Abstract
WRKY transcription factors play central roles in developmental processes and stress responses of wheat. Most WRKY proteins of the same group (Group III) have a similar function in abiotic stress responses in plants. TaWRKY46, a member of Group III, was up-regulated by PEG treatment. TaWRKY46-GFP fusion proteins localize to the nucleus in wheat mesophyll protoplasts. Overexpression of TaWRKY46 enhanced osmotic stress tolerance in transgenic Arabidopsis thaliana plants, which was mainly demonstrated by transgenic Arabidopsis plants forming higher germination rate and longer root length on 1/2 Murashige and Skoog (MS) medium containing mannitol. Furthermore, the expression of several stress-related genes (P5CS1, RD29B, DREB2A, ABF3, CBF2, and CBF3) was significantly increased in TaWRKY46-overexpressing transgenic Arabidopsis plants after mannitol treatment. Taken together, these findings proposed that TaWRKY46 possesses vital functions in improving drought tolerance through ABA-dependent and ABA-independent pathways when plants are exposed to adverse osmotic conditions. TaWRKY46 can be taken as a candidate gene for transgenic breeding against osmotic stress in wheat. It can further complement and improve the information of the WRKY family members of Group III.
1. Introduction
Drought is one of the most hazardous environmental abiotic stressors, which adversely influences the growth and development of wheat (Triticum aestivum L.) [1,2]. It has caused an average of 13.7% loss in cereal production worldwide over the past few decades [3]. In Northwest China, wheat is cultivated under rain-fed conditions and encounters severe water shortage because of the low rainfall [4]. Uncovering the sophisticated underlying mechanisms that allow wheat to adapt to various detrimental stresses has vital significance for the improvement of wheat drought tolerance [5,6]. Several transcription factor (TF) families, such as WRKY, MYB, and NAC, have been reported to play key roles in response to drought stress in plants.
WRKY TFs mediate signal transduction pathways by regulating the transcription of target DNA and eventually leading to stress tolerance in plants [7,8,9,10,11]. The WRKY family is characterized by the WRKY domain, which comprises a short conserved sequence WRKYGQK at the N-terminal and a zinc finger motif at the C-terminal [12,13]. According to the number of WRKY domains and zinc-finger structures, the WRKY family members are divided into three groups. Two WRKY domains and a C2H2 (CX4-5CX22-23HXH) zinc finger exist in Group I. Group II a-e and Group III contain one WRKY domain and a C2H2 motif or a C2HC (CX7CX23HXC) motif, respectively [14]. The WRKY domain controls transcription by specifically binding to the upstream W-box sequence of the target genes [15,16].
As a hexaploid plant, the wheat genome consists of the A, B, and D genomes with 15,966 Mb in size [17,18]. In a previous study, 171 WRKY TFs were identified in wheat [19]. Less than one-third of wheat TaWRKYs have been functionally analyzed so far. Recent studies have emphasized the involvement of TaWRKY TFs in various biotic and abiotic stress responses and plant growth and development processes, such as pathogen defense [20], high salt stress [21,22], extreme temperature [23,24], drought stress [21,22,25,26] and senescence [15].
The WRKY members of Group III have been identified as participants in abiotic stress responses in plants. AtWRKY46 played roles in regulating plant responses to osmotic stress and stomatal movement [27]. GhWRKY33 functioned as a negative regulator in drought stress response and participates in the ABA signaling pathway [28]. The overexpression of VlWRKY48 enhanced plant drought tolerance by multiple mechanisms. Additionally, VlWRKY48 controlled the resistance to powdery mildew infection [29]. The function of the WRKY members that belonged to Group III is conserved. Whether or not the wheat WRKY members of Group III play a role in abiotic stress responses is still unclear.
In our previous work, 45 WRKY TFs which belonged to Group III were identified in wheat [19]. To assess the function of wheat Group III members, a Group III gene TaWRKY46 (TRAES3BF051200110CFD_t1) is cloned in this study. Our results showed that overexpression of TaWRKY46 promoted the germination rate and boosted the root growth in TaWRKY46-overexpressing Arabidopsis plants under osmotic stress. Overexpression of TaWRKY46 also induced stress-related genes in transgenic Arabidopsis plants, which suggested that TaWRKY46 played an important role in enhancing osmotic tolerance through ABA-dependent and ABA-independent pathways. This study provides more information for further investigation into the underlying mechanisms of TaWRKY genes that are involved in osmotic-related regulatory networks.
2. Results
2.1. Identification of TaWRKY46 and its Transcription Profiles under Various Stress Conditions
TaWRKY46 comprised the splicing of three exons and two introns. The length of wheat WRKY46 was 1062 base pairs (bp). The sizes of the introns were 83 bp and 94 bp, respectively (Figure 1A). TaWRKY46 and its orthologous from other plant species were used to construct a phylogenetic tree. The result showed that TaWRKY46 had the closest relationship with Hv1065C13 (AK357930.1) from Hordeum vulgare due to its high identity (Figure 1B). The TaWRKY46 protein had 294 amino acid residues. A total of 71 amino acid residues (24% of sequence) had been modelled with 100% confidence by the single highest scoring template. The image of the tertiary structure of TaWRKY46 was colored by rainbow from N to C terminus. It had four β-sheets which were displayed in the tertiary structure. The model dimensions of tertiary structures were also given in detail (Figure 1C). Multiple sequence alignment revealed that all WRKYs had the conserved WRKYGQK motif and amino acid residues. TaWRKY46 was assigned to Group III, due to the existence of one WRKY domain and a C2HC zinc finger motif (Figure 1D).
