Encoding Information into Polyethylene Glycol Using an Alcohol-Isocyanate “Click” Reaction
Abstract
1. Introduction
2. Results and Discussion
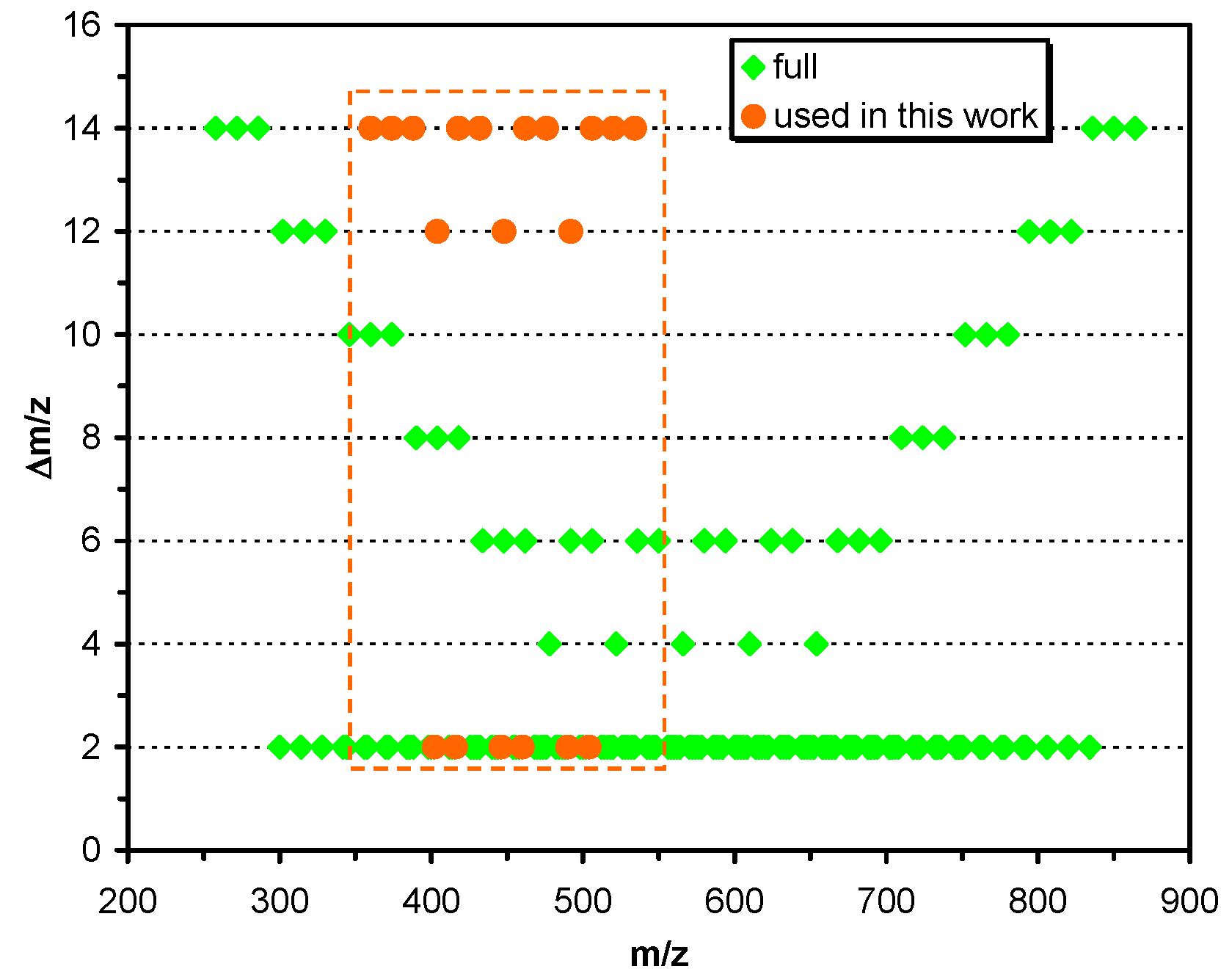
2.1. Theoretical Requirement for Encoding Oligomers Composed of Two Different Homologous Series
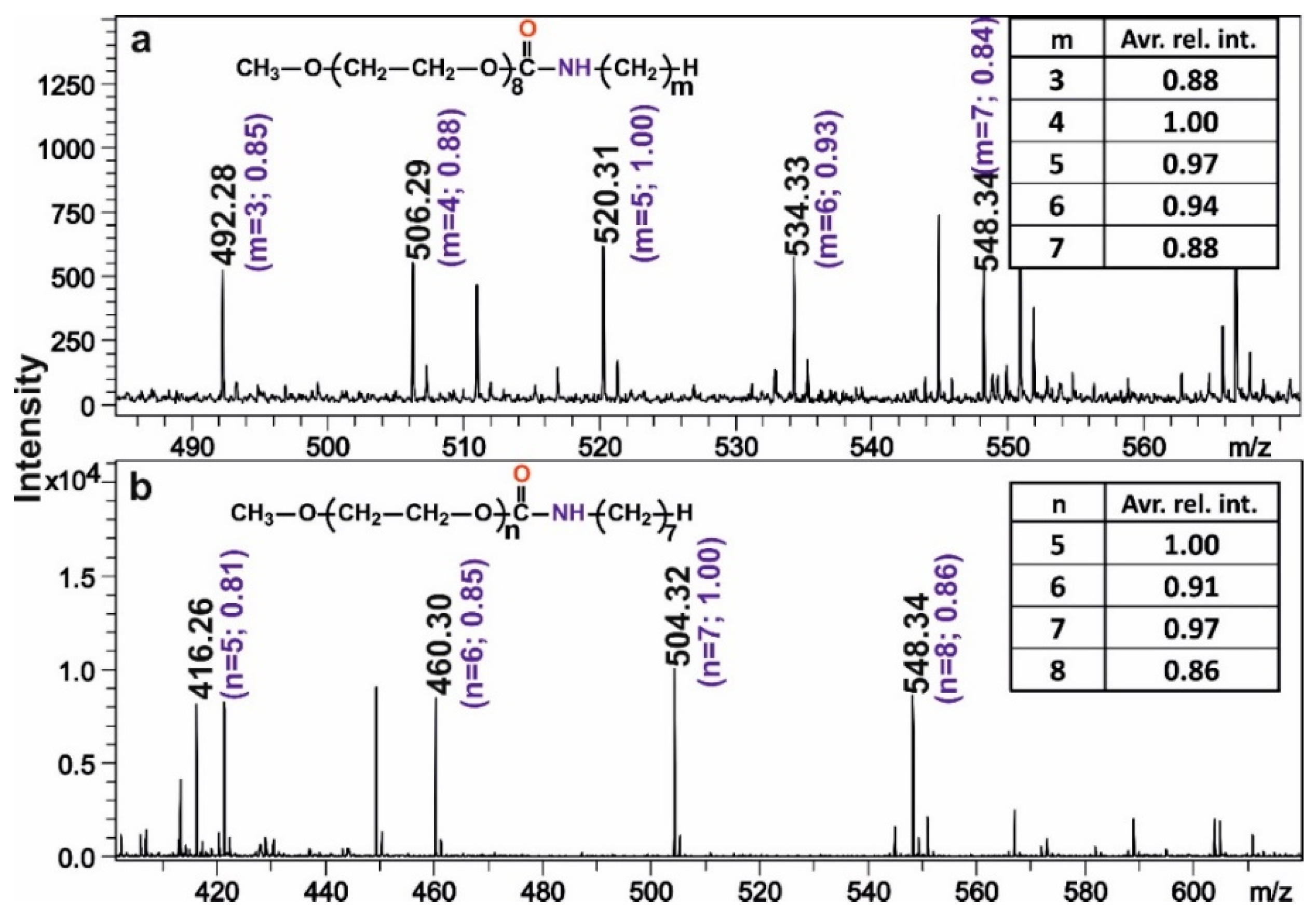
2.2. The Relative Ion Intensities of mPEG-OCONHR Oligomers
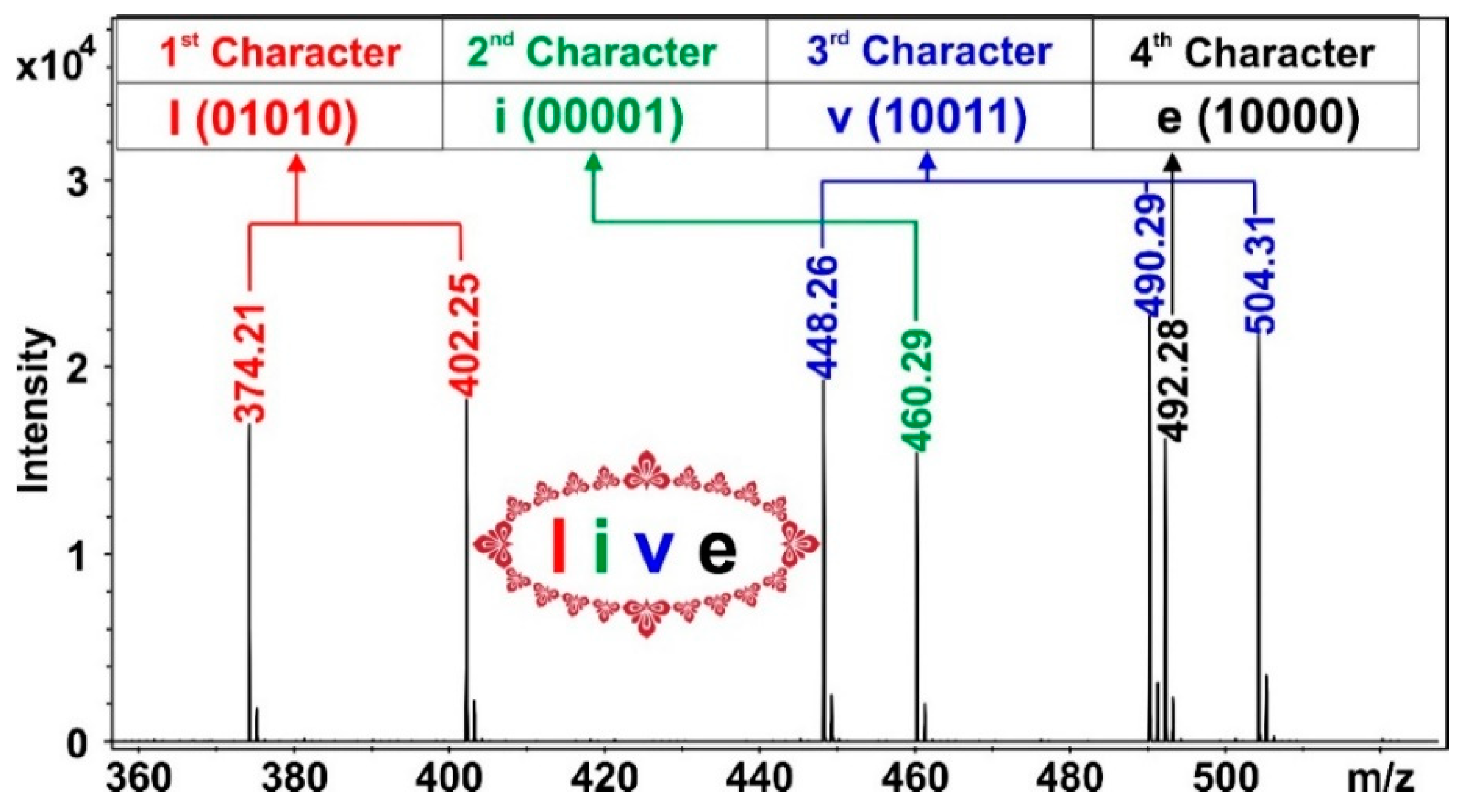
2.3. Storing Information in mPEG-OCONHR Oligomers
3. Experimental Section
3.1. Chemicals
3.2. Preparation of Encoding Oligomers
3.3. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
3.4. Reading Information by MALDI-TOF MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lape, P.V.; O’Connor, S.; Burningham, N. Rock art: A potential source of information about past maritime technology in the south-east Asia-Pacific region. Int. J. Naut. Archaeol. 2007, 36, 238–253. [Google Scholar] [CrossRef]
- Domenici, V.; Domenici, D. Talking Knots of the Inka. Archaeology 1996, 6, 199649. [Google Scholar]
- Colquhoun, H.; Lutz, J.F. Information-containing macromolecules. Nat. Chem. 2014, 6, 455–456. [Google Scholar] [CrossRef] [PubMed]
