Abstract
The plant-special SHI-RELATED SEQUENCE (SRS) family plays vital roles in various biological processes. However, the genome-wide analysis and abiotic stress-related functions of this family were less reported in soybean. In this work, 21 members of soybean SRS family were identified, which were divided into three groups (Group I, II, and III). The chromosome location and gene structure were analyzed, which indicated that the members in the same group may have similar functions. The analysis of stress-related cis-elements showed that the SRS family may be involved in abiotic stress signaling pathway. The analysis of expression patterns in various tissues demonstrated that SRS family may play crucial roles in special tissue-dependent regulatory networks. The data based on soybean RNA sequencing (RNA-seq) and quantitative Real-Time PCR (qRT-PCR) proved that SRS genes were induced by drought, NaCl, and exogenous abscisic acid (ABA). GmSRS18 significantly induced by drought and NaCl was selected for further functional verification. GmSRS18, encoding a cell nuclear protein, could negatively regulate drought and salt resistance in transgenic Arabidopsis. It can affect stress-related physiological index, including chlorophyll, proline, and relative electrolyte leakage. Additionally, it inhibited the expression levels of stress-related marker genes. Taken together, these results provide valuable information for understanding the classification of soybean SRS transcription factors and indicates that SRS plays important roles in abiotic stress responses.
1. Introduction
The members of SHI-RELATED SEQUENCE (SRS) transcription factor family as the plant-specific family play crucial roles in plant growth and development. LRP1 was the first member of the SRS family found in Arabidopsis [1]. The structure of the proteins encoded by the SRS transcription factor genes (SRSs) contains a conserved RING-like zinc-finger domain (CX2CX7CX4CX2C2X6C), and some proteins among them contain the IXGH domain [2]. The RING-like zinc-finger domain is one type of typical RING domain (C3H2C3 or C3HC4). The conserved RING domain was first found as a DNA-binding motif in animal Xenopus laevis [3]. This domain can bind to RNA, protein and lipid substrates, which showed that they possess multiple roles in many intracellular physiological and biochemical processes [4,5]. It is known that there is a cysteine-rich model coupling two zinc atoms in RING domain [6]. There are many types derived from typical RING domain, such as RING-V, RING-D, RING-S/T, RING-G and RING-C2 [6,7]. The IXGH domain contains acidic amino acids, which demonstrate that SRSs possessing IXGH domains are transcriptional activators [2,8].
In Arabidopsis, nine active members of the SHORT INTERNODES/STYLISH (SHI/STY) and SRS family were widely reported. The nine members possess both the RING-like zinc finger domain and IXGH domain. They play very important roles in the progress of plant growth. LRP1 was involved in chromatin modification and auxin signaling during lateral root (LR) development by may forming a complex with SHI, STY1, SRS3, SRS6, and SRS7 [8,9,10]. Interestingly, LRP1 as well as STY1 could regulate the expression of YUC4 in the process of auxin biosynthesis [8,11]. STY1 (SRS1) was reported to regulate auxin biosynthesis, affect apical-basal patterning of stamen, influence cell expansion and timing of flowering [12,13]. STY2 also promoted the formation of apical of stamen [14]. The mutant of AtSHI showed resembled phenotype of a mutant defective in gibberellin (GA) biosynthesis, demonstrating that AtSHI may be involved in the GA signaling pathway [15]. Overexpression of AtSHI in the ornamental Kalanchoë and the poinsettia caused compact phenotype of transgenic plants [16,17]. Interestingly, the members of the SHI/STY family had the redundant function in the progress of auxin biosynthesis probably by regulating the photomorphogenesis-related genes (HY5, BBX21, and BBX22) [11,13,18,19]. In addition to Arabidopsis, LjSTY1/2/3 acted as the targets of LjNFYA1 which played an important role during nodule differentiation in lotus japonicas [20]. In rice, OsSHI1 increased tiller number and diminished panicle size by modulating the transcriptional activity of IPA1 [21]. In barley, LKS2 and VRS2, two members of SHI/STY family, regulated awn elongation, pistil morphology, and inflorescence patterning [22,23]. However, the genome-wide analysis and the function of this family in soybean are still not well characterized.
