Gibberellin Signaling Repressor LlDELLA1 Controls the Flower and Pod Development of Yellow Lupine (Lupinus luteus L.)
Abstract
1. Introduction
2. Results
2.1. Cloning and Analysis of Full-Length cDNA Encoding L. luteus DELLA1 Protein
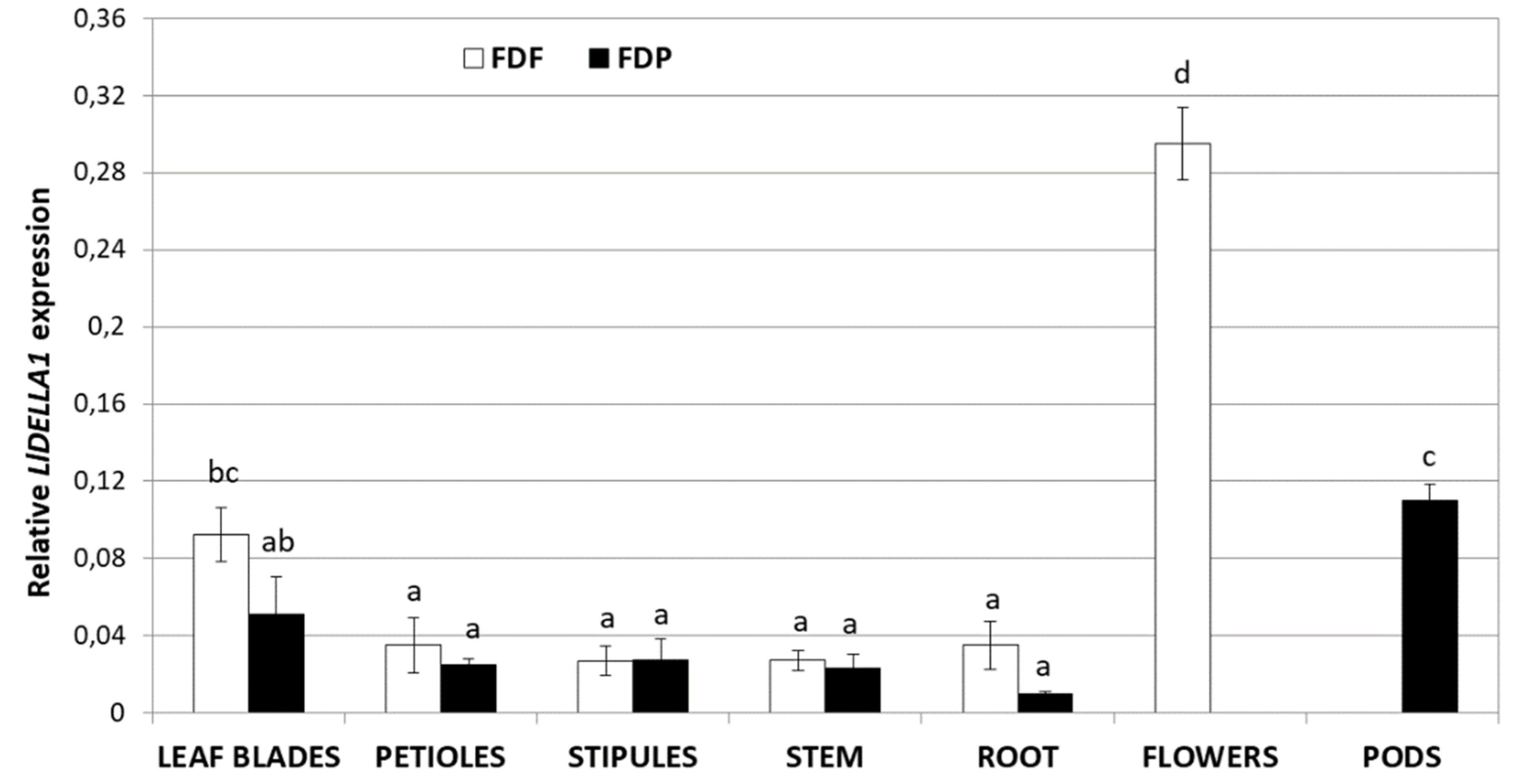
2.2. General Expression Profile of LlDELLA1 in Various Organs of L. luteus
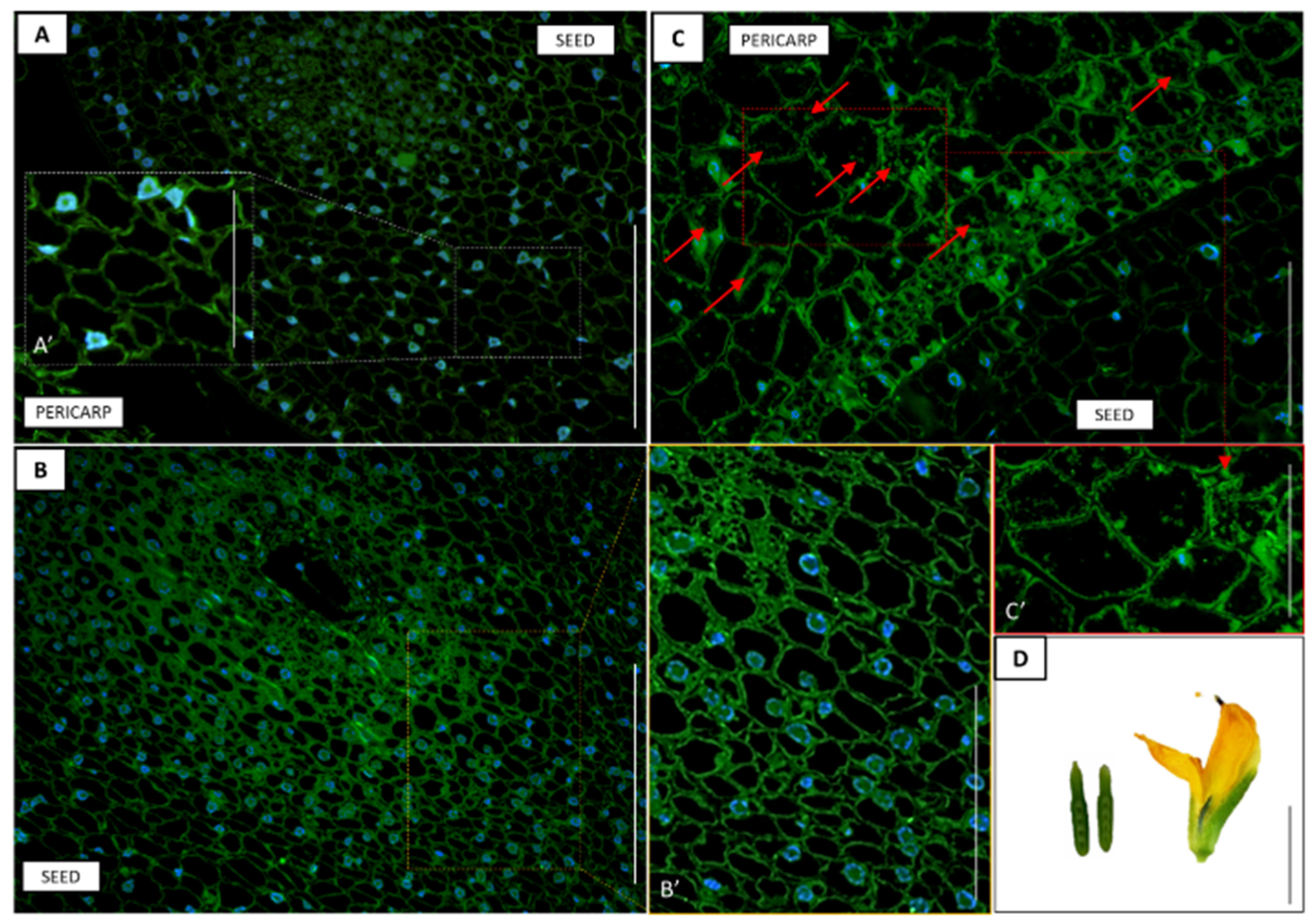
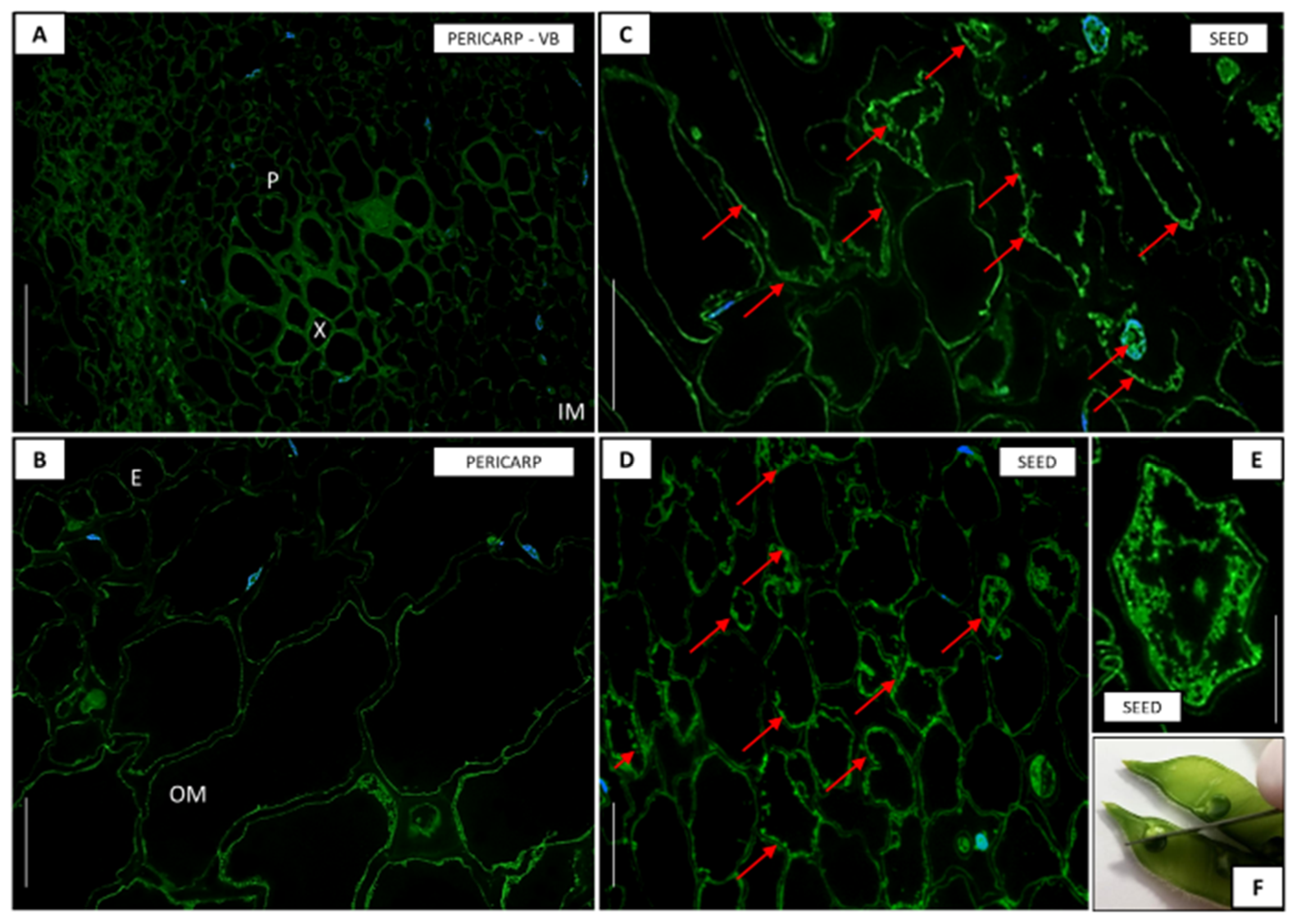
2.3. Histological Analyses
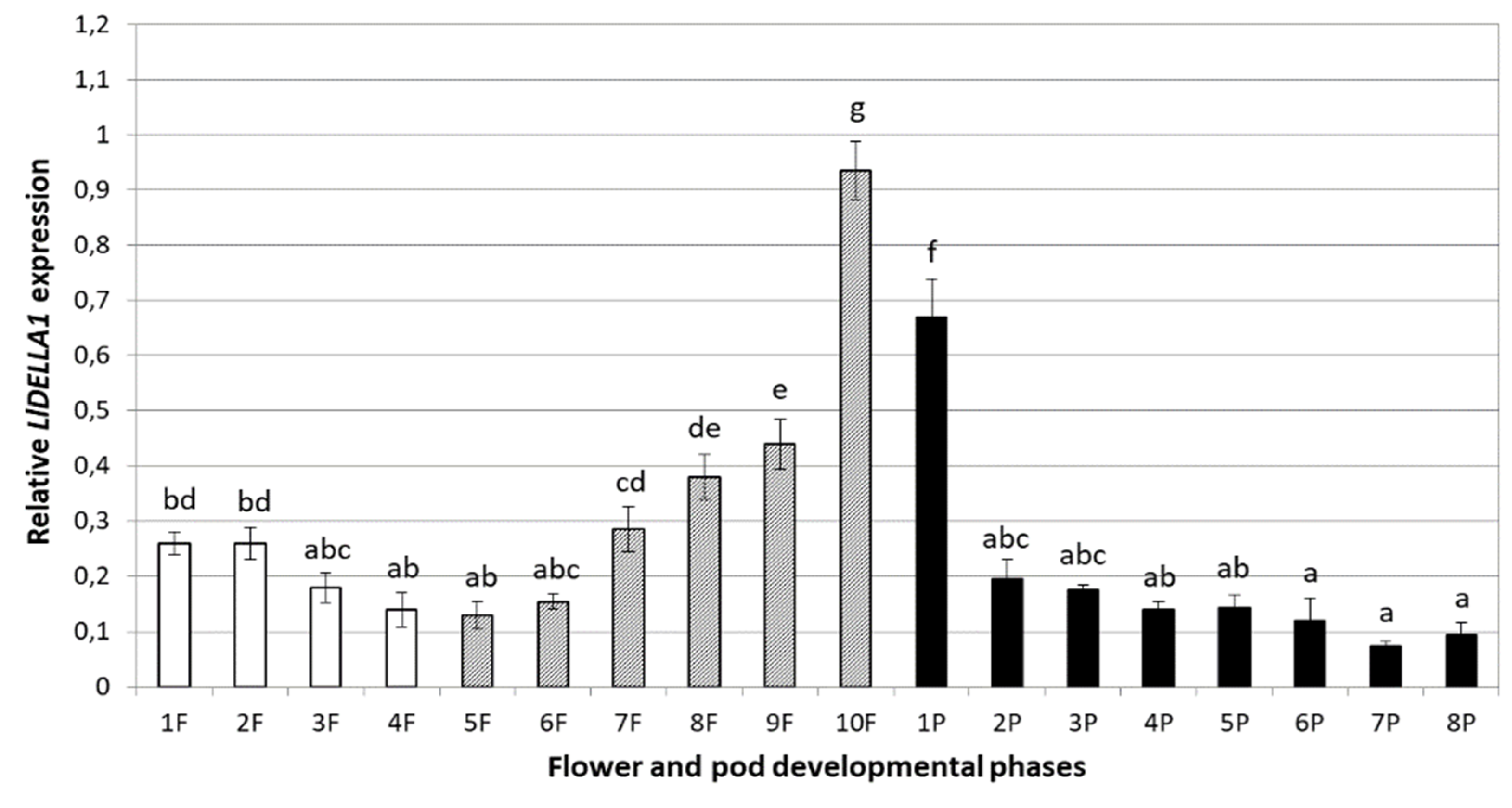
2.4. Expression Profile of LlDELLA1 during Generative Organ Development
2.5. Expression Profile of LlDELLA1 in Response to Phytohormone and Inhibitor Treatment
2.6. Immunolocalization of GA3
3. Discussion
