125. [Google Scholar] [CrossRef]
- Black, R.E.; Victora, C.G.; Walker, S.P.; Bhutta, Z.A.; Christian, P.; Mercedes, M.D.; Ezzati, M.; Grantham-McGregor, S.; Katz, J.; Martorell, R. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 2013, 382, 427–451. [Google Scholar] [CrossRef]
- Hambidge, M. Human Zinc Deficiency. J. Nutr. 2000, 130, 1344S–1349S. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Bailey, L.B.; Pietrzik, K.; Shane, B.; Holzgreve, W. Micronutrients and women of reproductive potential: Required dietary intake and consequences of dietary deficienty or excess. Part II—Vitamin D, Vitamin A, Iron, Zinc, Iodine, Essential Fatty Acids. J. Matern. Fetal Neonatal Med. 2011, 24, 1–24. [Google Scholar] [CrossRef]
- Wessells, K.R.; Singh, G.M.; Brown, K.H. Estimating the global prevalence of inadequate zinc intake from national food balance sheets: Effects of methodological assumptions. PLoS ONE 2012, 7, e50565. [Google Scholar] [CrossRef]
- Gibson, R.S.; Bailey, K.B.; Gibbs, M.; Ferguson, E.L. A review of phytate, iron, zinc, and calcium concentrations in plant-based complementary foods used in low-income countries and implications for bioavailability. Food Nutr. Bull. 2010, 31, 134–146. [Google Scholar] [CrossRef]
- Chen, C.L.; Cui, Y.; Cui, M.; Zhou, W.J.; Wu, H.L.; Ling, H.Q. A FIT-binding protein is involved in modulating iron and zinc homeostasis in Arabidopsis. Plant Cell Environ. 2018, 41, 1698–1714. [Google Scholar] [CrossRef]
- Carvalho, S.M.P.; Vasconcelos, M.W. Producing more with less: Strategies and novel technologies for plant-based food biofortification. Food Res. Int. 2013, 54, 961–971. [Google Scholar] [CrossRef]
- Rai, V.; Sanagala, R.; Sinilal, B.; Yadav, S.; Sarkar, A.K.; Dantu, P.K.; Jain, A. Iron availability affects phosphate deficiency-mediated responses, and evidence of cross-yalk with auxin and zinc in Arabidopsis. Plant Cell Physiol. 2015, 56, 1107–1123. [Google Scholar] [CrossRef]
- Yagai, S.; Miyatake, T.; Shimono, Y.; Tamiaki, H. Supramolecular structure of self-assembled synthetic zinc-13(1)-oxo-chlorins possessing a primary, secondary or tertiary alcoholic 3(1)-hydroxyl group: Visible spectroscopic and molecular modeling studies. Photochem. Photobiol. 2010, 73, 153–163. [Google Scholar] [CrossRef]
- Mandell, J.G.; Barbas, C.F. Zinc Finger Tools: Custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006, 34, W516–W523. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Stodart, B.J.; Cavanagh, C.; Mackay, M.; Morell, M.; Milgate, A.; Martin, P. Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop Pasture Sci. 2010, 61, 222–229. [Google Scholar] [CrossRef]