- De Silva, P.Y.; Ganegoda, G.U. New Trends of Digital Data Storage in DNA. Biomed. Res. Int. 2016. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.T.; Rutten, A.; Vaandrager, F.W.; Elemans, J.A.A.W.; Nolte, R.J.M. Encoding information into polymers. Nat. Rev. Chem. 2018, 2, 365–381. [Google Scholar]
- Cox, J.P. Long-term data storage in DNA. Trends Biotechnol. 2001, 19, 247–250. [Google Scholar] [CrossRef]
- Church, G.M.; Gao, Y.; Kosuri, S. Next-Generation Digital Information Storage in DNA. Science 2012, 337, 1628. [Google Scholar] [CrossRef]
- Anavy, L.; Vaknin, I.; Atar, O.; Amit, R.; Yakhini, Z. Data storage in DNA with fewer synthesis cycles using composite DNA letters. Nat. Biotechnol. 2019, 37, 1229–1239. [Google Scholar] [CrossRef]
- Meiser, L.C.; Antkowiak, P.L.; Koch, J.; Chen, W.D.; Kohll, A.X.; Stark, W.J.; Heckel, R.; Grass, R.N. Reading and writing digital data in DNA. Nat. Protoc. 2020, 15, 86–101. [Google Scholar] [CrossRef]
- Lutz, J.F.; Ouchi, M.; Liu, D.R.; Sawamoto, M. Sequence-Controlled Polymers. Science 2013, 341, 628–638. [Google Scholar] [CrossRef]
- Lutz, J.F. Coding Macromolecules: Inputting Information in Polymers Using Monomer-Based Alphabets. Macromolecules 2015, 48, 4759–4767. [Google Scholar] [CrossRef]