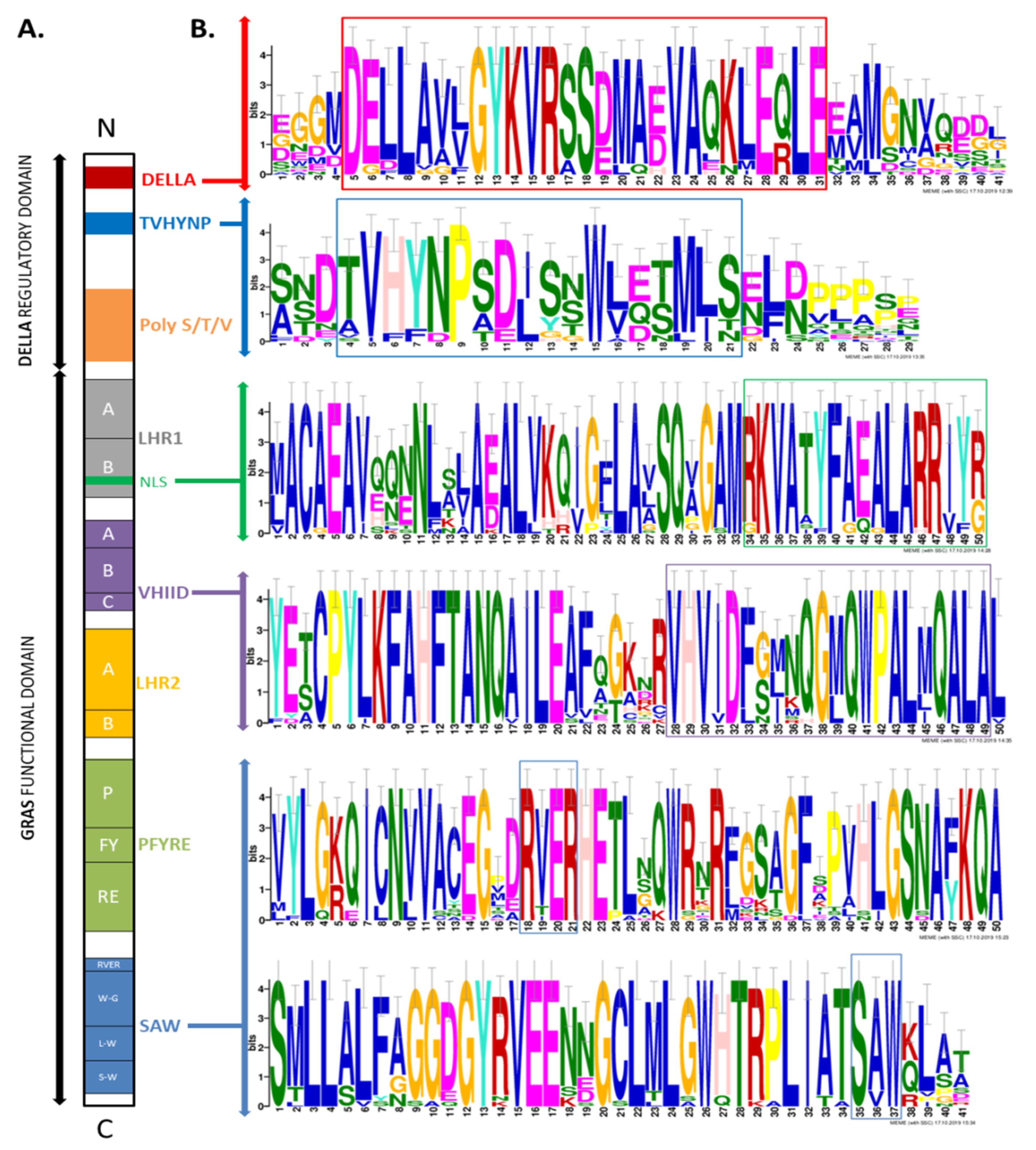
3.1. LlDELLA1 Encodes Protein Containing Characteristic and Conserved Motifs and Domains
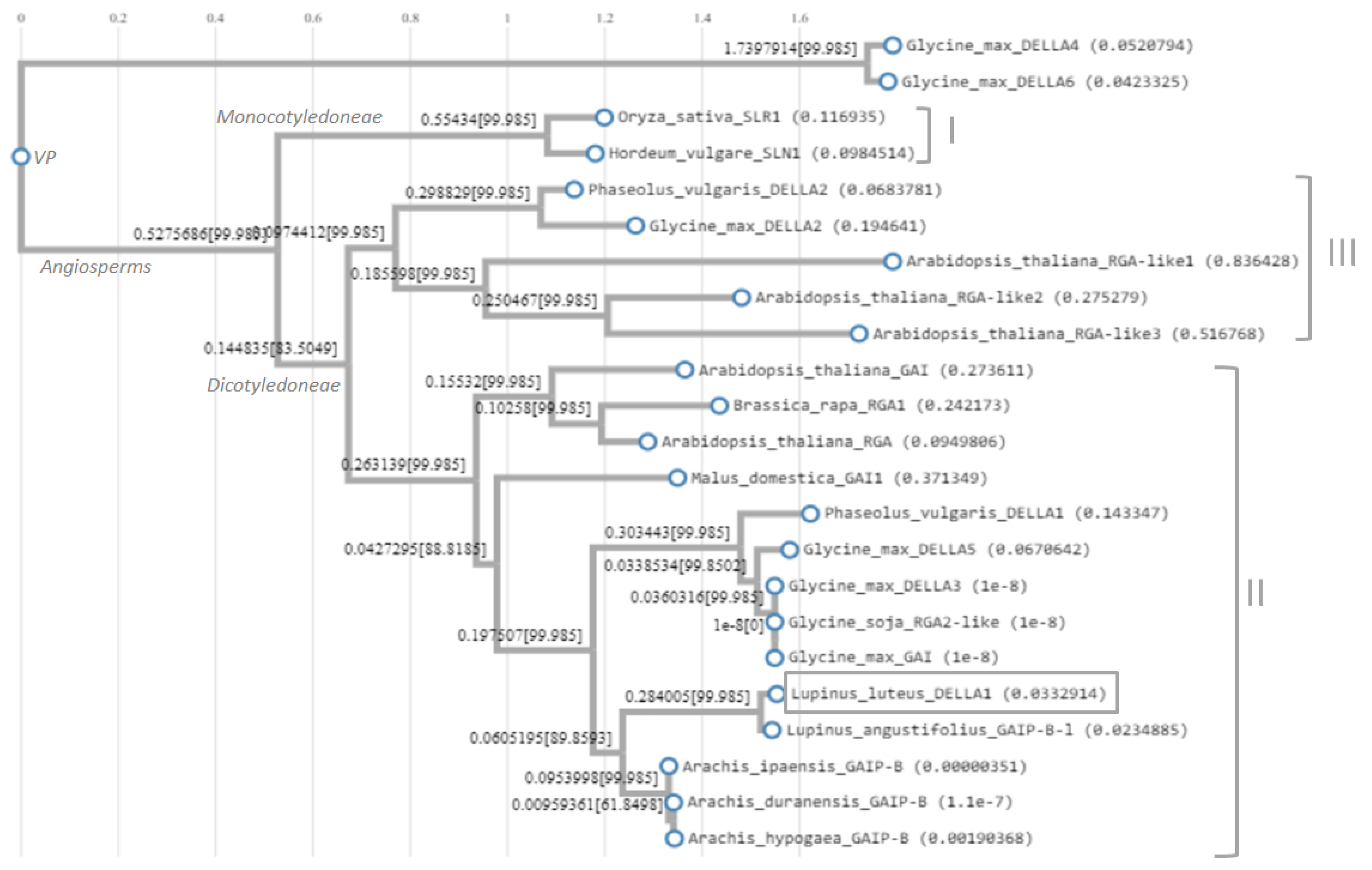
3.2. Nuclear Protein LlDELLA1 is Clustered within Group II with GAI and RGA
3.3. Generative Organs Contain the Highest Level of LlDELLA1 Transcripts
3.4. Self-Pollination Occurs when Flowers are Closed Followed by Pod Setting and Development
3.5. Fluctuating LlDELLA1 Expression Profile Ensures Proper Flower and Pod Development
3.6. In Yellow Lupine, LlDELLA1 is Considered as a GA Signaling Pathway Repressor
3.7. Accumulation of GA3 in Pericarp and Seed Depending on the Developmental Phase of Pods
4. Materials and Methods