Soybean is an important food and oil crop and its growth and productivity are easily susceptible to environmental stimuli [24,25]. Especially drought and salt stress have become one of the most abiotic stress problems for soybean development and growth [26,27,28,29,30,31]. One way to alleviate the yield loss is to discover the stress resistant genes to breed the robust soybean cultivar under the abiotic stress. In this study, we did a genome-wide analysis of soybean SRS family and investigated the potential functions in soybean response to various stimuli. We identified the function of GmSRS18 in drought and salt responses. These results will give us a novel understanding about the soybean SRS family and provide the candidate resistant gene for soybean breeding.
2. Results
2.1. Identification of SRS Transcription Factors in Soybean
To excavate all the members of the SRS family, we searched the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) and Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) based on the conserved RING-like zinc-finger domain (DUF702). Moreover, we identified 21 members in soybean and 11 members in Arabidopsis. We analyzed the basal characteristic of 21 members in soybean containing amino acid residues (aa), molecular mass (KD), PI, chromosome, domain location, and the best hit in Arabidopsis (Table 1). We named them GmSRS1~GmSRS21 based on their gene ID number. The number of amino acid residues (aa) ranges from 201 to 371, and the molecular mass (KD) ranges from 22323.7 to 41022.1. Interestingly, the characteristics of their proteins were mainly neutral and alkaline except GmSRS19 (PI 5.765). Meanwhile, we found their corresponding members with the highest homology in Arabidopsis (Table 1).

Table 1.
The characteristic information of the SHI-RELATED SEQUENCE (SRS) transcription factors.
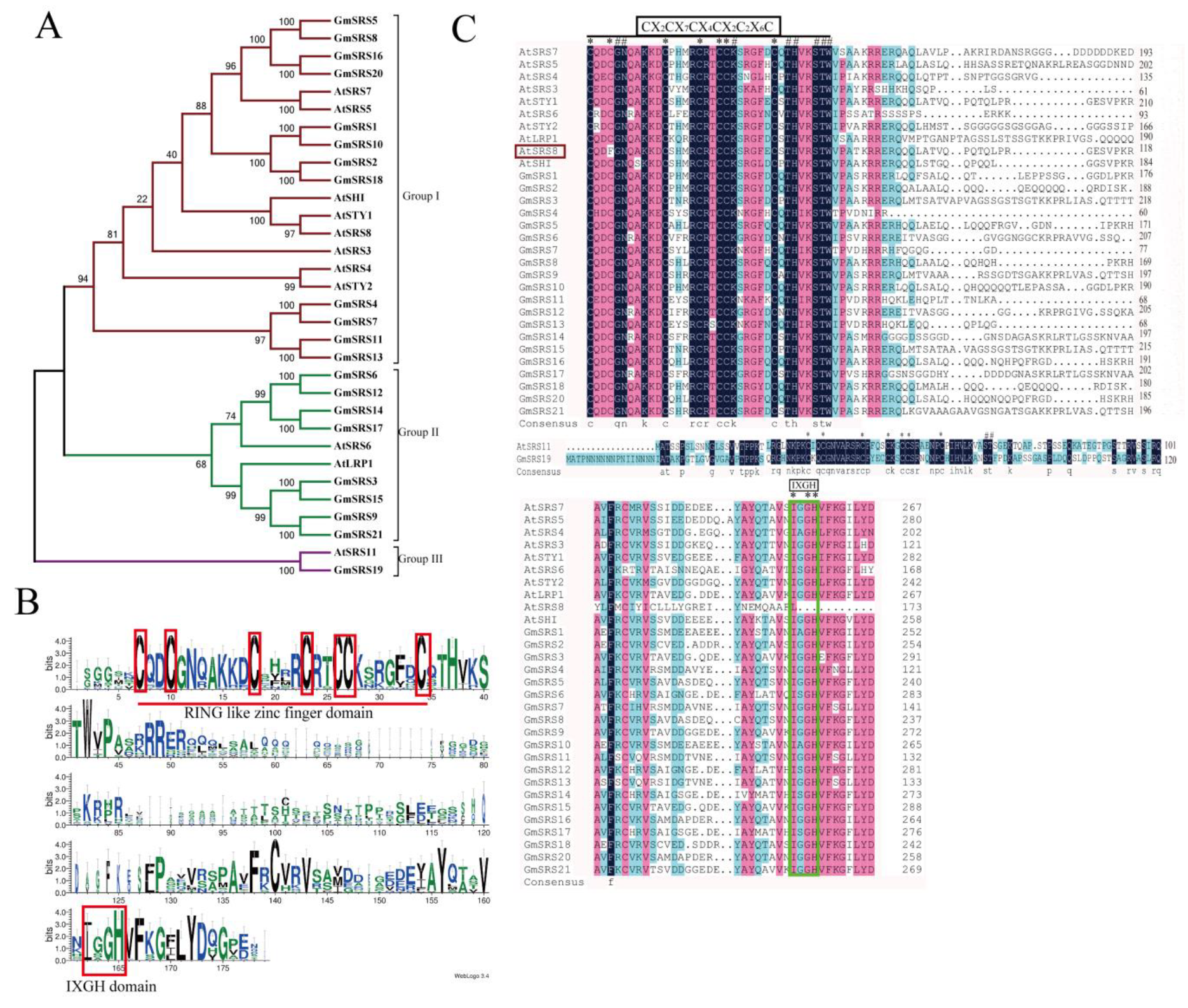
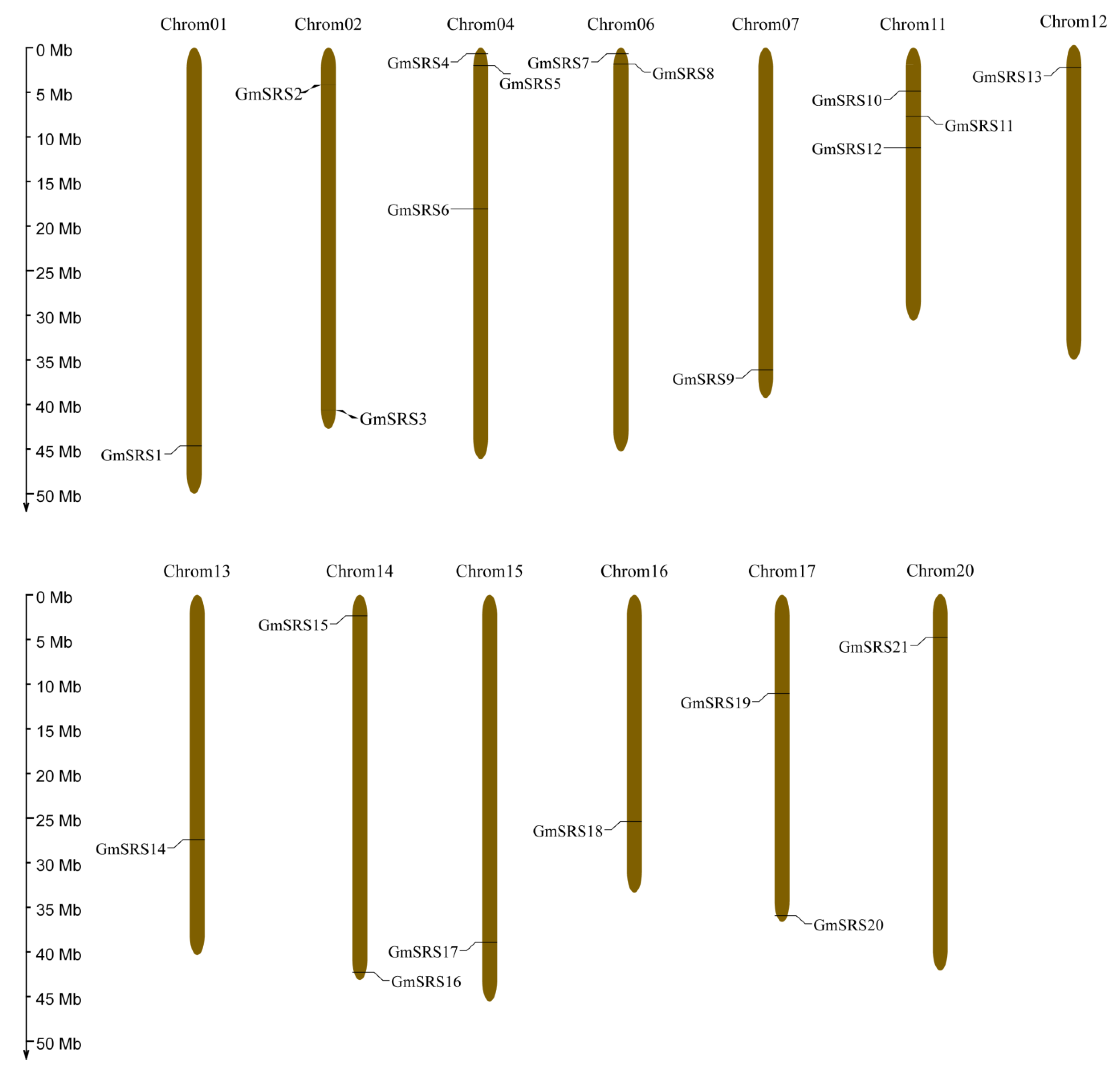
To better realize the members of SRS family, we conducted the phylogenetic tree, which could easily evaluate the phylogenetic relationships of SRSs between soybean and Arabidopsis. We divided them into three groups (I, II, and III) (Figure 1A). In Arabidopsis, AtSRS11 was in Group III, AtLRP1 and AtSRS6 in Group II, and others in Group I. The result was consistent with previous report that AtLRP1 and AtSRS6 had the highest homologous relationship [32]. We found that all members of this family contained the RING-like zinc finger domain (CX2CX7CX4CX2C2X6C) through sequence alignment, but the second cysteine residues of AtSRS8 mutate to phenylalanine residues (Figure 1B,C). Moreover, they had the IXGH domain except AtSRS11 and GmSRS19, which both belong to Group III (Figure 1C). The 21 members localized on 13 chromosomes in soybean (Figure 2).
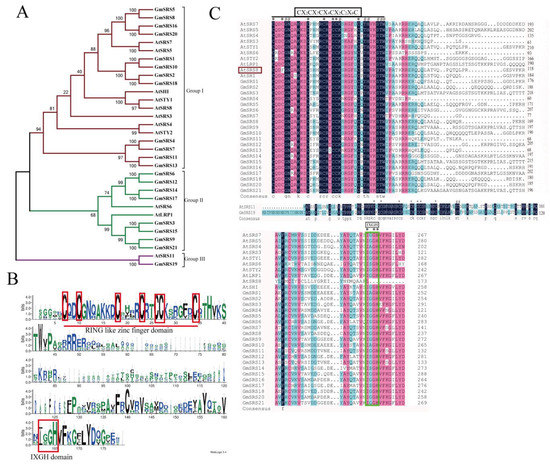
Figure 1.
Phylogenetic relationship and predicted structures of SRS family proteins in soybean and Arabidopsis. (A) Neighbor-joining phylogenetic tree of the SRS family in soybean and Arabidopsis. (B) The conserved RING-like zinc finger domain (CX2CX7CX4CX2C2X6C) and the IXGH domain. The predicted domain sequence was obtained from the website PlantTFDB v5.0 (http://planttfdb.cbi.pku.edu.cn/) (C) Alignment of SRS proteins in soybean and Arabidopsis.
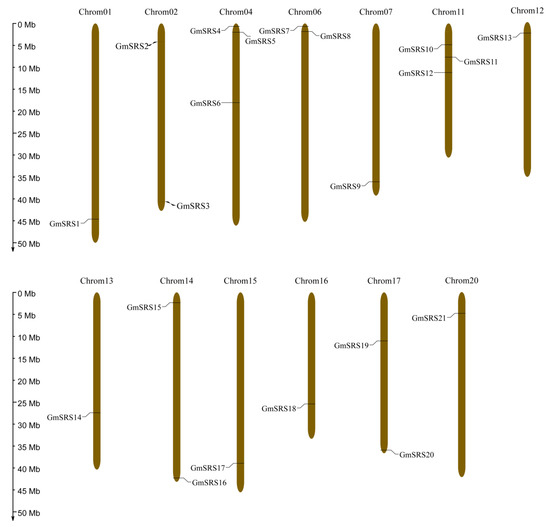
Figure 2.
Chromosomal locations of soybean SRS genes. The ratio on the left represents the chromosome length.
2.2. Analysis of Gene Structure and Cis-Acting Elements
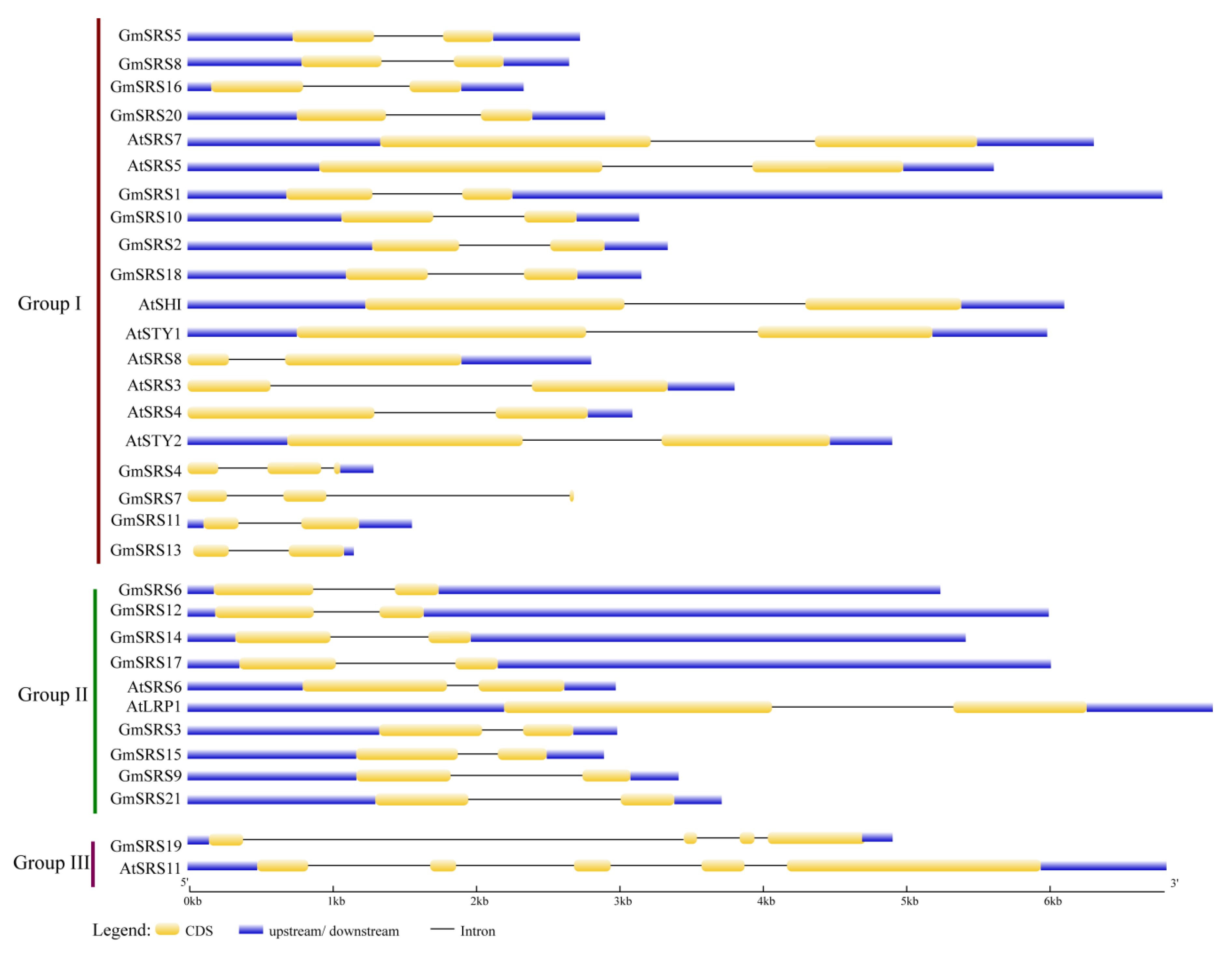
To better understand the structure, we drew the intron-exon model based on the genome sequence of soybean and Arabidopsis SRS genes. In Group I and II, they had two exons and one intron except GmSRS4 and Gm SRS7 which had three exons and two introns. In Group III, they had at least four exons and three introns (Figure 3). These may demonstrate that the members of Group I and II had the similar functions and GmSRS19 and AtSRS11 in Group III also had similar functions.