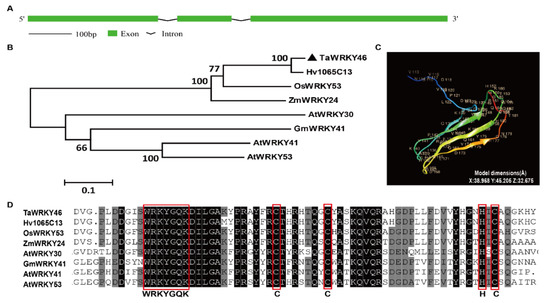
Figure 1.
Sequence analysis of TaWRKY46. (A) A schematic representation showing the gene structure of TaWRKY46. (B) Phylogenetic relationship of TaWRKY46 with its orthologous in other plant species. The accession numbers of the WRKYs are listed in Supplementary Table S1. (C) Tertiary structure of TaWRKY46 protein. A total of 71 amino acid residues were modelled with 100% confidence by the single highest scoring template. Image was colored by rainbow from N to C terminus. The number and capital letter represent the order and abbreviation of amino acids. (D) Sequence alignment of TaWRKY46 and its homologs. Black and grey background represent 100 and 75% similarity, respectively. The completed WRKY domain and C2HC zinc finger motif are circled in red boxes.
To dissect potential functions, the expression of TaWRKY46 was investigated in different tissues and various stress conditions by qRT-PCR. The results showed that TaWRKY46 was expressed in all organs, with the higher and lower transcriptional levels in the leaf and root, respectively (Figure 2A). Transcription of TaWRKY46 was not affected by low-temperature, but was repressed by high-temperature and exogenous ABA treatments (Figure 2D–F). The TaWRKY46 gene was slightly induced by NaCl at a maximum level of about 1.48-fold at 3 h (Figure 2C). TaWRKY46 was markedly induced after PEG treatment, and increased by 3.57-fold at 3 h (Figure 2B).
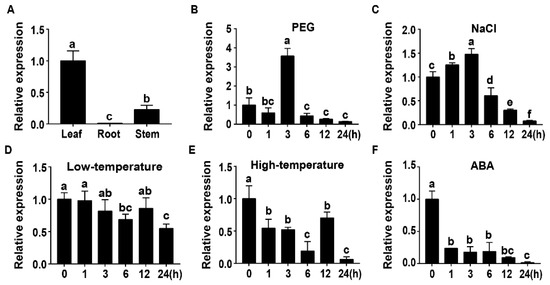
Figure 2.
Transcription profiles of TaWRKY46. (A) Organ expression assay of TaWRKY46 in different wheat organs (leaf, root, and stem). Transcription profiles of TaWRKY46 under 20% PEG 6000 (B); 200 mM NaCl (C); 4 °C (D); 42 °C (E); 100 μM ABA (F) treatments in wheat leaves. The transcriptional level at time point 0 (for the multiple stress experiments) and the leaf (for the organ expression assay) was defined as 1.0.
2.2. Subcellular Location of TaWRKY46
The full coding region of TaWRKY46 was cloned into the pTF486-GFP vector under control of the CaMV35S promoter and transformed into wheat leaf protoplasts. The empty vector was used as a control. The GFP signal of 35S:TaWRKY46-GFP was exclusively observed in the nucleus, whereas GFP signal of the control was discovered in the cytoplasm and nucleus (Figure 3). Therefore, TaWRKY46 likely functioned as a transcription factor in the nucleus.
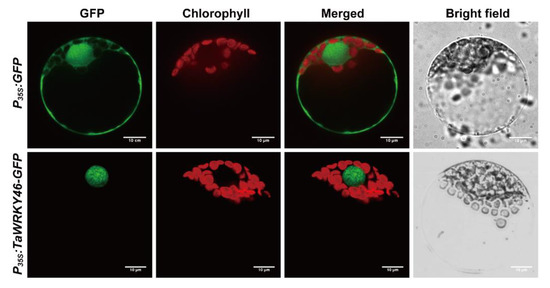
Figure 3.
Subcellular localization of the TaWRKY46. P35S:TaWRKY46-GFP and P35S:GFP control vectors were transiently expressed in wheat leaf protoplasts. Scale bar = 10 μm.
2.3. Transcriptional Activation Assay of TaWRKY46 in Yeast
Transcriptional activation of TaWRKY46 was detected with the GAL4 yeast expression system. Yeast strain AH109 was transformed with constructions pGBKT7-TaWRKY46 (1-294 aa), pGBKT7-TaWRKY46 (1-191 aa), and pGBKT7-TaWRKY46 (192-294 aa), and pGBKT7 was taken as a negative control. The yeast cells transformed with pGBKT7-TaWRKY46 (1-294 aa) and pGBKT7-TaWRKY46 (192-294 aa) grew well on the SD-W/H/A medium (Figure 4). Meanwhile, yeast cells transformed with pGBKT7-TaWRKY46 (1-191 aa) and empty vector pGBKT7 could only survive on the SD-W medium. The yeast cells transformed with pGBKT7-TaWRKY46 (1-294 aa) and pGBKT7-TaWRKY46 (192-294 aa) grew well on SD-W/H/A medium, and turned blue in the presence of X-α-gal. The results demonstrated that the C-terminal region of TaWRKY46 functioned as a transcriptional activator.