4.1. Plant Material, Growing Conditions, and Phytohormone/Inhibitor Treatments
4.2. Molecular Cloning of LlDELLA1 cDNA
4.3. Bioinformatic Analyses
4.4. Expression Analysis
4.5. Histological Studies and GA3 Immunolocalization
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAP | 6-benzylaminopurine |
| BSA | Bovine serum albumin |
| C | Connective |
| CCC | Chlorocholine chloride |
| D8/9 | DWARF8/9 |
| E | Epidermis |
| EDAC | N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride |
| En | Endosperm |
| F | Flower |
| FBPs | F-box proteins |
| FDF | Fully developed flowers |
| FDP | Fully developed pods |
| GA3 | Gibberellic acid |
| GAs | Gibberellins |
| GAI | GA Insensitive |
| GID1 | Gibberellin Insensitive Dwarf1 |
| GRAS | GAI RGA and SCR |
| IAA | Indole-3-acetic acid |
| IM | Inner mesocarp |
| LHR | Leucine heptad repeats |
| MeJA | Methyl jasmonate |
| NLS | Nuclear localization sequence/signal |
| OM | Outer mesocarp |
| P | Pod |
| PBS | Phosphate-buffered saline buffer |
| PDB | Protein Data Bank |
| Pe | Pericarp |
| poly S/T/V | Polymeric Ser/Thr/Val motifs |
| RGA | Repressor of GA1-3 |
| RGL | RGA-like |
| RHT-1 | Reduced Height-1 |
| SCF | SKP Cullin F-box complex |
| SCL | Scarecrow-like protein |
| SHR | Short-root |
| SCR | Scarecrow |
| SLN1 | Slender1 |
| SLR1 | Slender Rice1 |
| SLY | Sleepy |
| SNE | Sneezy |
| SRA | Sequence Read Archive |
| TFs | Transcriptional factors |
| TRs | Transcriptional regulators |
| VB | Vascular bundle |
| UPL | Universal Probe Library |
| X | Xylem |
References
- Prusiński, J. The legumes in European Union. ZPPNR 2010, 550, 11–19. [Google Scholar]
- Duranti, M.; Consonni, A.; Magni, C.; Sessa, F.; Scarafoni, A. The major proteins of lupin seed: Characterization and molecular properties for use as functional and nutraceutical ingredients. Food Sci. Tech. 2008, 19, 624. [Google Scholar] [CrossRef]
- Księżak, J.; Bojarszczuk, J. The productivity of selected species and cultivars of legumes grown for seeds in organic production system. 2019. Available online: https://cdn.intechopen.com/pdfs/64941.pdf (accessed on 3 March 2020).