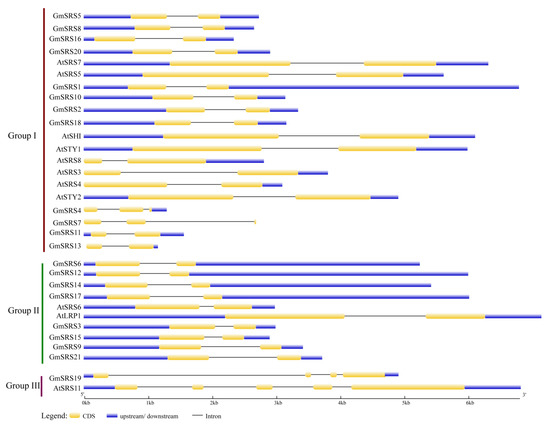
Figure 3.
Intron-exon structures of SRS genes in soybean and Arabidopsis. The GSDS online tool was used to produce the intron-exon structures. The exons, introns, and untranslated regions (UTRs) are indicated by the white boxes, black lines, and gray line, respectively.
The elements in the promoter of genes that can be recognized by transcription factors play crucial roles in progress of transcriptional control [33]. To predict the roles of SRSs in abiotic stress response, we analyzed the numbers of abiotic stress-related cis-elements, including ABA-responsive element (ABRE), E-BOX, GT-1, low-temperature responsive element (LTRE), drought responsive element MYB, and MYC, which were located on the promoters of SRS genes (Table 2). The promoters of SRS genes possess larger of ABRE, E-BOX, GT-1, MYB, and MYC elements. For example, every promoter had at least 16 GT-1, eight E-box, and eight MYC elements. Some promoters also had the temperature stress-related element LTRE. These results showed that the members may act as targets involving abiotic stress response in soybean.

Table 2.
The cis-acting elements in the promoters of soybean SRS genes.
2.3. Analysis of Expression Patterns in Various Tissues
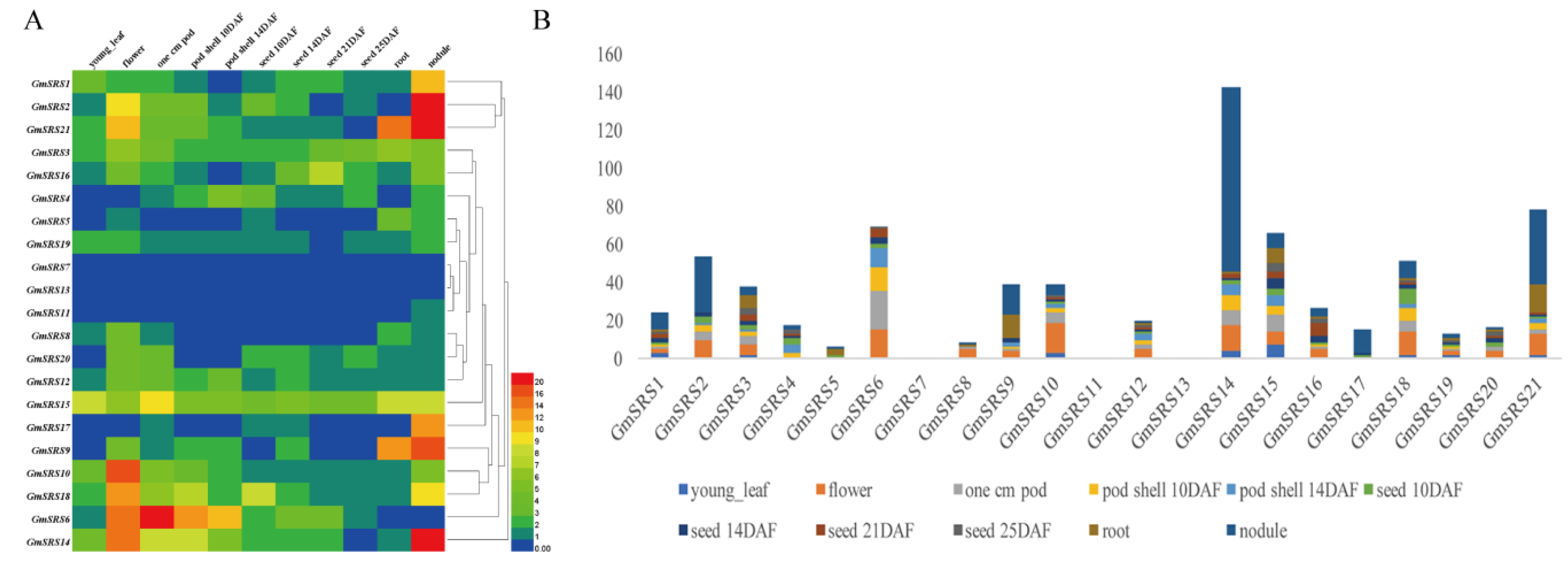
We searched the expression levels in 11 tissues containing young leaf, flower, 1 cm pod, pod shell 10 DAF, pod shell 14 DAF, seed 10 DAF, seed 14 DAF, seed 21 DAF, seed 25 DAF, root, and nodule based on the database SoyBase. The heat map shown that the members of SRS family expressed in leaf, flower, pod, pod shell, seed, root, and nodule. Except GmSRS7 and GmSRS13, others remain higher expression levels in different tissues (Figure 4A). Especially, the expression levels of GmSRS2, 9, 14, and 21 were higher than that of others in nodule (Figure 4B). Moreover, GmSRS6 was expressed primarily at the young leaf, flower, and pod, and GmSRS9 was expressed at the root and nodule. Previous reports demonstrated the SRS family played vital roles in timing of flower [12,13]. GmSRS2, 3, 6, 8, 9, 10, 12, 14, 15, 16, 18, 20, and 21 may all involved in the development of flower based on their expression in flower (Figure 4B) (Table S1).
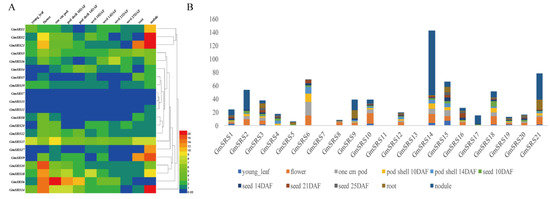
Figure 4.
Expression patterns of soybean SRS genes in different tissues. The heat map (A) and histogram of soybean SRS genes expression patterns (B) in different tissues. The names are on the left and the tissue names are on the top of the figure, and the different colors indicate express degree of gene.
2.4. RNA-Seq Analysis of Drought-, NaCl-, and ABA-responsive SRS Genes
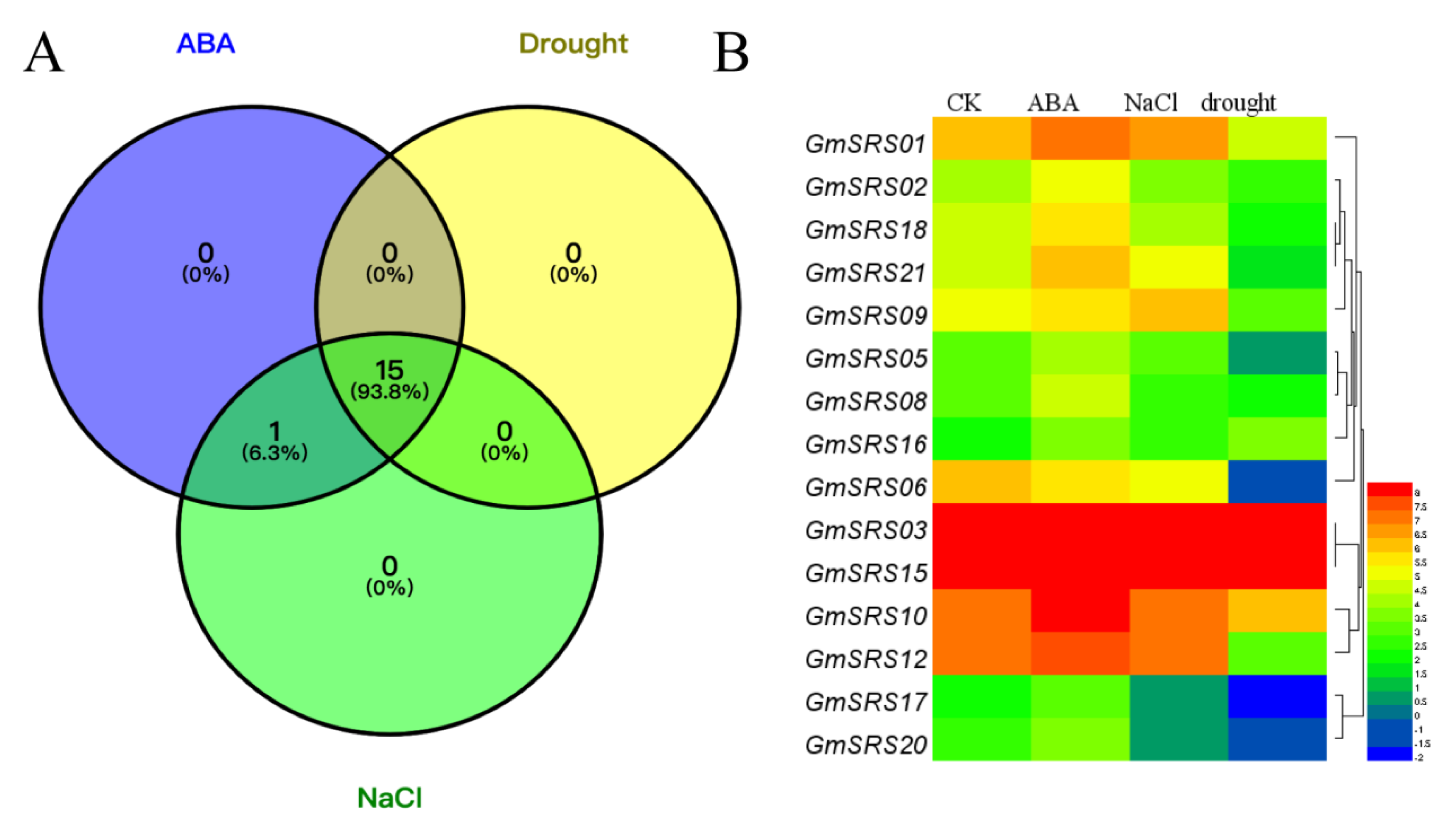
To investigate the functions of the SRS family in the abiotic stress response, we performed soybean transcriptome sequencing analysis under drought, NaCl, and exogenous ABA treatment (Table S2). Fifteen members of SRS family in soybean were induced under drought, NaCl, and exogenous ABA treatment, and one member (GmSRS11) was only induced by exogenous ABA (Figure 5A). The expression of GmSRS3 and GmSRS15 were still maintaining a high level. Moreover, the expression of GmSRS18 and GmSRS21 was increased under ABA treatment and reduced under drought treatment (Figure 5A). GmSRS6 was only negatively induced by drought and NaCl stresses. Although some members were induced by drought, NaCl, and exogenous ABA, the expression of them did not make a significant difference (Table S2).