- Laure, C.; Karamessini, D.; Milenkovic, O.; Charles, L.; Lutz, J.F. Coding in 2D: Using Intentional Dispersity to Enhance the Information Capacity of Sequence-Coded Polymer Barcodes. Angew. Chem. Int. Ed. Engl. 2016, 55, 10722–10725. [Google Scholar] [CrossRef] [PubMed]
- Konig, N.F.; Al Ouahabi, A.; Oswald, L.; Szweda, R.; Charles, L.; Lutz, J.F. Photo-editable macromolecular information. Nat. Commun. 2019, 10, 3774–3783. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Landuyt, A.; Espeel, P.; Devreese, B.; Dawyndt, P.; Du Prez, F. Multifunctional sequencedefined macromolecules for chemical data storage. Nat. Commun. 2018, 9, 4451–4459. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Guo, B.; Huang, Y. Toward the synthesis of sequence-controlled vinyl copolymers. Chem. Commun. 2011, 47, 1455–1457. [Google Scholar] [CrossRef]
- Wei, P.; Li, B.; Leon, A.D.; Pentzer, E. Beyond binary: Optical data storage with 0, 1, 2, and 3 in polymerfilms. J. Mat. Chem. C 2017, 5, 5780–5786. [Google Scholar] [CrossRef]
- Green, J.E.; Choi, J.W.; Boukai, A.; Bunimovich, Y.; Johnston-Halperin, E.; DeIonno, E.; Luo, Y.; Sheriff, B.A.; Xu, K.; Shin, Y.S.; et al. A 160-kilobit molecular electronic memory patterned at 1011 bits per square centimetre. Nature 2007, 445, 414–417. [Google Scholar] [CrossRef]