- The biology of Lupinus L. (lupin or lupine). 2013. Available online: http://www.ogtr.gov.au/internet/ogtr/publishing.nsf/Content/biologylupin2013-toc/$FILE/biologylupin2013-2.pdf. (accessed on 3 March 2020).
- Wilmowicz, E.; Frankowski, K.; Glazińska, P.; Sidłowska, M.; Marciniak, K.; Kopcewicz, J. The role of gibberellins in the regulation of flowering in plants. Kosmos 2011, 60, 129–140. [Google Scholar]
- Marciniak, K.; Świeżawska, B.; Kęsy, J.; Tretyn, A.; Kopcewicz, J. Gibberellins – perception and signal transduction in plants. Post. Biol. Kom. 2012, 39, 25–48. [Google Scholar]
- Davière, J.-M.; Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant. 2016, 9, 10–20. [Google Scholar] [CrossRef]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Hirano, K.; Asano, K.; Tsuji, H.; Kawamura, M.; Mori, H.; Kitano, H.; Ueguchi-Tanaka, H. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 2010, 22, 2680–2696. [Google Scholar] [CrossRef]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S.; Hongyu, X.; Ashikari, M.; Kitano, H.; Yamaguchi, I.; et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.; Maier, A.; Schwechheimer, C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Marti, C.; Orzáez, D.; Ellul, P.; Moreno, V.; Carbonell, J.; Granell, A. Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant. J. 2007, 52, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.Y.; Yang, Y. Characterization of grape Gibberellin Insensitive1 mutant alleles in transgenic Arabidopsis. Transgenic Res. 2012, 21, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef]
- Chandler, P.M.; Marion-Poll, A.; Ellis, M.; Gubler, F. Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 2002, 129, 181–190. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Winkler, R.G.; Freeling, M. Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf8 and Dwarf9. Planta 1994, 193, 341–348. [Google Scholar] [CrossRef]
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Mak, P.Y.A.; Martínez, E.C.; Sun, T.-P. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 1997, 146, 1087–1099. [Google Scholar]
- Wen, C.-K.; Chang, C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002, 16, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Wild, M.; Davière, J.-M.; Cheminant, S.; Regnault, T.; Baumberger, N.; Heintz, D.; Baltz, R.; Genschik, P.; Achard, P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012, 24, 3307–3319. [Google Scholar] [CrossRef] [PubMed]
- Dill, A.; Sun, T. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 2001, 159, 777–785. [Google Scholar] [PubMed]
- King, K.E.; Moritz, T.; Harberd, N.P. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 2001, 159, 767–776. [Google Scholar] [PubMed]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1164. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Weston, D.E.; Elliott, R.C.; Lester, D.R.; Rameau, C.; Reid, J.B.; Murfet, I.C.; Ross, J.J. The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol. 2008, 147, 199–205. [Google Scholar] [CrossRef]
- Floss, D.S.; Levy, J.G.; Lévesque-Tremblay, V.; Pumplin, N.; Harrison, M.J. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Pnasci. Usa. 2013, 110, E5025–E5034. [Google Scholar] [CrossRef]
- Chen, J.; Cheng, T.; Wang, P.; Tian, L.; Wang, G.; Luo, Y.; Wang, J.; Yang, L.; Shi, J. Genome-wide bioinformatics analysis of DELLA-family proteins from plants. Plant Omics 2013, 6, 201–207. [Google Scholar]
- Shen, Q.; Cui, J.; Fu, X.Q.; Yan, T.X.; Tang, K.X. Cloning and characterization of DELLA genes in Artemisia annua. GMR 2015, 14, 10037–10049. [Google Scholar] [CrossRef]
- Lu, J.; Yang, W.; Zhang, Q. Genome-wide identification and characterization of the DELLA subfamily in Prunus mume. J. Amer. Soc. Hort. Sci. 2015, 140, 223–232. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Sherif, S.; Abdulla, M.; Jayasankar, S. Plum fruit development occurs via gibberellin–sensitive and –insensitive DELLA repressors. Plos One 2017, 12, e0169440. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, K.; Turowski, T.; Wilmowicz, E.; Frankowski, K.; Kęsy, J.; Kopcewicz, J. Ubiquitin ligases involved in auxin, jasmonate and gibberellin signal transduction pathways. Post. Biol. Kom. 2010, 37, 409–432. [Google Scholar]