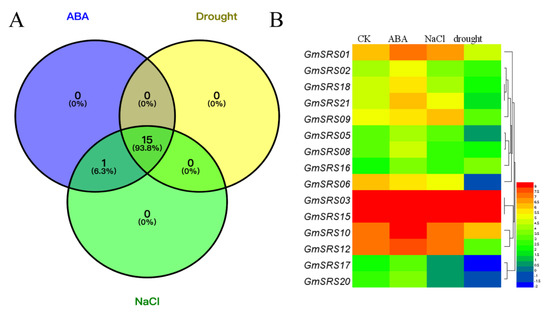
Figure 5.
Transcriptome analysis-based soybean RNA sequencing data. (A) The Venn diagram and (B) the heatmap of stress-related genes under drought, NaCl, and abscisic acid (ABA) treatment.
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis of Soybean SRS Genes
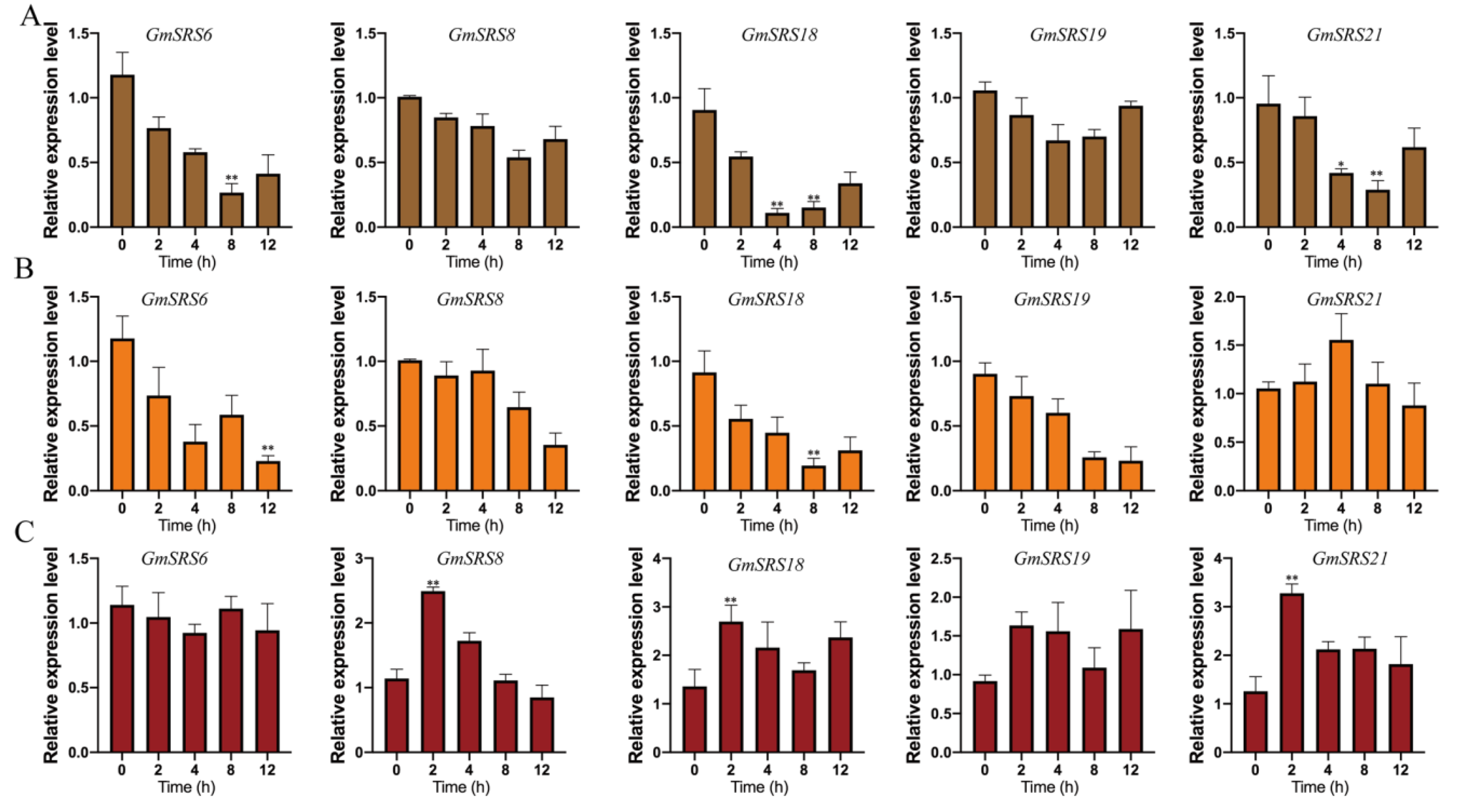
Phytohormone and environment stimuli severely affect crop growth and development. To investigate the potential roles of the soybean SRS genes in response to different stimuli, the expression patterns of these genes in soybean treated with ABA, NaCl, and drought were analyzed by qRT-PCR (Figure 6). We selected five members that were from Group I (GmSRS8 and GmSRS18), Group II (GmSRS6 and GmSRS21), and Group III (GmSRS19) for further analysis.
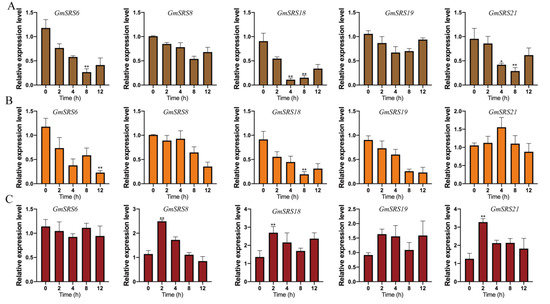
Figure 6.
Relative expression levels of soybean SRS genes under drought, NaCl, and exogenous ABA treatment. quantitative Real-Time PCR (qRT-PCR) analyses of plants treated with drought (A), NaCl (B), and ABA (C). * p < 0.05, ** p < 0.01.
2.6. Subcellular Localization of GmSRS18 in Arabidopsis Protoplasts
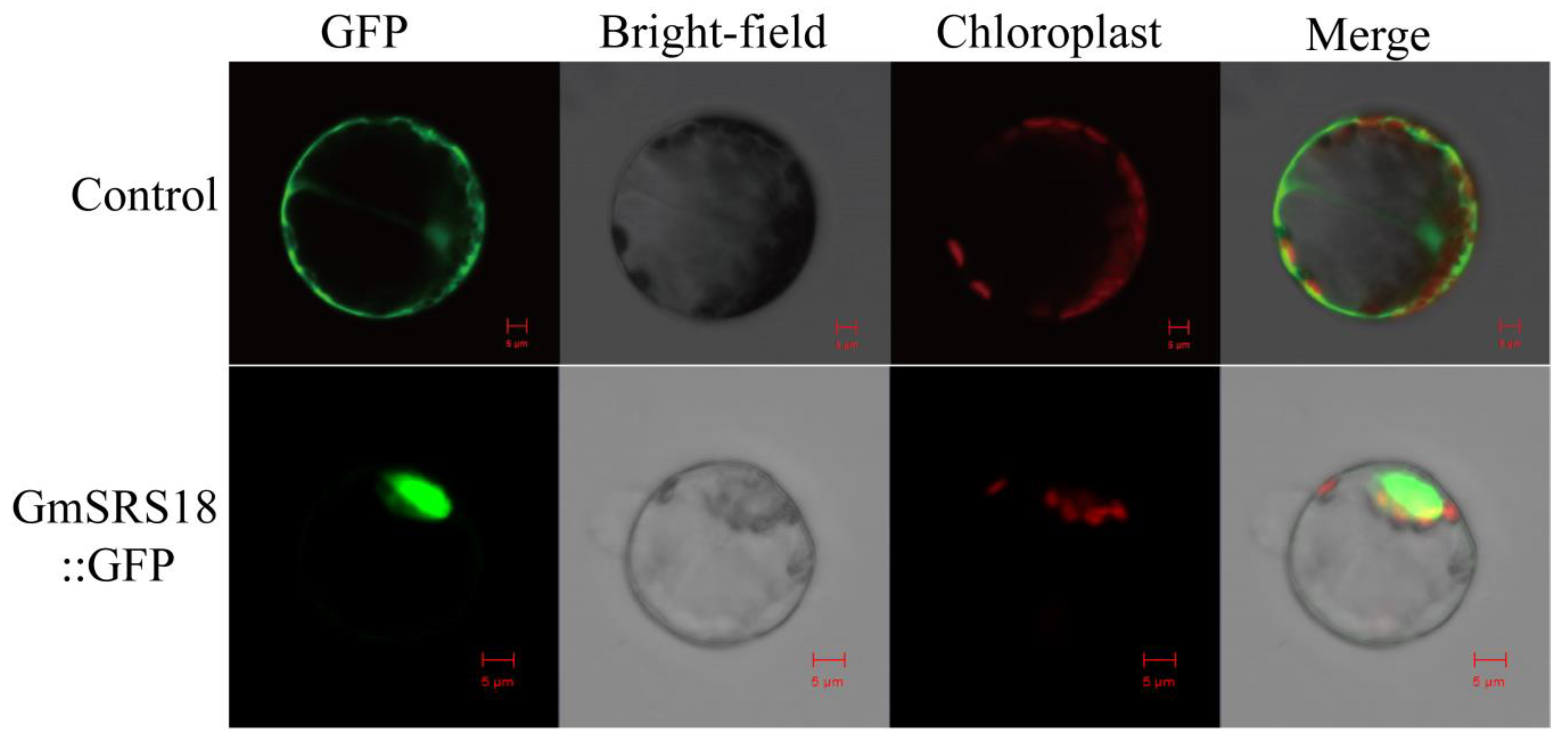
To investigate the functions of the SRS family, GmSRS18, which was from Group I and significantly induced by drought, NaCl, and ABA, was selected for further assay. Protein localization is closely related to its function [34,35]. The fused plasmid containing the full-length sequence of GmSRS18 and the Green fluorescent protein (GFP) tag was transformed into Arabidopsis protoplasts. The empty vector 16318GFP as the control was transformed into Arabidopsis protoplasts. And the results showed that the control protein was localized in the whole cell including cell membrane, cell cytoplasm, and cell nuclear and the protein encoded by GmSRS18 was only localized in cell nuclear (Figure 7), which suggested that GmSRS18 functioned mainly in the nucleus.