- Shewry, P.R.; Lafiandra, D.; Bedo, Z. Improving the nutritional quality and health benefits of wheat. Qual. Assur. Saf. Crop. Foods 2012, 4, 136. [Google Scholar] [CrossRef]
- Cheema, S.A.; Rehman, H.U.; Kiran, A.; Bashir, K.; Wakeel, A. Plant Micronutrient Use Efficiency, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 261–278. [Google Scholar]
- Velu, G.; Ortiz-Monasterio, I.; Cakmak, I.; Hao, Y.; Singh, R.P. Biofortification strategies to increase grain zinc and iron concentrations in wheat. J. Cereal Sci. 2014, 59, 365–372. [Google Scholar] [CrossRef]
- Andersson, M.S.; Saltzman, A.; Virk, P.S.; Pfeiffer, W.H. Progress update: Crop development of biofortified staple food crops under HarvestPlus. Food Agric. Nutr. Dev. 2017, 17, 11905–11935. [Google Scholar] [CrossRef]
- Distelfeld, A.; Cakmak, I.; Peleg, Z.; Ozturk, L. Multiple QTL-effects of wheat GPC-B1 locus on grain protein and micronutrient concentrations. Physiol. Plant. 2007, 129, 635–643. [Google Scholar] [CrossRef]
- Velu, G.; Tutus, Y.; Gomez-Becerra, H.F.; Hao, Y.; Demir, L.; Kara, R.; Crespo-Herrera, L.A.; Orhan, S.; Yazici, A.; Singh, R.P. QTL mapping for grain zinc and iron concentrations and zinc efficiency in a tetraploid and hexaploid wheat mapping populations. Plant Soil 2017, 411, 81–99. [Google Scholar] [CrossRef]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat x wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Rawat, N.; Chhuneja, P.; Neelam, K.; Aggarwal, R.; Randhawa, G.S.; Dhaliwal, H.S.; Keller, B.; Singh, K. Mapping of quantitative trait loci for grain iron and zinc concentration in diploid A Genome wheat. J. Hered. 2009, 100, 771–776. [Google Scholar] [CrossRef]
- Xu, Y.; An, D.; Liu, D.; Zhang, A.; Xu, H.; Li, B. Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Field Crops Res. 2012, 138, 57–62. [Google Scholar] [CrossRef]
- Srinivasa, J.; Arun, B.; Mishra, V.K.; Singh, G.P.; Velu, G.; Babu, R.; Vasistha, N.K. Zinc and iron concentration QTL mapped in a Triticum spelta × T. aestivum cross. Theor. Appl. Genet. 2014, 127, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.F.; Velu, G.; Pena, R.J.; Singh, S.; Singh, R.P. Genetic loci associated with high grain zinc concentration and pleiotropic effect on kernel weight in wheat (Triticum aestivum L.). Mol. Breed. 2014, 34, 1893–1902. [Google Scholar] [CrossRef]
- Genc, Y.; Verbyla, A.P.; Torun, A.A.; Cakmak, I.; Willsmore, K.; Wallwork, H.; McDonald, G.K. Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil 2008, 314, 49–66. [Google Scholar] [CrossRef]
- Flintgarcia, S.A.; Thornsberry, J.M.; And, E.S.; Buckler, I.V. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Cardon, L.R. The complex interplay among factors that influence allelic association. Nat. Rev. Genet. 2004, 5, 89–100. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29–38. [Google Scholar] [CrossRef]