- Sun, T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011, 21, 338–345. [Google Scholar] [CrossRef]
- Mutasa-Göttgens, E.; Hedden, P. Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 2009, 60, 1979–1989. [Google Scholar] [CrossRef]
- Marciniak, K.; Kućko, A.; Wilmowicz, E.; Świdziński, M.; Kęsy, J.; Kopcewicz, J. Photoperiodic flower induction in Ipomoea nil is accompanied by decreasing content of gibberellins. Plant Growth Regul. 2018, 84, 395–400. [Google Scholar] [CrossRef]
- de Jong, M.; Mariani, C.; Vriezen, W.H. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef]
- Williams, I.H. The pollination of white lupin. Acta Hort. 1991, 288, 469–472. [Google Scholar] [CrossRef]
- Kazimierska, E.M.; Kazimierski, T. Biology of flowering, embryological and caryological peculiarities. In Lupins, Geography, Classification, Genetic Resources and Breeding; Kurlovich, B.S., Ed.; OY International North Express: St. Petersburg, Russia—Pellosniemi, Finland, 2002; Chapter 8; pp. 205–239. [Google Scholar]
- Gao, Y.; Chen, J.; Zhao, Y.; Li, T.; Wang, M. Molecular cloning and expression analysis of a RGA-like gene responsive to plant hormones in Brassica napus. Mol. Biol. Rep. 2011, 39, 1957–1962. [Google Scholar] [CrossRef]
- Gubler, F.; Chandler, P.M.; White, R.G.; Llewellyn, D.J.; Jacobsen, J.V. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 2002, 129, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.B.; Ruan, M.B.; Cui, B.M.; Xu, N.F.; Lu, J.J.; Peng, M. Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum. Plant Growth Regul. 2009, 58, 35–45. [Google Scholar] [CrossRef]
- Itoh, H.; Ueguchi-Tanaka, M.; Sato, Y.; Ashikari, M.; Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 2002, 14, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Kouketu, E.; Katoh, H.; Aya, K.; Ueguchi-Tanaka, M.; Matsuoka, M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012, 71, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Hirano, Y.; Sun, T.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the scarecrow-like genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef]
- Foster, T.; Kirk, C.; Jones, W.T.; Allan, A.C.; Espley, R.; Karunairetnam, S.; Rakonjac, J. Characterization of the DELLA subfamily in apple (Malus x domestica Borkh.). Tree Genet. Genomes 2007, 3, 187–197. [Google Scholar] [CrossRef]
- Dill, A.; Jung, H.S.; Sun, T.P. The DELLA motif is essential for gibberellin-induced degradation of RGA. Pnasci Usa 2001, 98, 14162–14167. [Google Scholar] [CrossRef]
- Nelson, S.K.; Steber, C.M. Gibberellin hormone signal perception: Down-regulating DELLA repressors of plant growth and development. Ann. Plant Rev. 2016, 49, 153–188. [Google Scholar]
- Engstrom, E.M. Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal. Behav. 2011, 6, 850–854. [Google Scholar] [CrossRef][Green Version]
- Hirsch, S.; Kim, J.; Muñoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Gilday, A.D.; Halliday, K.J.; Graham, I.A. DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 2006, 16, 2366–2370. [Google Scholar] [CrossRef] [PubMed]
- de Lucas, M.; Daviere, J.M.; Rodriguez-Falcon, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blazquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, 46–60. [Google Scholar] [CrossRef]
- Marciniak, K.; Przedniczek, K. Comprehensive insight into gibberellin- and jasmonate-mediated stamen development. Genes (Basel) 2019, 10, 801. [Google Scholar] [CrossRef]
- Kittelson, P.M.; Maron, J.L. Outcrossing rate and inbreeding depression in the perennial yellow bush lupine, Lupinus arboreus (Fabaceae). Am. J. Bot. 2000, 87, 652–660. [Google Scholar] [CrossRef]
- Kurlovich, B.S. The history of lupin domestication. In Lupins, Geography, Classification, Genetic Resources and Breeding; Kurlovich, B.S., Ed.; OY International North Express: St. Petersburg, Russia—Pellosniemi, Finland, 2002; Chapter 5; pp. 147–164. [Google Scholar]
- Luckett, D. Lupini bean—a bitter contamination risk for sweet albus lupins. Industry & Investment NSW. 2010. Available online: http://www.dpi.nsw.gov.au/__data/assets/pdf_file/0003/186672/Lupini-bean-a-bitter-contamination-risk-for-lupins.pdf. (accessed on 3 March 2020).