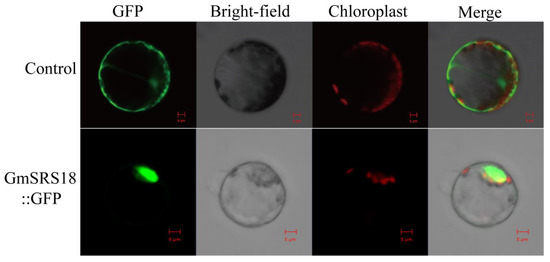
Figure 7.
Subcellular localization of the soybean GmSRS18 protein. The picture above is a control and below is GmSRS18 protein. Results were visualized with confocal microscopy 16 h after transformation. Scale bars = 5 μm.
2.7. Overexpression of GmSRS18 Conferred Drought and Salt sensitivity in Arabidopsis
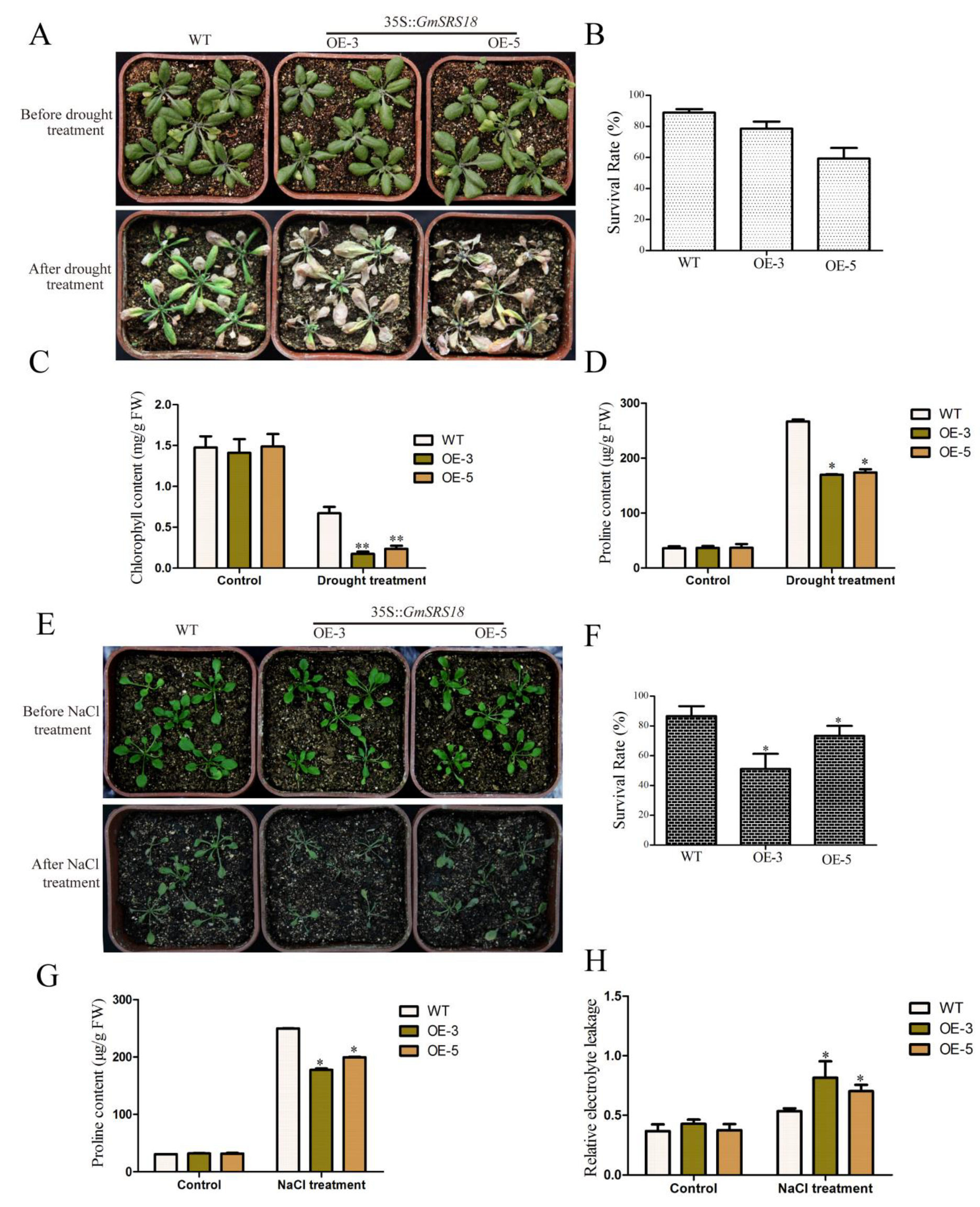
To investigate whether GmSRS18 plays important role in regulating plant response to drought and salt stresses, we generated transgenic Arabidopsis overexpressing GmSRS18 in wild type (WT, Col-0). The expression level of SRS18 in transgenic lines was investigated by qRT-PCR (Figure S1). Then, we selected two transgenic lines (OE-3 and OE-5) to identify their functions under drought and salt treatment. To examine the sensitivity of transgenic Arabidopsis to drought stress, 15-day-old seedlings containing WT and two transgenic lines were deprived water and the control lines were well watered. There was no significant difference among all lines when they were well watered. However, when they were deprived water, the seedlings of OE-3 and OE-5 lines exhibited more wilting and etiolated as compared with the WT seedlings (Figure 8A). To explain this difference visually, relevant physiological indexes were tested. The data analysis of the physiological indexes demonstrated that the two transgenic lines had a higher survival rate, lower chlorophyll content, and lower proline content than that of WT (Figure 8B–D).
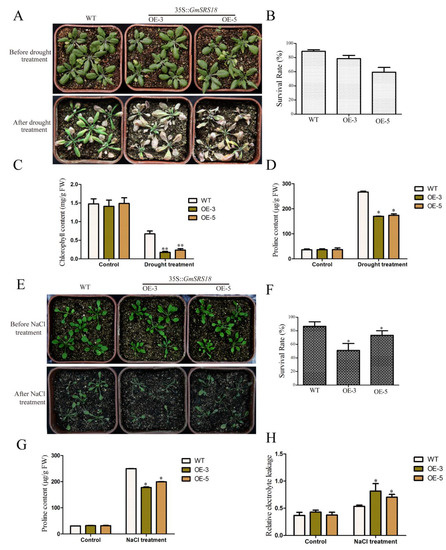
Figure 8.
Phenotype of GmSRS18 overexpression Arabidopsis under drought and salt treatment conditions. (A) The phenotype of wild type WT and transgenic lines (OE-3 and OE-5) under drought treatment. 15-day-old plants were subjected to drought stress. The drought related physiological index was measured, including survival rate (B), chlorophyll content (C), and proline content (D). (E) The phenotype of WT and transgenic lines under NaCl treatment. 10-day-old plants were subjected to NaCl stress. The salt stress-related physiological index was measured, including survival rate (F), proline content (G), and relative electrolyte leakage (H). The data are shown as mean ± Standard Deviation (SD) (n = 45). Independent t-tests demonstrated that there was significant difference (* p < 0.05, ** p < 0.01). Each experiment was repeated at least three times.
To further determine whether GmSRS18 is also sensitive to salt, 10-day-old seedlings of WT and two transgenic lines were treated by NaCl solution. In the absence of NaCl, WT and transgenic lines exhibited similar phenotype. However, in the presence of NaCl, transgenic lines were more wilting than WT (Figure 8E). The lower survival rate, lower chlorophyll content and higher relative electrolyte leakage of transgenic lines showed that transgenic Arabidopsis were more sensitive to NaCl than WT (Figure 8F–H). These results suggested that GmSRS18 was a negative regulator in the drought and salt stresses signaling pathway.
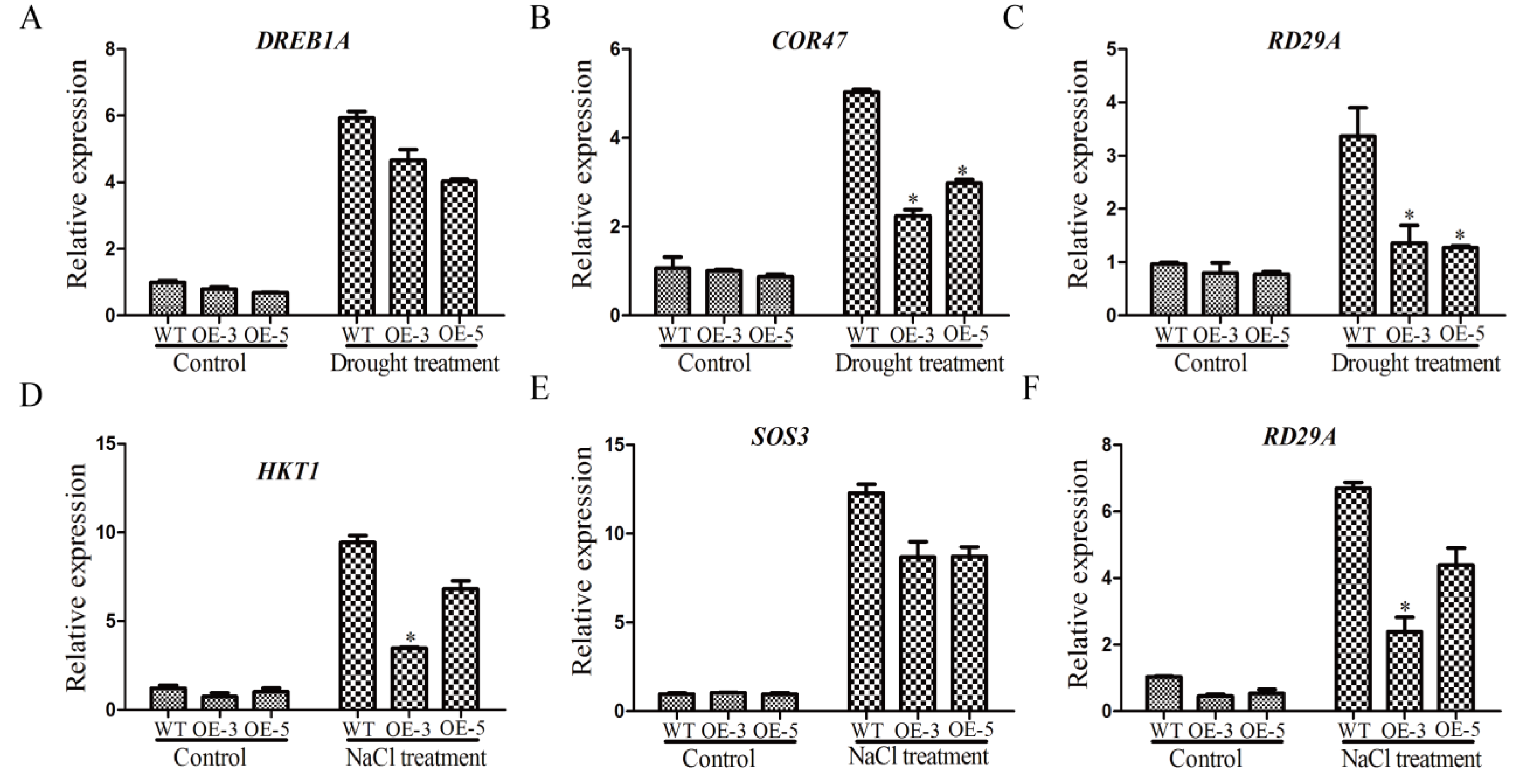
2.8. GmSRS18 Negatively Regulated Drought- and Salt-Related Gene Expression
The expression levels of stress-related genes were affected by drought and salt stresses [36]. To try to explore the function mechanism of GmSRS18, we examined the expression changes of several stress-related marker genes, including RD29A, DREB1A, COR47, HKT1, and SOS3. Under normal conditions, there was no significant difference in their expression levels between WT and transgenic lines (Figure 9). Under drought treatment, compared with control plants, the transcript levels of DREB1A, RD29A, and COR47 were lower in transgenic lines (Figure 9A–C). Likely, the transcript levels of HKT1, RD29A, and SOS3 in transgenic lines were lower than that of WT. These results showed that GmSRS18 may negatively regulate drought- and salt-related genes.