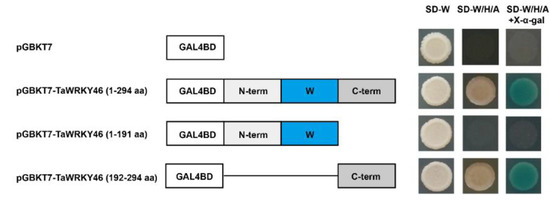
Figure 4.
Transcriptional activity assay of TaWRKY46 protein in yeast. A schematic diagram of fused vectors exhibiting the truncated segments of TaWRKY46 that were fused to the vector pGBKT7. The transformants were incubated on the SD-W, SD-W/H/A, and SD-W/H/A with X-α-gal medium.
2.4. Overexpression of TaWRKY46 Improved Osmotic Tolerance in Transgenic Arabidopsis Plants
The complementary DNA (cDNA) encoding TaWRKY46 was inserted into the pBI1304 vector under the control of the CaMV35S promoter. The PCR and qRT-PCR analysis showed that TaWRKY46 was detected in the two selected TaWRKY46-overexpressing Arabidopsis plants (OE-7 and OE-16), but not in the wild-type (WT) (Figure 5A,B). The water loss rate was evaluated at the indicated interval for both WT and TaWRKY46-overexpressing Arabidopsis plants. Compared with the WT plants, the detached leaves of TaWRKY46-overexpressing Arabidopsis plants possessed lower water loss rate (Figure 5C). The 24-day-old seedlings of WT and transgenic Arabidopsis plants were exposed to drought conditions for 25 days. All leaves of WT Arabidopsis plants were severely curled and turned yellow, while some leaves of the transgenic plants (OE-7 and OE-16) remained green (Figure 5D). After withholding water for 25 d, 17.34% of the WT plants survived while the survival rates of the transgenic Arabidopsis OE7 and OE16 lines were 61.13% and 63.33%, respectively (Figure 5E).
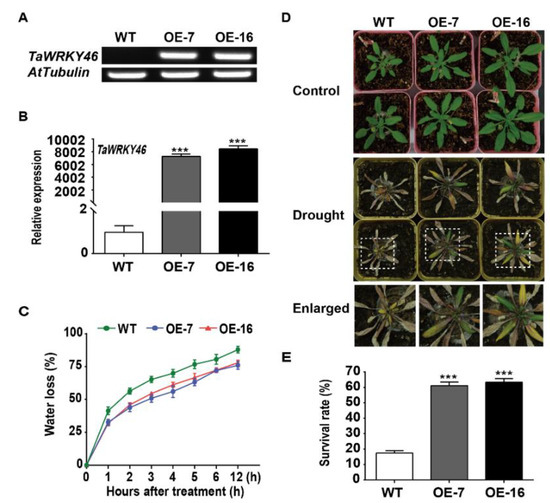
Figure 5.
The phenotype and tolerance assay of TaWRKY46-overexpressing Arabidopsis after drought treatment. Gene validation of TaWRKY46 in transgenic Arabidopsis lines by PCR (A) and qRT-PCR (B). (C) The water loss rate of the detached leaves. (D) The phenotype of TaWRKY46-overexpressing Arabidopsis plants after water withholding for 25 days. (E) The survival rate of TaWRKY46-overexpressing Arabidopsis plants after drought treatment. Asterisks indicate a significant difference between wild-type (WT) and transgenic Arabidopsis lines (*** p < 0.001).
To assess the osmotic stress tolerance of the transgenic Arabidopsis plants, WT and TaWRKY46-overexpressing Arabidopsis seeds were sown on 1/2 MS medium containing mannitol (0 mM, 150 mM, and 300 mM), and the seed germination rate was monitored for 4 days. Compared with WT, TaWRKY46-overexpressing transgenic Arabidopsis seeds grown on 1/2 MS medium containing mannitol showed remarkable differences in the germination rate (Figure 6A–C). More than 87.34% and 76.19% of TaWRKY46 transgenic seeds germinated in 1/2 MS medium supplemented with 150 mM and 300 mM mannitol, respectively, compared to 77.06% and 63.72% for WT seeds (Figure 6E,F). However, the seeds planted on the 1/2 MS medium without mannitol had no obvious difference between the WT and TaWRKY46-overexpressing transgenic Arabidopsis lines (Figure 6D). The results suggested that overexpression of TaWRKY46 enhanced osmotic tolerance during seed germination in transgenic Arabidopsis plants.
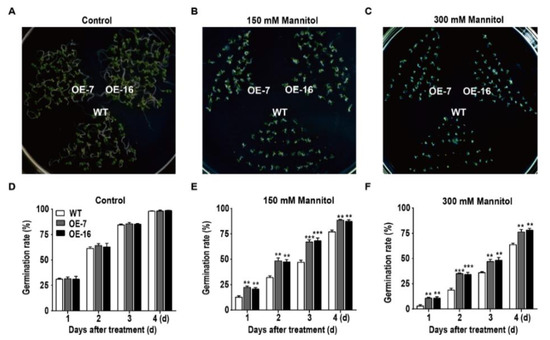
Figure 6.
The germination rate of overexpressing TaWRKY46 in Arabidopsis under osmotic stress. WT and transgenic Arabidopsis seeds were sown on 1/2 MS medium containing 0 mM (A,D), 150 mM (B,E), and 300 mM mannitol (C,F). Panels (A–C) are the photos of germination status on media after 4 days; (D–F) are the diagrams of germination rate calculated for 4 days. Asterisks indicate a significant difference between WT and transgenic Arabidopsis lines (**p < 0.01; ***p < 0.001).