- Govindan, V.; Prakash, S.R.; Leonardo, C.-H.; Philomin, J.; Susanne, D.; Ravi, V.; James, S.; Singh, S.V.; Singh, M.G.; Kumar, M.V. Genetic dissection of grain zinc concentration in spring wheat for mainstreaming biofortification in CIMMYT wheat breeding. Sci. Rep. 2018, 8, 13526. [Google Scholar]
- Alomari, D.Z.; Eggert, K.; Von Wirén, N.; Alqudah, A.M.; Polley, A.; Plieske, J.; Ganal, M.W.; Pillen, K.; Röder, M.S. Identifying Candidate Genes for Enhancing Grain Zn Concentration in Wheat. Front. Plant Sci. 2018, 9, 1313. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Govindan, V.; Stangoulis, J.; Hao, Y.; Singh, R.P. QTL mapping of grain Zn and Fe concentrations in two hexaploid wheat RIL populations with ample transgressive segregation. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Gopalareddy, K.; Mahendru, S.A.; Swati, C.; Kumar, A.A.; Kumar, S.S.; Bihari, S.R.; Prakash, J.J.; Pratap, S.G.; Singh, S.I.; Aimin, Z. Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0174972. [Google Scholar]
- Shivay, Y.S.; Kumar, D.; Prasad, R. Effect of zinc-enriched urea on productivity, zinc uptake and efficiency of an aromatic rice–wheat cropping system. Nutr. Cycl. Agroecosyst. 2008, 81, 229–243. [Google Scholar] [CrossRef]
- Liu, J.; Huang, L.; Wang, C.; Liu, Y.; Yan, Z.; Wang, Z.; Xiang, L.; Zhong, X.; Gong, F.; Zheng, Y.; et al. Genome-wide association study reveals novel genomic regions associated with high grain protein content in wheat lines derived from wild emmer wheat. Front. Plant Sci. 2019, 10, 464. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Descalsota, G.I.L.; Swamy, B.P.M.; Zaw, H.; Inabangan-Asilo, M.A.; Amparado, A.; Mauleon, R.; Chadha-Mohanty, P.; Arocena, E.C.; Raghavan, C.; Leung, H.; et al. Genome-wide association mapping in a rice MAGIC Plus population detects QTLs and genes useful for biofortification. Front. Plant Sci. 2018, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Velu, G.; Singh, R.P.; Espino, J.H.; Pena, R.J.; Arun, B.; Mahendru-Singh, A.; Mujahid, M.Y.; Sohu, V.S.; Mavi, G.S.; Crossa, J. Performance of biofortified spring wheat genotypes in target environments for grain zinc and iron concentrations. Field Crops Res. 2012, 137, 261–267. [Google Scholar] [CrossRef]
- Mackay, T.F.C.; Stone, E.A.; Ayroles, J.F. The genetics of quantitative traits: Challenges and prospects. Nat. Rev. Genet. 2009, 10, 565–577. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Velu, G.; Singh, R.P. Quantitative trait loci mapping reveals pleiotropic effect for grain iron and zinc concentrations in wheat. Ann. Appl. Biol. 2016, 169, 27–35. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Sun-Wada, G.H.; Wada, Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 1166–1172. [Google Scholar] [CrossRef]