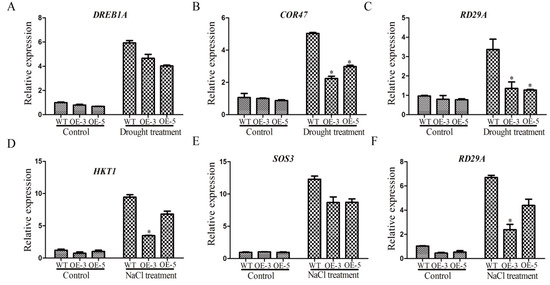
Figure 9.
The expression levels of stress-related marker genes. Drought stress-related marker genes were tested, including DREB1A (A), COR47 (B), and RD29A (C). Salt stress-related marker genes were tested, including HKT1 (D), SOS3 (E), and RD29A (F). Independent t-tests demonstrated that there was a significant difference (* p < 0.05).
3. Discussion
Transcription factors play vital roles in plant growth and development and many transcription factors have been identified to be involved in a series of progresses responding to abiotic and biotic stresses and signaling networks of plant hormones [37,38,39,40,41,42,43,44,45,46,47,48,49]. The annotation and structure analysis of many transcription factors in different plant species had been reported [50,51,52,53,54,55]. Ten members of SRS family were identified in Arabidopsis [1,14,15]. However, reports on the whole genome of SRS transcription factors family in soybean are relatively lacking. In this research, we first identified 21 members of SRS family in soybean and 11 members in Arabidopsis. Previous studies reported that SHI family also called SHI/STY family contained SHI, STY1, STY2, LRP1, SRS3, SRS4, SRS5, SRS6, SRS7, and SRS8 in Arabidopsis. In addition to this, we referred to AtSRS11 (AT1G32730), which also had the conserved RING-like zinc finger domain, as one member of SRS family. In this way, we could define that the main characteristic of this family is that it contains a conserved RING-like zinc finger domain. Thus, the novel Arabidopsis SRS family contained previous SHI family and new member (SRS11) and was assigned to three groups (eight members in Group I, two members in Group II and one member in Group III). The phylogenetic analysis showed that SRS family in soybean was also divided into three groups based on their amino acid sequences (12 members in Group I, eight members in Group II, and one member in Group III) (Figure 1A). Major members of SRS family in soybean and Arabidopsis contain both the RING-like zinc finger domain and the IXGH domain, and some only contain the RING-like zinc finger domain, including the members in Group III and GmSRS8 in Group I (Figure 1C). We hypothesized that the IXGH domains in some members of SRS family were lost during evolution.
Introns are important structures affecting the rate of transcription, nuclear export, and transcript stability and increase the efficiency of mRNA translation [56,57,58]. Although introns could not function as a binding site for transcription factors, they can increase the gene expression [57]. We analyzed the gene structure and found that the members located in the same subfamily have similar gene structure, which proved a presumption that the members in same group had the similar evolutionary relationship or function (Figure 3). As described in the analysis of cis-acting elements in the promoter of SRS genes, they contained many stress-related elements, such as ABRE, E-BOX, GT-1, LTRE, MYB, and MYC (Table 2). ABRE-binding proteins (AREBs/ABFs) can bind to ABRE element to be involved in ABA, dehydration, and high-salinity stress responses [59]. In rice, OSBZ8, an ABRE-binding factor, may regulate the salt tolerance in Indica rice [60]. AtHSFA7b positively regulated the salinity tolerance in Arabidopsis through binding to the E-BOX motif [61]. The E-BOX motif acts as an important component in salinity tolerance response. The MYB element played vital roles in drought, salt, ABA, and GA responses [62,63]. Likely, MYC responds to drought, salt, and ABA stresses through combining with MYC binding proteins [64]. GT-1 may be related to pathogen and salt signals [65]. LTRE element is critical in responding to low temperature stress [66]. The members of the SRS family in soybean contained a number of stress-related elements, which demonstrated that they have a great relationship with the plant stress response. We also found that 15 members were simultaneously induced by ABA, NaCl, and drought, and one member was induced by ABA and NaCl based on the soybean transcriptome sequencing (Figure 5). The expression of the selected five members was inhibited under drought and NaCl treatment (Figure 6A,B). However, their expression increased under exogenous ABA treatment (Figure 6C). These indicated that GmSRS18 regulated the drought and salt response by an ABA-dependent/independent pathway.
GmSRS18, induced by drought, NaCl, and exogenous ABA, was selected for functional identification in Arabidopsis. GmSRS18, as a transcription factor, was located in the cell nucleus, which showed that it functions in cell nucleus. Transcription factors were involved in the abiotic and biotic stress response in many plants [67,68,69,70,71,72]. Though there are few reports of SRS family about the abiotic stress response thus far, we demonstrated that GmSRS18 can increase the transgenic Arabidopsis sensitivity to drought and salt stresses. To further explain the function molecular mechanism of GmSRS18, the expression levels of drought- and salt-related marker genes were tested by qRT-PCR. RD29A was involved in drought and salt stresses response [73]. COR47 and DREB1A were associated with drought stress and HKT1 and SOS3 were associated with salt stress [74,75,76,77,78,79,80]. They all were increased by drought and NaCl treatment. However, their transcript levels in transgenic Arabidopsis were lower than that of WT, which showed that they may be inhibited by GmSRS18. The relationship between GmSRS18 and stress-related genes need further discussion and research, which can provide evidence for how GmSRSs can regulate drought and salt stresses response in plant.
4. Materials and Methods
4.1. The Search of All Members in SRS Family in the Soybean and Arabidopsis
The database PlantTFDB v5.0 (http://planttfdb.cbi.pku.edu.cn/) and phytozome V12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) (Glycine max Wm82.a2.v1 and Arabidopsis thaliana TAIR10) were used to search all members in SRS family based on the conserved RING-like zinc-finger domain (DUF702) [81,82]. We then combined the data from PlantTFDB V5.0 and phytozome V12.1.
4.2. Phylogenetic Analysis and Alignment of SRSs Sequences
The amino acid sequences of SRS members in soybean and Arabidopsis downloaded from database phytozome V12.1 were used to analysis by ClustalX (1.83) and MEGA7.0.21 [83]. The parameter of the neighbor-joining (NJ) algorithm in MEGA7.0.21 was set (No. of bootstrap replications: 1000). And the alignment of SRSs was conducted by DNAMAN V7 and the result calibration was done in Adobe Illustrator CS5 V15.0.0.
4.3. Chromosome Locations of Soybean SRS Genes
We obtained the location information of SRS genes in soybean from the database phytozome V12.1. Moreover, we generated Chromosome locations using online tool MG2C v2.1 (http://mg2c.iask.in/mg2c_v2.1/). The picture calibration was done in Adobe Illustrator CS5 V15.0.0.
4.4. Gene Structure and cis-acting Element Analysis
The genome and CDS sequences of SRS genes in soybean and Arabidopsis download from phytozome V12.1 and the exon-intron substructure map was generated by the online tool Gene Structure Display Server 2.0 (GSDS http://gsds.cbi.pku.edu.cn/) [84]. The 2000 bp promoter before initial codon ATG was also download from phytozome V12.1 and was evaluated using Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/) [85]. The cis-acting element were analyzed by the online tool NEW PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace) [86].
4.5. The Expression Patterns Analysis of SRS Genes in Different Tissues
To analysis the expression of SRS genes in different tissues, we downloaded the Affimetrix soybean gene chip in SoyBase (http://www.soybase.org/soyseq/). Additionally, we generated the heat map containing expression levels in different tissues by using the software Heml 1.0 [87].
4.6. Plant Materials and Stress Treatments
Soybean Tiefeng 8 and Arabidopsis Colombia-0 (Col-0) were used in this research. Moreover, soybean seeds were grown in soil (nutrient soil: vermiculite 1:1) in a chamber at 25 °C with a 16 h light and 8 h dark photoperiod. Two-week-old seedlings were treated by various stimuli for Quantitative Real-time analysis. The treatment method was modified on the basis of previous reports [24]. For the drought treatment, the seedlings were transferred to vermiculite and dehydrated for 0, 2, 4, 8, and 12 h. For the NaCl treatment, the solution containing 200 mM NaCl was poured into the pot. For exogenous ABA treatment, the solution containing 100 μM ABA was sprayed on the soybean seedlings. The seedlings treated by drought, NaCl, and exogenous ABA were harvested at 0, 2, 4, 8, and 12 h. The detached samples were immediately thrown into liquid nitrogen, then, the samples were used to extract the RNA or stored at -80 °C refrigerator until RNA extraction.
4.7. Soybean RNA-seq and Quantitative Real-time PCR (qRT-PCR)
The experimental methods of RNA-seq have been reported by our laboratory [88]. The leaves of 10-day-old soybean seedlings were treated by natural drought (5 h), 100 mM NaCl (1 h), and 100 μM exogenous ABA (1 h) for RNA-seq. We obtained all SRS genes from RNA-seq.