TaWRKY46-overexpressing Arabidopsis and WT seeds were grown vertically on 1/2 MS medium at 22 °C. For the mannitol treatment, 5-old-day seedlings were then grown on 1/2 MS medium supplemented with mannitol (0 mM, 150 mM, and 300 mM) in the same growth chamber for 10 days. WT plants grew a little better on 1/2 MS medium without mannitol than the TaWRKY46-overexpressing Arabidopsis plants (Figure 7A). In contrast, the overexpression lines grew noticeably better on the mannitol medium than those of the WT (Figure 7B,C). The root length results further confirmed the promoted tolerance of the overexpression of TaWRKY46 in transgenic Arabidopsis plants. The root lengths of the TaWRKY46-overexpressing Arabidopsis plants were longer than those of WT plants under mannitol treatments, although mannitol stress repressed the growth of both TaWRKY46-overexpressing Arabidopsis and WT plants (Figure 7D–F). However, the root lengths of WT and TaWRKY46-overexpressing Arabidopsis plants had no differences under 100 and 200 mM NaCl treatments (Supplementary Figure S1).
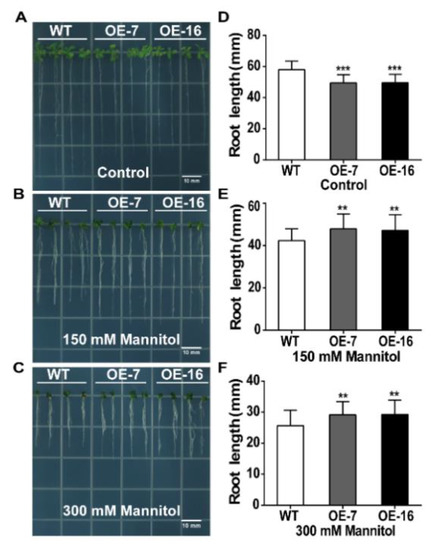
Figure 7.
Osmotic tolerance of TaWRKY46-overexpressing Arabidopsis plants. (A–C) The phenotype of seedlings after treatments with 0 mM, 150 mM, and 300 mM mannitol for 10 days. (D–F) The root length statistics of seedlings after treatments with 0 mM,150 mM, and 300 mM mannitol for 10 days. Asterisks indicate a significant difference between WT and transgenic Arabidopsis lines (** p < 0.01; *** p < 0.001). Scale bar = 10 mm.
2.5. TaWRKY46 Regulates the Expression of Stress-Related Genes Under Osmotic Stress in Arabidopsis Plants
The transcriptional levels of stress-related genes were detected in the 15-day-old WT and TaWRKY46-overexpressing transgenic Arabidopsis seedlings after mannitol stress. We selected six genes listed below for this experiment: Δ-1-pyrroline-5-carboxylate synthetase 1 (P5CS1), dehydration-responsive 29B (RD29B), dehydration-response element-binding protein 2A (DREB2A), ABA-response element (ABRE) binding factor 3 (ABF3), C repeat/dehydration-responsive element-binding factor 2 (CBF2), and CBF3. Compared with the WT plants, the six stress-related genes analyzed were prominently up-regulated in the TaWRKY46-overexpressing transgenic Arabidopsis lines when seedlings were exposed to 150 mM and 300 mM mannitol treatments (Figure 8). The results demonstrated that the overexpression of TaWRKY46 enhanced osmotic tolerance by inducing the expression of some stress-related genes in transgenic Arabidopsis plants.
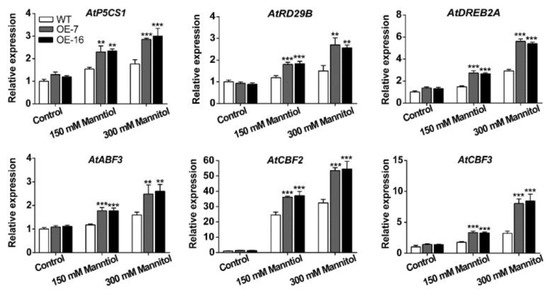
Figure 8.
The transcriptional levels of stress-related genes in TaWRKY46-overexpressing Arabidopsis plants after 0, 150 and 300 mM mannitol treatments for 10 days. Asterisks indicate a significant difference between WT and transgenic Arabidopsis lines (** p < 0.01; *** p < 0.001).
3. Discussion
WRKY TFs are one of the largest families of transcriptional regulators in plants, and have multiple developmental and physiological functions in response to environmental stresses [30]. In plants such as rice [31], soybean [16,32], cotton [33], Brachypodium distachyon [34,35], maize [36], and wheat [37], an especially large number of WRKY proteins have been identified in recent years.
In the present study, multiple sequence alignment showed that TaWRKY46 possessed the conserved WRKYGQK domain and a C2HC (CX7CX23HXC) motif (Figure 1D). The TaWRKY46-GFP fusion protein was exclusively localized to the nucleus of wheat protoplasts in a transient expression assay (Figure 3). It was in agreement with its putative role as a transcription factor and similar to previous reports on some other WRKY TFs [38,39]. Transcriptional activation analysis illuminated that the sequence of TaWRKY46 has transcriptional activation activity (Figure 4). It was in accordance with a senescence-associated transcription factor TaWRKY7 [15]. The transcriptional activation of truncated segments of TaWRKY46 illuminated that the C-terminal sequence of TaWRKY46 has transcriptional activation activity.