- Bickel-Sandkotter, S.; Wagner, V.; Schumann, D. ATP-synthesis in archaea: Structure-function relations of the halobacterial A-ATPase. Photosynth. Res. 1998, 57, 335–345. [Google Scholar] [CrossRef]
- Rosado, A.; Schapire, A.L.; Bressan, R.A.; Harfouche, A.L.; Hasegawa, P.M.; Valpuesta, V.; Botella, M.A. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 2006, 142, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Neumann, J. Effects of serine/threonine protein phosphatases on ion channels in excitable membranes. Physiol. Rev. 2000, 80, 173–210. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Carman, G.M. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 2008, 47, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Waters, B.M.; Cristobal, U.; Jorge, D.; Grusak, M.A. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 2009, 60, 4263–4274. [Google Scholar] [CrossRef] [PubMed]
- Mortel, V.D.; Schat, H.; Moerland, P.D.; Themaat, E.; Ent, S.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2007, 31, 301–324. [Google Scholar] [CrossRef]
- Fan, M.S.; Zhao, F.J.; Susan, F.T.; Poulton, P.R.; Dunham, S.J.; Steve, P.M. Evidence of decreasing mineral density in wheat grain over the last 160 years. J. Trace Elem. Med. Biol. 2008, 22, 315–324. [Google Scholar] [CrossRef]
- Garvin, D.F.; Welch, R.M.; Finley, J.W. Historical shifts in the seed mineral micronutrient concentration of US hard red winter wheat germplasm. J. Sci. Food Agric. 2006, 86, 2213–2220. [Google Scholar] [CrossRef]
- Borrill, P.; Connorton, J.; Balk, J.; Miller, T.; Sanders, D.; Uauy, C. Biofortification of wheat grain with iron and zinc: Integrating novel genomic resources and knowledge from model crops. Front. Plant Sci. 2014, 5, P21. [Google Scholar] [CrossRef]
- Gong, Z.; Lei, Z.; Chen, Z.; Gao, Y.; Zeng, S.; Zhang, G.; Xiao, D.; Li, S. Chinese soil taxonomy. Bull. Chin. Acad. Sci. 2007, 27, 36–39. [Google Scholar]
- Zhang, L.; Li, L.; Pan, G. Variation of Cd, Zn and Se contents of polished rice and the potential health risk for subsistence-diet farmers from typical areas of south China. Environ. Sci. 2009, 30, 2792–2797. [Google Scholar]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhang, N.; Fan, X.; Zhang, W.; Zhao, C.; Yang, L.; Pan, R.; Chen, M.; Han, J.; Zhao, X.; et al. Utilization of a Wheat660K SNP array-derived high-density genetic map for high-resolution mapping of a major QTL for kernel number. Sci. Rep. 2017, 7, 3788. [Google Scholar] [CrossRef] [PubMed]