The total RNA was extracted from the treated sample following the instructions of the Fast plant RNApure Kit (ZOMANBIO, Beijing, China). The cDNA synthesis was conducted by using the kit (Transcript One-step gDNA Removal and cDNA Synthesis SuperMix) (TRANS, Beijing, China) as previously described [89]. The ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA) was used to detect the expression patterns of SRS genes, and then, the data was dealt by the software Graphpad Prism 8 and was analyzed as previously described [85,86]. The primers used for qRT-PCR were designed by the software Primer Premier 5.0 (Table S3).
4.8. Subcellular Localization Analysis of GmSRS18
The full-length CDS sequence of GmSRS18 was cloned from soybean cDNA. Then, the CDS with BamH I enzyme site was ligated to the vector 16318GFP, which contained green fluorescent protein (GFP) tags and was controlled by the CaMV35S promoter. As described previously, the subcellular localization of GmSRS18 and the control protein GFP were conducted by the Arabidopsis protoplasts. After 16 h of protoplast incubation, the sample was monitored by confocal microscopy (LSM700; CarlZeiss, Oberkochen, Germany) [85].
4.9. Functional analysis of GmSRS18 in Arabidopsis
To generate the transgenic Arabidopsis, the full-length CDS sequence of GmSRS18 was ligated into the pCAMBIA1302 vector under control of the CaMV 35S promoter. Referring to the previous Arabidopsis transformation method (floral dipping method), we generated the transgenic Arabidopsis [24]. The transgenic lines were selected by the antibiotics hygromycin (30 mg/L) and T3 generation lines (OE-3 and OE-5) were used as the following experimental material.
For drought resistant assessment, the 15-day-old seedlings of WT and transgenic lines were deprived water, and the control lines were well watered. After 10 days, we measured the survival rate, chlorophyll content, and proline content. For salt resistant ability, the 10-day-old seedlings of WT and transgenic lines were watered with 200 mM NaCl solution and the control lines were well watered. After four days, we measured the survival rate, relative electrolyte leakage, and proline content.
4.10. Physiological Measurements
The chlorophyll and proline content were measured based on the method described previously with slight modification [90,91]. The leaf samples (0.1g) were transferred into the mix solution (50% alcohol and 50% acetone). The mixtures were incubated in darkness for 12 h, then centrifuged at 5000 g for 15 min at 4 °C. The instrument Varioskan Lux (Thermo scientific, Waltham, MA, USA) was used to photometrically quantified photometrically at 663 and 645 nm. The proline content was measured according to the method described previously [92]. The proline detecting kit (Comin, Beijing, China) provided the necessary reagents and instructions.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/5/1810/s1.
Author Contributions
Data curation, S.-P.Z., L.-L.G., and X.-Z.Z.; Formal analysis, S.-P.Z.; Funding acquisition, W.-J.Z.; Investigation, S.-P.Z.; Project administration, W.-J.Z.; Resources, X.-Z.Z.; Software, X.-Y.S.; Supervision, Visualization, and Writing—original draft, S.-P.Z.; Writing—review and editing, W.-J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the National Transgenic Key Project of the Ministry of Agriculture of China (2018ZX0800909B).
Acknowledgments
We thank Lijuan Qiu from Crop Science, Chinese Academy of Agricultural Sciences for providing us with soybean seeds (Tiefeng 8).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABA | abscisic acid |
| qRT-PCR | quantitative Real-Time PCR |
| WT | wild type |
| ABRE | ABA-responsive element |
| GFP | Green fluorescent protein |
References
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar] [PubMed]
- Fridborg, I.; Kuusk, S.; Robertson, M.; Sundberg, E. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and Barley. Plant Physiol. 2001, 127, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Kaulen, H.; Pognonec, P.; Gregor, P.D.; Roeder, R.G. The Xenopus B1 factor is closely related to the mammalian activator USF and is implicated in the developmental regulation of TFIIIA gene expression. Mol. Cell. Biol. 1991, 11, 412–424. [Google Scholar] [CrossRef]
- Elenbaas, B.; Dobbelstein, M.; Roth, J.; Shenk, T.; Levine, A.J. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol. Med. 1996, 2, 439–451. [Google Scholar] [CrossRef]
- Berg, J.M.; Shi, Y. The galvanization of biology: A growing appreciation for the roles of zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef]
- Stone, S.L.; Hauksdottir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, S.; Liu, S.; Yu, M.; Su, H.; Shu, H.; Li, X. Global analysis of ankyrin repeat domain C3HC4-type RING finger gene family in plants. PLoS ONE 2013, 8, e58003. [Google Scholar] [CrossRef]
- Singh, S.; Yadav, S.; Singh, A.; Mahima, M.; Singh, A.; Gautam, V.; Sarkar, A.K. Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. Cell Mol. Biol. 2020, 101, 87–100. [Google Scholar] [CrossRef]
- De Smet, I.; Vassileva, V.; De Rybel, B.; Levesque, M.P.; Grunewald, W.; Van Damme, D.; Van Noorden, G.; Naudts, M.; Van Isterdael, G.; De Clercq, R.; et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 2008, 322, 594–597. [Google Scholar] [CrossRef]
- De Rybel, B.; Audenaert, D.; Xuan, W.; Overvoorde, P.; Strader, L.C.; Kepinski, S.; Hoye, R.; Brisbois, R.; Parizot, B.; Vanneste, S.; et al. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat. Chem. Biol. 2012, 8, 798–805. [Google Scholar] [CrossRef]
- Eklund, D.M.; Staldal, V.; Valsecchi, I.; Cierlik, I.; Eriksson, C.; Hiratsu, K.; Ohme-Takagi, M.; Sundstrom, J.F.; Thelander, M.; Ezcurra, I.; et al. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 2010, 22, 349–363. [Google Scholar] [CrossRef]
- Staldal, V.; Cierlik, I.; Chen, S.; Landberg, K.; Baylis, T.; Myrenas, M.; Sundstrom, J.F.; Eklund, D.M.; Ljung, K.; Sundberg, E. The Arabidopsis thaliana transcriptional activator STYLISH1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol. Biol. 2012, 78, 545–559. [Google Scholar] [CrossRef]
- Sohlberg, J.J.; Myrenas, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef]
- Kuusk, S.; Sohlberg, J.J.; Long, J.A.; Fridborg, I.; Sundberg, E. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 2002, 129, 4707–4717. [Google Scholar]
- Fridborg, I.; Kuusk, S.; Moritz, T.; Sundberg, E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 1999, 11, 1019–1032. [Google Scholar] [CrossRef]
- Lutken, H.; Jensen, L.S.; Topp, S.H.; Mibus, H.; Muller, R.; Rasmussen, S.K. Production of compact plants by overexpression of AtSHI in the ornamental Kalanchoe. Plant Biotechnol. J. 2010, 8, 211–222. [Google Scholar] [CrossRef]
- Islam, M.A.; Lutken, H.; Haugslien, S.; Blystad, D.R.; Torre, S.; Rolcik, J.; Rasmussen, S.K.; Olsen, J.E.; Clarke, J.L. Overexpression of the AtSHI gene in poinsettia, Euphorbia pulcherrima, results in compact plants. PLoS ONE 2013, 8, e53377. [Google Scholar] [CrossRef]
- Baylis, T.; Cierlik, I.; Sundberg, E.; Mattsson, J. SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana. New Phytol. 2013, 197, 737–750. [Google Scholar] [CrossRef]
- Yuan, T.T.; Xu, H.H.; Zhang, Q.; Zhang, L.Y.; Lu, Y.T. The COP1 Target SHI-RELATED SEQUENCE5 directly activates photomorphogenesis-promoting genes. Plant Cell 2018, 30, 2368–2382. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shrestha, A.; Zhong, S.; Miri, M.; Austin, R.S.; Sato, S.; Ross, L.; Huebert, T.; Tromas, A.; Torres-Jerez, I.; et al. Lotus japonicus NF-YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Mol. Plant Microbe Interact. 2016, 29, 950–964. [Google Scholar] [CrossRef]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef]
- Yuo, T.; Yamashita, Y.; Kanamori, H.; Matsumoto, T.; Lundqvist, U.; Sato, K.; Ichii, M.; Jobling, S.A.; Taketa, S. A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J. Exp. Bot. 2012, 63, 5223–5232. [Google Scholar] [CrossRef]
- Zhao, S.P.; Xu, Z.S.; Zheng, W.J.; Zhao, W.; Wang, Y.X.; Yu, T.F.; Chen, M.; Zhou, Y.B.; Min, D.H.; Ma, Y.Z.; et al. Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant Sci. 2017, 8, 905. [Google Scholar] [CrossRef]
- Zhao, S.P.; Lu, D.; Yu, T.F.; Ji, Y.J.; Zheng, W.J.; Zhang, S.X.; Chai, S.C.; Chen, Z.Y.; Cui, X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. PPB 2017, 119, 132–146. [Google Scholar] [CrossRef]
- Chen, L.; Fang, Y.; Li, X.; Zeng, K.; Chen, H.; Zhang, H.; Yang, H.; Cao, D.; Hao, Q.; Yuan, S.; et al. Identification of soybean drought-tolerant genotypes and loci correlated with agronomic traits contributes new candidate genes for breeding. Plant Mol. Biol. 2020, 102, 109–122. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, M.; Guo, J.; Wang, Y.; Min, D.; Jiang, Q.; Ji, H.; Huang, C.; Wei, W.; Xu, H.; et al. Overexpression of the soybean (Glycine max) DRE-binding transcription factor GmDREB1 enhanced drought stress tolerance of transgenic wheat in the field. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.W.; Bian, X.H.; Shen, M.; Xiao, J.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X.; et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca(2+) signaling pathways in transgenic soybean. Plant J. 2019, 100, 384–398. [Google Scholar] [CrossRef]
- Wang, D.; Liu, Y.X.; Yu, Q.; Zhao, S.P.; Zhao, J.Y.; Ru, J.N.; Cao, X.Y.; Fang, Z.W.; Chen, J.; Zhou, Y.B.; et al. Functional analysis of the soybean GmCDPK3 gene responding to drought and salt stresses. Int. J. Mol. Sci. 2019, 20, 5909. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.F.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Wei, W.L.; Xu, Z.S. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar] [CrossRef]
- Kuusk, S.; Sohlberg, J.J.; Magnus Eklund, D.; Sundberg, E. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef]