The WRKY TF members are known to be a vital section in drought stress and the ABA signal pathway [30,40]. The activation of the ABA signal transduction pathway also elevated plant drought tolerance by repressing transpiration, stomata opening, and the growth retardation factors [41]. But the expression patterns of TaWRKY46 seemed to be inconsistent under PEG and ABA treatments (Figure 2B,F). The underlying mechanism will need further exploration.
The structural conservation of WRKY proteins determines the functional specificity in regulating gene expression. Investigations showed that most WRKY proteins of the same group have a similar function in many plant species. Gene expression of TaWRKY46, which belonged to Group III, was induced in the leaf under PEG treatment (Figure 2B). It implied that TaWRKY46 might play a key role in response to osmotic stress. It was similar to the expression profiles of AtWRKY46 and AtWRKY63. AtWRKY46 plays important functions in responses to osmotic stress and stomatal movement [27]. AtWRKY63, a member of Group III, mediated responses to exogenous ABA and drought stress in Arabidopsis [42]. All these currently available data suggested that they may possess conserved functions in abiotic stress signaling.
The water loss rate of the detached leaves has been widely used to reflect drought tolerance in plants. The detached leaves of TaWRKY46-overexpressing Arabidopsis plants exhibited lower rates of water loss compared to the WT plants (Figure 5C). TaWRKY46-overexpressing Arabidopsis plants had the higher survival rate after drought treatment, and possessed the higher germination rate at different concentrations of mannitol (Figure 5E and Figure 6). Further phenotypic analysis showed that overexpression of TaWRKY46 altered the length of roots in transgenic Arabidopsis plants under osmotic stress (Figure 7). All the results suggested that overexpression of TaWRKY46 enhanced osmotic tolerance in transgenic Arabidopsis plants. Similar findings were also verified in some other WRKY transcription factors [22]. However, a remarkable difference was not observed in the root length of WT and TaWRKY46-overexpressing Arabidopsis plants after 100 mM and 200 mM NaCl treatments (Supplementary Figure S1). It was mainly because of the low induction of TaWRKY46 gene expression under NaCl treatment.
To gain further insight into the mechanism of action of TaWRKY46 in osmotic stress at the molecular level, six stress-related genes were detected at the transcriptional level. P5CS1 is one of the core enzymes involved in proline biosynthesis. Proline is a widely distributed osmolyte that regulates the osmotic balance [43]. Herein, we found that P5CS1 was greatly induced in transgenic Arabidopsis plants (Figure 8). It indicated that AtP5CS1 was possibly regulated by TaWRKY46. There was also a possibility that the resistance ability was given by WRKY SNPs mutations [44]. Fortunella crassifolia WRKY40 could also specifically bind to and activate the promoter of FcP5CS1 under salt stress [45]. The promoted expression of P5CS1 could lead to the accumulation of osmolytes that can maintain the osmotic capacity, thus sustaining the water content.
ABA, a vital hormone, is involved in plant gene regulation networks by an ABA-dependent or ABA-independent pathway when plants combat adverse environmental stresses [5,46]. The expression of ABF3 and RD29B (ABA-dependent) and DREB2A, CBF2, and CBF3 (ABA-independent) genes were enhanced in TaWRKY46-overexpressing transgenic Arabidopsis plants (Figure 8). It suggested that TaWRKY46 appeared to be a regulator involved both in ABA-dependent and ABA-independent pathways. Similar findings were also found in the functional characterizations of TaWRKY2, TaWRKY19, TaWRKY93, and TaMYB73 transcription factors [21,22,47]. It was partly because that a single WRKY TF can be involved in regulating several seemingly disparate processes [30,44]. These results were in accordance with previous findings that plants promoted their responses to abiotic stresses by enhancing the expression of stress-related genes [48,49,50,51,52,53]. Therefore, inducing stress-related genes may be the dominant mechanism to enhancing osmotic stress tolerance. If TaWRKY46 could directly bind to the upstream region of these genes is an idea that needs to be further explored, which will illuminate the underlying molecular mechanism of TaWRKY46 involved in the osmotic stress signal pathway in wheat.
4. Materials and Methods
4.1. Hydroponic Wheat and Stress Treatments
Winter wheat variety Zhengyin 1 (drought-sensitive) was utilized in the experiment. The seeds were sterilized in 75% ethanol (v/v) for 10 min, rinsed six times with distilled water, and germinated in a glass dish with wet filter paper (Zhejiang, China) for 36 h in the dark. The germinated seeds were transferred into pots filled with 1/2 Hoagland solution (Qingdao, China, Supplementary Figure S2A). The pots were cultured in a growth chamber with a day/night temperature of 25 °C/20 °C and 16 h/8 h photoperiod at 180 µmol m−2 s−1 illumination. The 1/2 Hoagland solution was refreshed every 2 days. When the second leaf fully expanded, 8-day-old seedlings (Supplementary Figure S2B) were treated with 1/2 Hoagland solution containing 20% PEG 6000, 200 mM NaCl, and 100 μΜ ABA for 24 h under the same chamber. Low- and high-temperature treatments were conducted at 4 °C and 42 °C chambers for 24 h, respectively. Seedlings cultivated in free 1/2 Hoagland solution were taken as a control. For the multiple stress experiments, the leaves were sampled at 0, 1, 3, 6, 12, and 24 h after PEG, NaCl, ABA, and low- and high-temperature treatments. For the organ expression analysis, the leaf, root, and stem were collected from control seedlings at 0 h. All the samples were immersed immediately into liquid nitrogen and stored at −80 °C for RNA extraction. The specific gene primers were listed in Supplementary Table S2.