- Panagiotou, O.A.; Evangelou, E.; Ioannidis, J.P.A. Genome-wide significant associations for variants with minor allele frequency of 5% or less--an overview: A HuGE review. Am. J. Epidemiol. 2010, 172, 869–889. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.S.; Regnaut, S.J.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Vanraden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef]
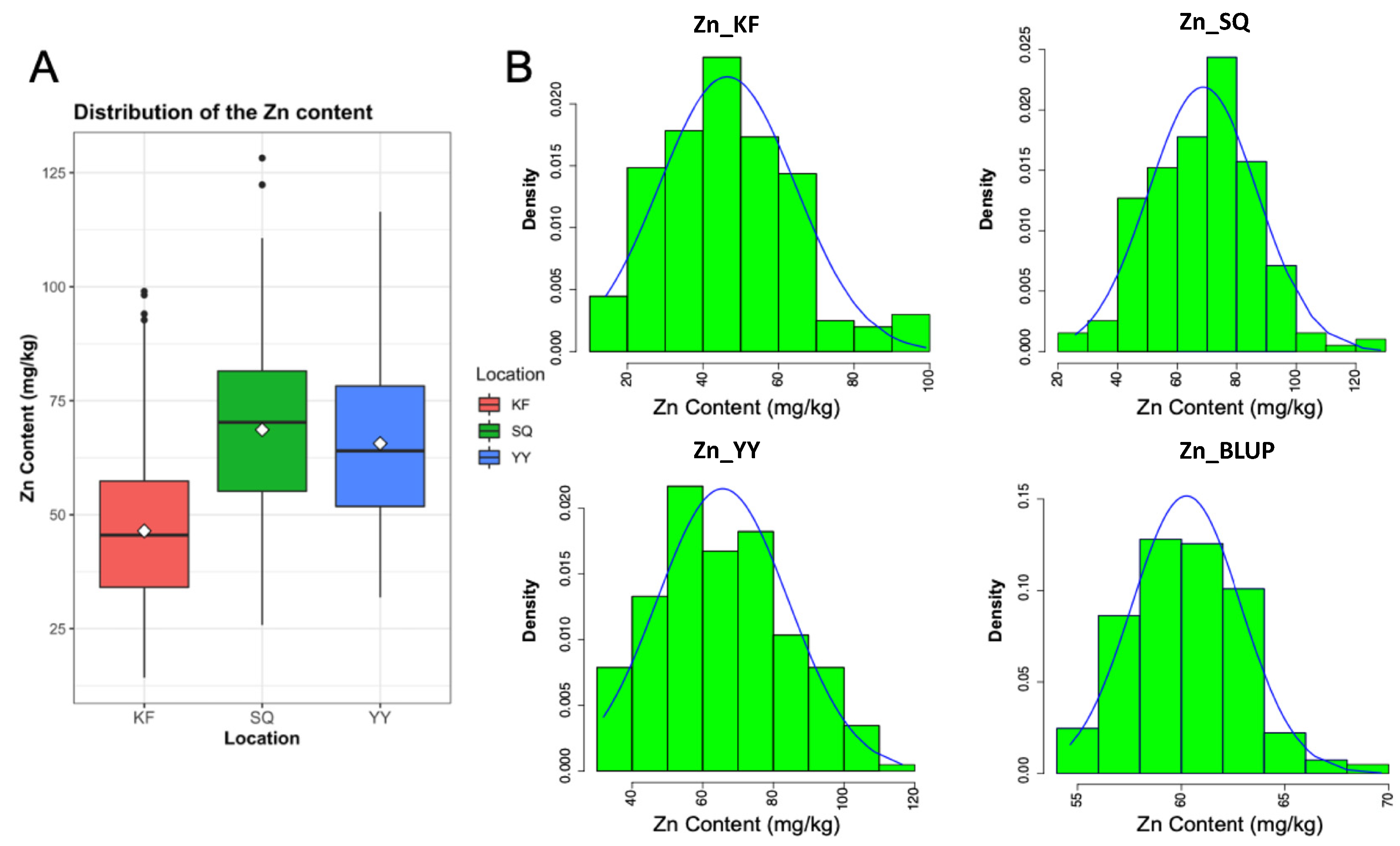
| Location | Trait | Mean ± SD 1 (mg/kg) | Range (mg/kg) | Kurt 2 | Skew 3 |
|---|---|---|---|---|---|
| Yuanyang (YY) | Zn content | 65.63 ± 18.57 | 31.87–116.47 | −0.61 | 0.31 |
| Kaifeng (KF) | Zn content | 46.45 ± 17.98 | 14.24–99.00 | 0.14 | 0.53 |
| Shangqiu (SQ) | Zn content | 68.63 ± 18.23 | 25.83–128.22 | 0.01 | 0.08 |
| BLUP | Zn content | 60.24 ± 2.63 | 54.65–69.65 | 0.20 | 0.35 |
| ID 1 | Chromosome | Interval Range (bp) | No. of SNPs | Location | Peak SNP 2 | Position (bp) 3 | p Value 4 | R2 (%) 5 |
|---|---|---|---|---|---|---|---|---|
| 1 | 1A | 574,477,246–574,479,186 | 3 | KF | AX-110606195 | 574,477,470 | 3.27 × 10−5 | 10.26 |
| 2 | 1A | 592,315,138 | 1 | YY | AX-108995328 | 592,315,138 | 3.55 × 10−5 | 12.27 |
| 3 | 1B | 564,909,314 | 1 | YY | AX-110038787 | 564,909,314 | 1.03 × 10−5 | 13.53 |
| BLUP | AX-110038787 | 564,909,314 | 4.33 × 10−5 | 13.78 | ||||
| 4 | 1B | 665,798,565–668,100,871 | 18 | KF | AX-111144645 | 668,002,032 | 2.69 × 10−7 | 15.44 |
| 5 | 1D | 16,132,987 | 1 | SQ | AX-110529533 | 16,132,987 | 1.46 × 10−5 | 12.64 |
| 6 | 1D | 478,184,386 | 1 | KF | AX-110828223 | 478,184,386 | 8.47 × 10−5 | 9.27 |
| 7 | 2B | 154,930,484 | 1 | YY | AX-110620516 | 154,930,484 | 8.62 × 10−5 | 11.37 |
| 8 | 2B | 787,099,530 | 1 | KF | AX-109490599 | 787,099,530 | 5.82 × 10−5 | 9.66 |
| 9 | 2D | 1,638,170 | 1 | SQ | AX-94598102 | 1,638,170 | 9.19 × 10−5 | 10.70 |
| 10 | 2D | 519,133,490–650,654,168 | 3 | YY | AX-94583825 | 582,025,967 | 5.40 × 10−5 | 11.84 |
| KF | AX-94466886 | 650,654,168 | 2.75 × 10−5 | 10.44 | ||||
| 11 | 3A | 219,103,987–260,492,627 | 5 | KF | AX-108914831 | 235,844,032 | 4.80 × 10−5 | 9.86 |
| 12 | 3B | 376,625,452 | 1 | SQ | AX-110922471 | 376,625,452 | 2.09 × 10−6 | 14.75 |
| YY | AX-110922471 | 376,625,452 | 5.69 × 10−8 | 19.07 | ||||
| 13 | 3B | 779,542,533 | 1 | YY | AX-94567805 | 779,542,533 | 7.58 × 10−5 | 11.50 |
| 14 | 3D | 40,526,440 | 1 | KF | AX-94729264 | 40,526,440 | 1.24 × 10−6 | 13.76 |
| SQ | AX-94729264 | 40,526,440 | 4.45 × 10−10 | 24.55 | ||||
| YY | AX-94729264 | 40,526,440 | 3.69 × 10−10 | 24.77 | ||||
| BLUP | AX-94729264 | 40,526,440 | 2.32 × 10−5 | 14.40 | ||||
| 15 | 3D | 515,115,709–519,527,578 | 3 | BLUP | AX-111858412 | 515,115,709 | 3.18 × 10−5 | 14.08 |
| 16 | 4A | 669,454,046–699,571,654 | 2 | SQ | AX-108851891 | 669,454,046 | 5.27 × 10−5 | 11.28 |
| KF | AX-108912427 | 699,571,654 | 4.47 × 10−7 | 14.87 | ||||