- Shen, Q.; Huang, H.; Zhao, Y.; Xie, L.; He, Q.; Zhong, Y.; Wang, Y.; Wang, Y.; Tang, K. The transcription factor Aabzip9 positively regulates the biosynthesis of artemisinin in Artemisia annua. Front. Plant Sci. 2019, 10, 1294. [Google Scholar] [CrossRef]
- Lin, C.H.; Chen, C.Y. Characterization of the dual subcellular localization of lilium LsGRP1, a plant class II glycine-rich protein. Phytopathology 2014, 104, 1012–1020. [Google Scholar] [CrossRef]
- De la Fuente van Bentem, S.; Vossen, J.H.; Vermeer, J.E.; de Vroomen, M.J.; Gadella, T.W., Jr.; Haring, M.A.; Cornelissen, B.J. The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol. 2003, 133, 702–712. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Rahim, M.A.; Resentini, F.; Dalla Vecchia, F.; Trainotti, L. Effects on plant growth and reproduction of a peach R2R3-MYB transcription factor overexpressed in Tobacco. Front. Plant Sci. 2019, 10, 1143. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhang, C.L.; Wang, G.L.; Wang, Y.X.; Qi, C.H.; Zhao, Q.; You, C.X.; Li, Y.Y.; Hao, Y.J. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 2019, 19, 362. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, Y.; Guo, Y.; Chen, X.; Sun, Y.; Yang, J.; Ye, N. Identification, expression, and putative target gene analysis of nuclear factor-Y (NF-Y) transcription factors in tea plant (Camellia sinensis). Planta 2019, 250, 1671–1686. [Google Scholar] [CrossRef]
- Lai, X.; Daher, H.; Galien, A.; Hugouvieux, V.; Zubieta, C. Structural basis for plant MADS transcription factor oligomerization. Comput. Struct. Biotechnol. J. 2019, 17, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Xu, N.; Wu, Q.; Yu, B.; Li, X.; Chen, R.; Huang, J. Rice transcription factor OsMADS57 regulates plant height by modulating gibberellin catabolism. Rice (New York) 2019, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.; Jiang, H.; Tang, B.; Zhang, M.; Li, Z.; Yin, Y. The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Liang, H.; Chen, G.; Li, F.; Wang, Y.; Liao, C.; Hu, Z. The bHLH transcription factor SlPRE2 regulates tomato fruit development and modulates plant response to gibberellin. Plant Cell Rep. 2019, 38, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yu, H.; Yuan, R.; Yang, Y.; An, F.; Qin, G. Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. iScience 2019, 15, 611–622. [Google Scholar] [CrossRef]
- Yin, X.; Liu, X.; Xu, B.; Lu, P.; Dong, T.; Yang, D.; Ye, T.; Feng, Y.Q.; Wu, Y. OsMADS18, a membrane-bound MADS-box transcription factor, modulates plant architecture and the abscisic acid response in rice. J. Exp. Bot. 2019, 70, 3895–3909. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, L.; Sun, H.; Zhao, X.; Liu, F.; Cai, H.; Fu, Y.; Sun, X.; Gu, P.; Zhu, Z.; et al. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant 2019, 12, 1075–1089. [Google Scholar] [CrossRef]
- Fasani, E.; DalCorso, G.; Costa, A.; Zenoni, S.; Furini, A. The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol. Biol. 2019, 99, 517–534. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Z.; Xiong, T.; Xiong, X.; Wu, X.; Guan, C.; Xiao, G. Corrigendum to “The CCCH-type transcription factor BnZFP1 is a positive regulator to control oleic acid levels through the expression of diacylglycerol O-acyltransferase 1 gene in Brassica napus” [Plant Physiol. Biochem. 132 (November 2018) 633–640]. Plant Physiol. Biochem. 2019, 135, 611. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, Q.; Shi, Y.; Sun, Z.; Zhang, Q.; Hou, S.; Wu, R.; Jiang, L.; Zhao, X.; Guo, Y. Genome-wide analysis of the NAC domain transcription factor gene family in Theobroma cacao. Genes 2019, 11, 35. [Google Scholar] [CrossRef]
- Wan, R.; Liu, J.; Yang, Z.; Zhu, P.; Cao, Q.; Xu, T. Genome-wide identification, characterisation and expression profile analysis of DEAD-box family genes in sweet potato wild ancestor Ipomoea trifida under abiotic stresses. Genes Genom. 2020, 42, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhou, Q.; Wu, L.; Li, F.; Liu, B.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification and characterization of the ALOG gene family in Petunia. BMC Plant Biol. 2019, 19, 600. [Google Scholar] [CrossRef]
- Li, J.; Gao, X.; Sang, S.; Liu, C. Genome-wide identification, phylogeny, and expression analysis of the SBP-box gene family in Euphorbiaceae. BMC Genom. 2019, 20 (Suppl. 9), 912. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Wang, C.; Shi, H.H.; Wang, B.; Wang, J.X.; Liu, Y.S.; Ma, J.Y.; Tian, S.Y.; Zhang, Y.W. Genome-wide analysis of ethylene-response factor family in adzuki bean and functional determination of VaERF3 under saline-alkaline stress. Plant Physiol. Biochem. 2019, 147, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hua, X.; Zhong, W.; Yuan, Y.; Wang, Y.; Wang, Z.; Ming, R.; Zhang, J. Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Laxa, M.; Muller, K.; Lange, N.; Doering, L.; Pruscha, J.T.; Peterhansel, C. The 5’UTR intron of Arabidopsis GGT1 aminotransferase enhances promoter activity by recruiting RNA polymerase II. Plant Physiol. 2016, 172, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell B 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Wallace, E.W.J.; Beggs, J.D. Extremely fast and incredibly close: Cotranscriptional splicing in budding yeast. RNA 2017, 23, 601–610. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Mukherjee, K.; Choudhury, A.R.; Gupta, B.; Gupta, S.; Sengupta, D.N. An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol. 2006, 6, 18. [Google Scholar] [CrossRef]
- Zang, D.; Wang, J.; Zhang, X.; Liu, Z.; Wang, Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019, 70, 5355–5374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int. J. Biol. Macromol. 2019, 145, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.F.; Sun, C.Q.; Fu, Y.C.; Qian, X.Y.; Yang, J.S.; Wang, X.K. Isolation and analysis of a novel MYC gene from rice. Yi Chuan Xue Bao Acta Genet. Sin. 2005, 32, 393–398. [Google Scholar] [PubMed]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef]
- Maestrini, P.; Cavallini, A.; Rizzo, M.; Giordani, T.; Bernardi, R.; Durante, M.; Natali, L. Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 2009, 166, 1544–1556. [Google Scholar] [CrossRef] [PubMed]
- Gastaldi, V.; Lucero, L.E.; Ferrero, L.V.; Ariel, F.D.; Gonzalez, D.H. Class I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 2020. [Google Scholar] [CrossRef]
- Ma, X.; Su, P.; Yin, C.; Lin, X.; Wang, X.; Gao, Y.; Patil, S.; War, A.R.; Qadir, A.; Tian, Y.; et al. The roles of FoxO transcription factors in regulation of bone cells function. Int J. Mol. Sci. 2020, 21, 692. [Google Scholar] [CrossRef]
- Petrella, R.; Caselli, F.; Roig-Villanova, I.; Vignati, V.; Chiara, M.; Ezquer, I.; Tadini, L.; Kater, M.M.; Gregis, V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020. [Google Scholar] [CrossRef]
- Turton, K.L.; Meier-Stephenson, V.; Badmalia, M.D.; Coffin, C.S.; Patel, T.R. Host transcription factors in hepatitis B virus RNA synthesis. Viruses 2020, 12, 160. [Google Scholar] [CrossRef]
- Wei, X.; Lu, W.; Mao, L.; Han, X.; Wei, X.; Zhao, X.; Xia, M.; Xu, C. ABF2 and MYB transcription factors regulate feruloyl transferase FHT involved in ABA-mediated wound suberization of kiwifruit. J. Exp. Bot. 2020, 71, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.B.; Shang, G.D.; Pan, Y.; Xu, Z.G.; Zhou, C.M.; Mao, Y.B.; Bao, N.; Sun, L.; Xu, T.; Wang, J.W. AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 2020, 32, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gener. Genet. MGG 1993, 236, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nov. Found. Symp. 2001, 236, 176–186. [Google Scholar]
- Kurkela, S.; Borg-Franck, M. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 19, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, W.; Guo, Y. Arabidopsis SOS3 plays an important role in salt tolerance by mediating calcium-dependent microfilament reorganization. Plant Cell Rep. 2013, 32, 139–148. [Google Scholar] [CrossRef]
- Shohan, M.U.S.; Sinha, S.; Nabila, F.H.; Dastidar, S.G.; Seraj, Z.I. HKT1;5 transporter gene expression and association of amino acid substitutions with salt tolerance across rice genotypes. Front. Plant Sci. 2019, 10, 1420. [Google Scholar] [CrossRef]
- Nawaz, I.; Iqbal, M.; Hakvoort, H.W.J.; de Boer, A.H.; Schat, H. Analysis of Arabidopsis thaliana HKT1 and Eutrema salsugineum/botschantzevii HKT1;2 promoters in response to salt stress in Athkt1:1 mutant. Mol. Biotechnol. 2019, 61, 442–450. [Google Scholar] [CrossRef]
- Ali, A.; Maggio, A.; Bressan, R.A.; Yun, D.J. Role and functional differences of HKT1-Type transporters in plants under salt stress. Int. J. Mol. Sci. 2019, 20, 1059. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na(+) exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Knudsen, S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Deng, W.K.; Wang, Y.B.; Liu, Z.X.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in Soybean. Int J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Xu, Z.S.; Xia, L.Q.; Chen, M.; Cheng, X.G.; Zhang, R.Y.; Li, L.C.; Zhao, Y.X.; Lu, Y.; Ni, Z.Y.; Liu, L.; et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007, 65, 719–732. [Google Scholar] [CrossRef]
- Sanchez-Zabala, J.; Gonzalez-Murua, C.; Marino, D. Mild ammonium stress increases chlorophyll content in Arabidopsis thaliana. Plant Signal. Behav. 2015, 10, e991596. [Google Scholar] [CrossRef]
- Yamakawa, S.; Matsubayashi, Y.; Sakagami, Y.; Kamada, H.; Satoh, S. Promotive effects of the peptidyl plant growth factor, phytosulfokine-alpha, on the growth and chlorophyll content of Arabidopsis seedlings under high night-time temperature conditions. Biosci. Biotechnol. Biochem. 1999, 63, 2240–2243. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Lv, T.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.; Huang, Z.; Xiao, L.; Engineer, C.; Kim, T.H.; et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).