- Al Ouahabi, A.; Amalian, J.A.; Charles, L.; Lutz, J.F. Mass spectrometry sequencing of long digital polymers facilitated by programmed inter-byte fragmentation. Nat. Commun. 2017, 8, 967–975. [Google Scholar] [CrossRef]
- Zydziak, N.; Konrad, W.; Feist, F.; Afonin, S.; Weidner, S.; Barner-Kowollik, C. Coding and decoding libraries of sequence-defined functional copolymers synthesized via photoligation. Nat. Commun. 2016, 7, 13672. [Google Scholar] [CrossRef]
- Mutlu, H.; Lutz, J.F. Reading Polymers: Sequencing of Natural and Synthetic Macromolecules. Angew. Chem. Int. Ed. 2014, 53, 13010–13019. [Google Scholar] [CrossRef]
- Kennedy, E.; Arcadia, C.E.; Geiser, J.; Weber, P.M.; Rose, C.; Rubenstein, B.M.; Rosenstein, J.K. Encoding information in syntheticmetabolomes. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, B.J.; Ten, A.S.; Fink, M.J.; Morey, S.; Preston, D.J.; Mrksich, M.; Whitesides, G.M. Storage of Information Using Small Organic Molecules. ACS Cent. Sci. 2019, 5, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Horský, J.; Walterová, Z. Fingerprint Multiplicity in MALDI-TOF Mass Spectrometry of Copolymers. Macromol. Symp. 2014, 339, 9–16. [Google Scholar] [CrossRef]