4.2. Gene Isolation and Bioinformatics Analysis of TaWRKY46
The complete coding sequence of TaWRKY46 was amplified by PCR using specific primers (Supplementary Table S2). The PCR product was ligated to the pMD19-T vector (TaKaRa, Dalian, China) and then sequenced by the company (Sangon Biotech, Shanghai, China). The exon–intron structure of TaWRKY46 was obtained by the Gene Structure Display Server 2.0. TaWRKY46 and its highest homologies in other species including monocotyledonous grasses (barley, rice, and maize), and dicotyledonous soybean and Arabidopsis, were selected to generate a phylogenetic tree. The tree was constructed by MEGA 6 software using the Neighbor-Joining (NJ) method with 1000 bootstrap replicates. The accession numbers of the WRKYs were listed in Supplementary Table S1. The tertiary structure of TaWRKY46 protein was acquired through the Phyre2 tool (http://www.sbg.bio.ic.ac.uk/phyre2) and further edited by Chimera 1.11.2 software. Multiple sequence alignment was performed by DANMAN software (Lynnon Biosoft, San Ramon, California, USA).
4.3. Subcellular Localization Analysis of TaWRKY46
The cDNA encoding full-length TaWRKY46 without the stop codon was inserted into the pTF486-GFP vector [54] after it was cloned by PCR using specific primers (Supplementary Table S2) with BamHI restriction site. Wheat leaf protoplasts were isolated from 10-day-old seedlings according to the method of Shan [55]. pTF486-TaWRKY46-GFP was transformed into protoplasts by the PEG-mediated method and then incubated at 22 °C for 12 h in the dark. Empty vector was used as a control. GFP fluorescence signals were visualized using a laser confocal microscope (Andor, Belfast, UK) at an excitation wavelength of 488 nm.
4.4. Analysis of Transcriptional Activation in Yeast
The TaWRKY46 protein had 294 amino acid residues. The 1-191 amino acid residues contained the completed WRKY domain and C2HC zinc finger motif. The 191th amino acid position was chosen for truncation. The full-length coding sequence and truncated segments of TaWRKY46 were fused to the pGBKT7 vector [56] with EcoRI and BamHI restriction sites. The specific primers were listed in Supplementary Table S2. The various constructions were named pGBKT7-TaWRKY46 (1-294 aa), pGBKT7-TaWRKY46 (1-191 aa), and pGBKT7-TaWRKY46 (192-294 aa), respectively. Empty vector pGBKT7 was taken as a control. These constructions were introduced into the yeast strain AH109 by the lithium acetate method. The yeast cells were grown on selective medium (SD) medium without tryptophan (SD-W) and screened by PCR. The positive clones were cultured on the SD-W, SD without tryptophan, histidine, and adenine (SD-W/H/A) and SD-W/H/A with X-α-D-Galactosidase (X-α-gal) medium at 30 °C. The images were photographed after 3 d to detect the transcriptional activation.
4.5. Plant Transformation and Generation of TaWRKY46-Overexpressing Arabidopsis Plants
The complete coding sequence of TaWRKY46 was ligated into a binary vector pBI1304 [57] with NcoI and SpeI restriction sites under control of Cauliflower mosaic virus 35S (CaMV35S) promoter using the specific primers (Supplementary Table S2). The construction was introduced into Agrobacterium tumefaciens strain GV3101 by the freeze–thaw method. The floral dip method was used for generating transgenic Arabidopsis lines [58]. T1 plants were screened on 1/2 MS medium containing 1% sucrose, 1% agar, and 25 μg ml−1 hygromycin. T1 seedlings were confirmed by PCR and selfed through two more generations to generate T3 transgenic progeny. Sixteen independent transgenic T3 lines were acquired, and the transcriptional level of TaWRKY46 in the two selected overexpression (OE) lines was validated by PCR and qRT-PCR.
4.6. Water Loss Rate and Survive Rate Assays
Wild-type (WT) and TaWRKY46-overexpressing Arabidopsis seeds were sterilized in 10% (v/v) sodium hypochlorite (NaClO) for 10 min, rinsed six times with sterilized distilled water and vernalized at 4 °C for 3 days. Arabidopsis was grown on Jiffy-7-Peat Pellets (Jiffy Group) and cultivated in a growth chamber kept at 22 °C at 80 µmol m−2 s−1 illumination in a 16 h/8 h day/night cycle. For water loss rate assay, the detached leaves from 24-day-old WT and TaWRKY46-overexpressing Arabidopsis T3 seedlings were placed on filter papers at room temperature for 0, 1, 2, 3, 4, 5, 6, and 12 h, and the mass of the leaves was weighted at the indicated interval. Three replicates were performed. The 24-day-old seedlings were subjected to water withholding for 25 days, and 60 seedlings from each line were used to determine the survival rate.
4.7. Seed Germination Rate Assay
For the seed germination rate assay, WT and TaWRKY46-overexpressing transgenic Arabidopsis T3 seeds were sterilized and grown on 1/2 MS medium supplemented with 0 mM, 150 mM, and 300 mM mannitol for 4 days. The seed germination rate was recorded every day. At least 50 seeds were sown for one replicate of three parallel replicates. Three independent experiments of germination rate assay were performed. Measurements were repeated three times.