| SQ | AX-108912427 | 699,571,654 | 1.14 × 10−9 | 23.42 | ||||
| YY | AX-108912427 | 699,571,654 | 1.92 × 10−9 | 22.87 | ||||
| BLUP | AX-108912427 | 699,571,654 | 8.02 × 10−5 | 13.17 | ||||
| 17 | 4B | 13,996,819 | 1 | SQ | AX-89748062 | 13,996,819 | 1.81 × 10−5 | 12.42 |
| 18 | 5A | 42,281,607 | 1 | BLUP | AX-94932868 | 42,281,607 | 6.31 × 10−5 | 13.41 |
| 19 | 5A | 549,409,584–552,208,450 | 23 | KF | AX-111463331 | 549,576,304 | 4.61 × 10−6 | 12.33 |
| 20 | 5A | 650,240,330 | 1 | SQ | AX-110931014 | 650,240,330 | 9.82 × 10−7 | 15.59 |
| YY | AX-110931014 | 650,240,330 | 1.08 × 10−6 | 15.89 | ||||
| BLUP | AX-110931014 | 650,240,330 | 3.39 × 10−5 | 14.02 | ||||
| 21 | 5B | 57,158,679–57,493,343 | 3 | BLUP | AX-110398218 | 57,493,343 | 3.92 × 10−5 | 13.87 |
| 22 | 5B | 78,708,064 | 1 | KF | AX-112289745 | 78,708,064 | 2.47 × 10−6 | 13.00 |
| SQ | AX-112289745 | 78,708,064 | 1.55 × 10−5 | 12.58 | ||||
| YY | AX-112289745 | 78,708,064 | 3.29 × 10−7 | 17.16 | ||||
| 23 | 5B | 407,053,365–412,175,187 | 41 | BLUP | AX-86176241 | 411,929,890 | 3.15 × 10−5 | 14.09 |
| 24 | 5B | 693,033,900 | 1 | KF | AX-110443373 | 693,033,900 | 8.15 × 10−6 | 11.72 |
| 25 | 6A | 613,482,310 | 1 | KF | AX-94961930 | 613,482,310 | 1.72 × 10−5 | 10.93 |
| 26 | 6B | 462,555,585 | 1 | YY | AX-111084964 | 462,555,585 | 5.22 × 10−5 | 11.88 |
| 27 | 6B | 708,670,301 | 5 | KF | AX-109538092 | 708,670,301 | 2.09 × 10−5 | 10.73 |
| 28 | 6D | 464,120,129 | 1 | KF | AX-110536000 | 464,120,129 | 5.33 × 10−5 | 9.75 |
| 29 | 7A | 261,687,749 | 1 | SQ | AX-111012263 | 261,687,749 | 8.04 × 10−6 | 13.28 |
| YY | AX-111012263 | 261,687,749 | 3.21 × 10−6 | 14.74 |
| ID 1 | Chromosome | Identified Loci in Current Study | Position (bp) 2 | Near Locus Previously Reported in the Same Chromosome | Candidate Genes (Closest/Nearby) | Annotation |
|---|---|---|---|---|---|---|
| 1 | 1B | AX-110038787 | 564,909,314 | QGZn.sar_1BTSK [19] | TraesCS1B02G337400 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| 2 | 3B | AX-110922471 | 376,625,452 | QGZn.cimmyt-3B_1P2 [31] | TraesCS3B02G252000 | CTP synthase |
| 3 | 3D | AX-94729264 | 40,526,440 | -- 3 | TraesCS3D02G078500 | NAC domain-containing protein |
| 4 | 4A | AX-108912427 | 669,454,046 | QGZn.iari-4A [32] | TraesCS4A02G428900 | V-type proton ATPase |
| gwm397-gwm269 [19] | ||||||
| 5 | 5A | AX-110931014 | 650,240,330 | Xgwm291-Xgwm410 [33] | TraesCS5A02G475200 | Heavy metal transport/detoxification superfamily protein |
| Xbarc223.1-Xswes157 [22] | ||||||
| 6 | 5B | AX-112289745 | 78,708,064 | QGZn.cimmyt-5B_P2 [31] | TraesCS5B02G069200 | Serine/threonine-protein kinase/Kinase family protein |
| 7 | 7A | AX-111012263 | 261,687,749 | GZn.pau-7A [21] | TraesCS7A02G263700 | Basic helix-loop-helix transcription factor |