- Tanaka, H.; Leon-Garcia, A. Efficient run-length encodings. IEEE Trans. Inf. Theory 1982, 28, 880–890. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagy, L.; Kuki, Á.; Nagy, T.; Vadkerti, B.; Erdélyi, Z.; Kárpáti, L.; Zsuga, M.; Kéki, S. Encoding Information into Polyethylene Glycol Using an Alcohol-Isocyanate “Click” Reaction. Int. J. Mol. Sci. 2020, 21, 1318. https://doi.org/10.3390/ijms21041318
Nagy L, Kuki Á, Nagy T, Vadkerti B, Erdélyi Z, Kárpáti L, Zsuga M, Kéki S. Encoding Information into Polyethylene Glycol Using an Alcohol-Isocyanate “Click” Reaction. International Journal of Molecular Sciences. 2020; 21(4):1318. https://doi.org/10.3390/ijms21041318
Chicago/Turabian StyleNagy, Lajos, Ákos Kuki, Tibor Nagy, Bence Vadkerti, Zoltán Erdélyi, Levente Kárpáti, Miklós Zsuga, and Sándor Kéki. 2020. "Encoding Information into Polyethylene Glycol Using an Alcohol-Isocyanate “Click” Reaction" International Journal of Molecular Sciences 21, no. 4: 1318. https://doi.org/10.3390/ijms21041318
APA StyleNagy, L., Kuki, Á., Nagy, T., Vadkerti, B., Erdélyi, Z., Kárpáti, L., Zsuga, M., & Kéki, S. (2020). Encoding Information into Polyethylene Glycol Using an Alcohol-Isocyanate “Click” Reaction. International Journal of Molecular Sciences, 21(4), 1318. https://doi.org/10.3390/ijms21041318