- Gillaspy, G.; Ben-David, H.; Gruissem, W. Fruits: A developmental perspective. Plant Cell 1993, 5, 1439–1451. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Sherif, S.; El Kayal, W.; Mahboob, A.; Abubaker, K.; Ravindran, P.; Jyothi-Prakash, P.A.; Kumar, P.P.; Jayasankar, S. Characterization of gibberellin-signalling elements during plum fruit ontogeny defines the essentiality of gibberellin in fruit development. Plant Mol. Biol. 2014, 84, 399–413. [Google Scholar] [CrossRef]
- Serrani, J.C.; Sanjua, R.; Ruiz-Rivero, O.; Fos, M.; Garcıa Martinez, J.L. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007, 145, 246–257. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Green, R.; Nilsson, O.; Sussman, M.R.; Weigel, D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 1998, 10, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Pharis, R.P. Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 1999, 77, 944–954. [Google Scholar]
- Vivian-Smith, A.; Koltunow, A.M. Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 1999, 121, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J. Plant hormones: Biosynthesis, signal transduction, action! 3rd ed.; Springer: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin signaling: A theme and variations on DELLA repression. Plant. Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Silverstone, A.L.; Jung, H.S.; Dill, A.; Kawaide, H.; Kamiya, Y.; Sun, T.P. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant. Cell 2001, 13, 1555–1566. [Google Scholar] [CrossRef]
- Rieu, I.; Eriksson, S.; Powers, S.J.; Gong, F.; Griffiths, J.; Woolley, L.; Benlloch, R.; Nilsson, O.; Thomas, S.G.; Hedden, P.; et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 2008, 20, 2420–2436. [Google Scholar] [CrossRef]
- Srinivasan, A.; Morgan, D.G. Growth and development of the pod wall in spring rape (Brassica napus) as related to the presence of seeds and exogenous phytohormones. J. Agric. Sci. 1996, 127, 487–500. [Google Scholar] [CrossRef]
- Groot, S.P.C.; Bruinsma, J.; Karssen, C.M. The role of endogenous gibberellin in seed and fruit development of tomato: Studies with a gibberellin-deficient mutant. Physiol. Plant 1987, 71, 184–190. [Google Scholar] [CrossRef]
- Chandler, P.M.; Robertson, M. Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol. 1999, 120, 623–632. [Google Scholar] [CrossRef]
- Bennett, E.J.; Roberts, J.A.; Wagstaff, C. The role of the pod in seed development: Strategies for manipulating yield. New Phytol. 2011, 190, 838–853. [Google Scholar] [CrossRef]
- Poznan Plant Breeding Sp. z o.o. Information on yellow lupine cv. Taper. Available online: http://phr.pl/wp-content/uploads/2017/07/Taper.pdf (accessed on 3 March 2020).
- Marciniak, K.; Kućko, A.; Wilmowicz, E.; Świdziński, M.; Przedniczek, K.; Kopcewicz, J. Gibberellic acid affects the functioning of flower abscission zone in Lupinus luteus by cooperation with ethylene precursor, however independently of abscisic acid. J. Plant Physiol. 2018, 229, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995, 3, 21–29. [Google Scholar]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, 526–531. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucl. Acids Res. 2004, 32, 500–502. [Google Scholar] [CrossRef]
- Glazinska, P.; Wojciechowski, W.; Kulasek, M.; Glinkowski, W.; Marciniak, K.; Klajn, N.; Kesy, J.; Kopcewicz, J. De novo transcriptome profiling of flowers, flower pedicels and pods of Lupinus luteus (yellow lupine) reveals complex expression changes during organ abscission. Front. Plant Sci. 2017, 8, 641. [Google Scholar] [CrossRef]
- Wilmowicz, E.; Kućko, A.; Ostrowski, M.; Panek, K. INFLORESCENCE DEFICIENT IN ABSCISSION-like is an abscission-associated and phytohormone-regulated gene in flower separation of Lupinus luteus. Plant Growth Regul. 2018, 85, 91–100. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marciniak, K.; Przedniczek, K. Gibberellin Signaling Repressor LlDELLA1 Controls the Flower and Pod Development of Yellow Lupine (Lupinus luteus L.). Int. J. Mol. Sci. 2020, 21, 1815. https://doi.org/10.3390/ijms21051815
Marciniak K, Przedniczek K. Gibberellin Signaling Repressor LlDELLA1 Controls the Flower and Pod Development of Yellow Lupine (Lupinus luteus L.). International Journal of Molecular Sciences. 2020; 21(5):1815. https://doi.org/10.3390/ijms21051815
Chicago/Turabian StyleMarciniak, Katarzyna, and Krzysztof Przedniczek. 2020. "Gibberellin Signaling Repressor LlDELLA1 Controls the Flower and Pod Development of Yellow Lupine (Lupinus luteus L.)" International Journal of Molecular Sciences 21, no. 5: 1815. https://doi.org/10.3390/ijms21051815
APA StyleMarciniak, K., & Przedniczek, K. (2020). Gibberellin Signaling Repressor LlDELLA1 Controls the Flower and Pod Development of Yellow Lupine (Lupinus luteus L.). International Journal of Molecular Sciences, 21(5), 1815. https://doi.org/10.3390/ijms21051815