4.8. Stress Tolerance Assay and Expression Analysis of Stress-Related Genes in TaWRKY46-Overexpressing Arabidopsis Plants
WT and TaWRKY46-overexpressing Arabidopsis seeds were surface sterilized and planted on 1/2 MS medium containing 1% sucrose and 1% agar after 3 days of stratification at 4 °C. The plates were cultured vertically in a growth chamber at 22 °C under continuous illumination at 80 µmol m−2 s−1. For the salt stress assay, 5-day-old seedlings were transferred to 1/2 MS medium supplemented with 0, 100 and 200 mM NaCl. For the osmotic stress assay, 5-day-old seedlings were transferred to 1/2 MS medium supplemented with 0 mM, 150 mM, and 300 mM mannitol. The root lengths were measured using the software ImageJ after 10 days treatment. 15-day-old WT and transgenic Arabidopsis seedlings were collected from the 1/2 MS medium under 0, 150 and 300 mM mannitol treatments for further gene expression analysis. The expression of stress-related genes was detected by qRT-PCR. The specific primers were listed in Supplementary Table S2. Measurements were repeated three times.
4.9. RNA Extraction and qRT-PCR Analysis
Total RNA was isolated using the Trizol extraction protocol (Invitrogen, Carlsbad, California, USA) following the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 1 μg total RNA by the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China), and the quantitative real-time PCR (qRT-PCR) was carried out using SYBR® Premix Ex Taq™ II (TaKaRa, Dalian, China) on a CFX96 Touch RealTime PCR Detection System (BioRad, Hercules, California, USA). The specific gene primers were designed by Primer Premier 6.0 software and listed in Supplementary Table S2. The relative gene expression was calculated through the formula 2-ΔΔCT method [59]. Three biological and three technical replicates were performed in gene expression analysis. Three independent experiments were performed.
4.10. Statistical Analysis
Microsoft Office Excel 2013 was first used to analyze the data, and the mean values were calculated from three parallel experiments with their standard errors. Duncan’s method was performed using the SPSS Statistics 20.0 software. GraphPad Prism 7, Adobe Photoshop CC, and Adobe Illustrator software were used to generate charts.
5. Conclusions
A wheat Group III gene, TaWRKY46, was up-regulated by PEG treatment. Overexpression of TaWRKY46 activated the stress-related genes, increased the germination rate, and promoted the root growth in transgenic Arabidopsis plants under osmotic stress. These findings indicated that TaWRKY46 was a new positive regulator of osmotic stress. It played an important role in improving osmotic tolerance through ABA-dependent and ABA-independent pathways when plants were exposed to osmotic stress.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/4/1321/s1, Supplementary Materials consist of two tables and two figures. Supplementary Table S1: The accession numbers of the WRKYs used to construct phylogenetic tree in Figure 1. Supplementary Table S2: List of primers used in this study. F and R represent the forward and reverse primers from 5’ end to 3’ end. Supplementary Figure S1: Salt stress assay of TaWRKY46-overexpressing Arabidopsis plants. (A-C) The root length statistics of seedlings after treatments with 0 mM,100 mM, and 200 mM NaCl for 10 days. Lowercase letters represent significance at p < 0.05 between WT and transgenic Arabidopsis lines under the same condition, as determined by Duncan’s multiple range test. Supplementary Figure S2: The growth status of wheat seedlings. The overview of hydroponic wheat in plastic pots filled with 1/2 Hoagland solution (A) and the phenotype of 8-day-old seedlings before various treatments (B).
Author Contributions
X.L. and J.L. conceived and designed research. X.L. conducted experiments. X.L., Y.T., C.Z., and L.Z. analyzed the data. X.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No.31971835), Agricultural Science, Technology Innovation of Shaanxi Province Key Project (China, 2016NY-135), the Science and Technology Projects from Yangling Demonstration Zone of China (2018SF-05) and the National Key Research and Development Program of China (2017YFE0114000).
Acknowledgments
We thank Yanhua Dou, Shuangxi Zhang, and Ruihong Wang, College of Life Sciences, Northwest A & F University, for providing the WT Arabidopsis seed, Nicotiana benthamiana seed, and pBI1304 vector, respectively. We also thank the Teaching and Research Core Facility at the College of Life Sciences, NWAFU for support in this work.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Abbreviations
| ABF | ABA-response element-binding factor |
| CBF | C repeat/dehydration-responsive element-binding factor |
| GFP | green fluorescent protein |
| OE | overexpression |
| P5CS | Δ-1-pyrroline-5-carboxylate synthetase |
| RD | dehydration-responsive |
| TFs | transcription factors |
| WT | wild type |
References
- Abebe, T.; Melmaiee, K.; Berg, V.; Wise, R.P. Drought response in the spikes of barley: Gene expression in the lemma, palea, awn, and seed. Funct. Integr. Genomics 2010, 10, 191–205. [Google Scholar] [CrossRef]
- Bi, H.; Luang, S.; Li, Y.; Bazanova, N.; Morran, S.; Song, Z.; Perera, M.A.; Hrmova, M.; Borisjuk, N.; Lopato, S. Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 2016, 67, 5363–5380. [Google Scholar] [CrossRef] [PubMed]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liang, Z.S.; Xu, X.; Li, S.H.; Jing, J.H.; Monneveux, P. Relationships between carbon isotope discrimination and leaf morphophysiological traits in spring-planted spring wheat under drought and salinity stress in Northern China. Aust. J. Agric. Res. 2008, 59, 941–949. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Baloglu, M.C.; Inal, B.; Kavas, M.; Unver, T. Diverse expression pattern of wheat transcription factors against abiotic stresses in wheat species. Gene 2014, 550, 117–122. [Google Scholar] [CrossRef]