| SNP_id | Chr. | Allele Type | Phenotype Value | Allele Number | Allele Percentage (%) | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Superior | Inferior | Superior | Inferior | Superior | Inferior | Superior | Inferior | Zn_YY | Zn_SQ | Zn_KF | Zn_BLUP | ||
| AX-110038787 | 1B | CC | GC | 60.79 | 58.78 | 148 | 55 | 72.91 | 27.09 | 2.90 × 10−7 | 9.36 × 10−4 | 4.74 × 10−1 | 7.42 × 10−7 |
| AX-110922471 | 3B | AA | GG | 60.77 | 59.16 | 53 | 86 | 38.13 | 61.87 | 2.93 × 10−6 | 7.28 × 10−4 | 2.87 × 10−1 | 6.39 × 10−4 |
| AX-94729264 | 3D | CT | CC | 61.27 | 59.43 | 90 | 113 | 44.33 | 55.67 | 7.11 × 10−14 | 4.09 × 10−14 | 6.03 × 10−7 | 3.31 × 10−7 |
| AX-108912427 | 4A | AG | GG | 61.24 | 59.43 | 90 | 111 | 44.78 | 55.22 | 1.33 × 10−13 | 1.33 × 10−13 | 7.85 × 10−7 | 6.32 × 10−7 |
| AX-110931014 | 5A | AA | CC | 60.90 | 59.20 | 17 | 94 | 15.32 | 84.68 | 1.68 × 10−2 | 5.89 × 10−2 | 4.80 × 10−1 | 1.81 × 10−2 |
| AX-112289745 | 5B | AG | GG | 60.98 | 59.49 | 92 | 95 | 49.20 | 50.80 | 9.72 × 10−11 | 9.95 × 10−8 | 1.35 × 10−5 | 1.22 × 10−4 |
| AX-111012263 | 7A | GG | AG | 60.96 | 59.22 | 94 | 86 | 52.22 | 47.78 | 2.23 × 10−10 | 4.53 × 10−10 | 5.08 × 10−5 | 2.71 × 10−6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Shi, X.; Zhao, G.; Qin, M.; Ibba, M.I.; Wang, Y.; Li, W.; Yang, P.; Wu, Z.; Lei, Z.; et al. Identification of Novel Genomic Regions and Superior Alleles Associated with Zn Accumulation in Wheat Using a Genome-Wide Association Analysis Method. Int. J. Mol. Sci. 2020, 21, 1928. https://doi.org/10.3390/ijms21061928
Zhou Z, Shi X, Zhao G, Qin M, Ibba MI, Wang Y, Li W, Yang P, Wu Z, Lei Z, et al. Identification of Novel Genomic Regions and Superior Alleles Associated with Zn Accumulation in Wheat Using a Genome-Wide Association Analysis Method. International Journal of Molecular Sciences. 2020; 21(6):1928. https://doi.org/10.3390/ijms21061928
Chicago/Turabian StyleZhou, Zhengfu, Xia Shi, Ganqing Zhao, Maomao Qin, Maria Itria Ibba, Yahuan Wang, Wenxu Li, Pan Yang, Zhengqing Wu, Zhensheng Lei, and et al. 2020. "Identification of Novel Genomic Regions and Superior Alleles Associated with Zn Accumulation in Wheat Using a Genome-Wide Association Analysis Method" International Journal of Molecular Sciences 21, no. 6: 1928. https://doi.org/10.3390/ijms21061928
APA StyleZhou, Z., Shi, X., Zhao, G., Qin, M., Ibba, M. I., Wang, Y., Li, W., Yang, P., Wu, Z., Lei, Z., & Wang, J. (2020). Identification of Novel Genomic Regions and Superior Alleles Associated with Zn Accumulation in Wheat Using a Genome-Wide Association Analysis Method. International Journal of Molecular Sciences, 21(6), 1928. https://doi.org/10.3390/ijms21061928