- Budak, H.; Kantar, M.; Kurtoglu, K.Y. Drought tolerance in modern and wild wheat. Sci. World J. 2013, 2013, 1–16. [Google Scholar] [CrossRef]
- Chen, W.J.; Zhu, T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004, 9, 591–596. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; et al. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 2005, 17, 944–956. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Li, M.; Zhao, D.; Yang, J.F.; Fu, J.D.; Xu, Z.S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of a-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Yousfi, F.E.; Makhloufi, E.; Marande, W.; Ghorbel, A.W.; Bouzayen, M.; Berges, H. Comparative analysis of WRKY genes potentially involved in salt stress responses in Triticum turgidum L. ssp. durum. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, M.; Song, Q.; Zhao, L.; Wang, G.; Zhou, C. Identification and function analyses of senescence-associated WRKYs in wheat. Biochem. Biophys. Res. Commun. 2016, 474, 761–767. [Google Scholar] [CrossRef]
- Wu, J.; Chen, J.; Wang, L.; Wang, S. Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Bhalla, P.L. Genetic engineering of wheat—Current challenges and opportunities. Trends Biotechnol. 2006, 24, 305–311. [Google Scholar] [CrossRef]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Ning, P.; Liu, C.; Kang, J.; Lv, J. Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ 2017, 5, 1–24. [Google Scholar] [CrossRef]
- Kage, U.; Yogendra, K.N.; Kushalappa, A.C. TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Liu, X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, F.; Tian, W.; Guo, Z.; Chen, X.; Xu, X.; Shang, H.; Hu, X. The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. PLoS ONE 2017, 12, e0181963. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, J.; Li, Y.; Rong, X.; Sun, J.; Sun, T.; Li, M.; Wang, L.; Feng, Y.; Chai, R.; et al. Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Wang, N.N.; Xu, S.W.; Sun, Y.L.; Liu, D.; Zhou, L.; Li, Y.; Li, X.B. The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Guo, R.; Wang, Y.; Guo, C.; Li, Z.; Chen, Z.; Gao, H.; Wang, X. Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tissue Organ. Cult. 2017, 132, 359–370. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Xu, H.; Watanabe, K.A.; Zhang, L.; Shen, Q.J. WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 2016, 23, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Langum, T.J.; Boken, A.K.; Rushton, D.L.; Boomsma, D.D.; Rinerson, C.I.; Rabara, J.; Reese, R.N.; Chen, X.; et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genomics 2012, 13, 270. [Google Scholar] [CrossRef]
- Wen, F.; Zhu, H.; Li, P.; Jiang, M.; Mao, W.; Ong, C.; Chu, Z. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 2014, 21, 327–339. [Google Scholar] [CrossRef]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Yang, J.F.; Li, M.; Xu, Z.S.; Fu, J.D. The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, S.; Meng, C.; Qin, L.; Kong, L.; Xia, G. WRKY transcription factors in wheat and their induction by biotic and abiotic stress. Plant Mol. Biol. Rep. 2013, 31, 1053–1067. [Google Scholar] [CrossRef]
- Yang, L.; Ye, C.; Zhao, Y.; Cheng, X.; Wang, Y.; Jiang, Y.Q.; Yang, B. An oilseed rape WRKY-type transcription factor regulates ROS accumulation and leaf senescence in Nicotiana benthamiana and Arabidopsis through modulating transcription of RbohD and RbohF. Planta 2018, 247, 1323–1338. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Fei, X.; Hou, L.; Shi, J.; Yang, T.; Liu, Y.; Wei, A. Patterns of drought response of 38 WRKY transcription factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 2018, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.-K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zheng, Y.; Luo, L.; Yang, Y.; Hu, X.; Kong, X. Functional FRIGIDA allele enhances drought tolerance by regulating the P5CS1 pathway in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 495, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Mondini, L.; Nachit, M.; Porceddu, E.; Pagnotta, M.A. Identification of SNP mutations in DREB1, HKT1, and WRKY1 genes involved in drought and salt stress tolerance in durum wheat (Triticum turgidum L. var durum). OMICS 2012, 16, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Hua, Z.M.; Yang, X.; Fromm, M.E. Activation of the NaCl- and drought-induced RD29A and RD29B promoters by constitutively active Arabidopsis MAPKK or MAPK proteins. Plant Cell Environ. 2006, 29, 1761–1770. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Choi, Y.-S.; Kim, Y.-M.; Hwang, O.-J.; Han, Y.-J.; Kim, S.Y.; Kim, J.-I. Overexpression of Arabidopsis ABF3 gene confers enhanced tolerance to drought and heat stress in creeping bentgrass. Plant Biotechnol. Rep. 2012, 7, 165–173. [Google Scholar] [CrossRef]
- Bihmidine, S.; Lin, J.; Stone, J.M.; Awada, T.; Specht, J.E.; Clemente, T.E. Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 2013, 237, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Hadi, F.; Gilpin, M.; Fuller, M.P. Identification and expression analysis of CBF/DREB1 and COR15 genes in mutants of Brassica oleracea var. botrytis with enhanced proline production and frost resistance. Plant Physiol. Biochem. 2011, 49, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, J.; Jia, M.; Xu, N.; Liang, S.; Shao, J.; Qi, Y.; Liu, X.; An, L.; Yu, F. Balance between cytosolic and chloroplast translation affects leaf variegation. Plant Physiol. 2018, 176, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Wang, Z.; Cheng, X.; Li, F.; Mei, F.; Chen, N.; Kang, Z. Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G.; et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).