Analysis of Aldo–Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula
Abstract
1. Introduction
2. Results
2.1. Identification of AKR Genes in Medicago Truncatula
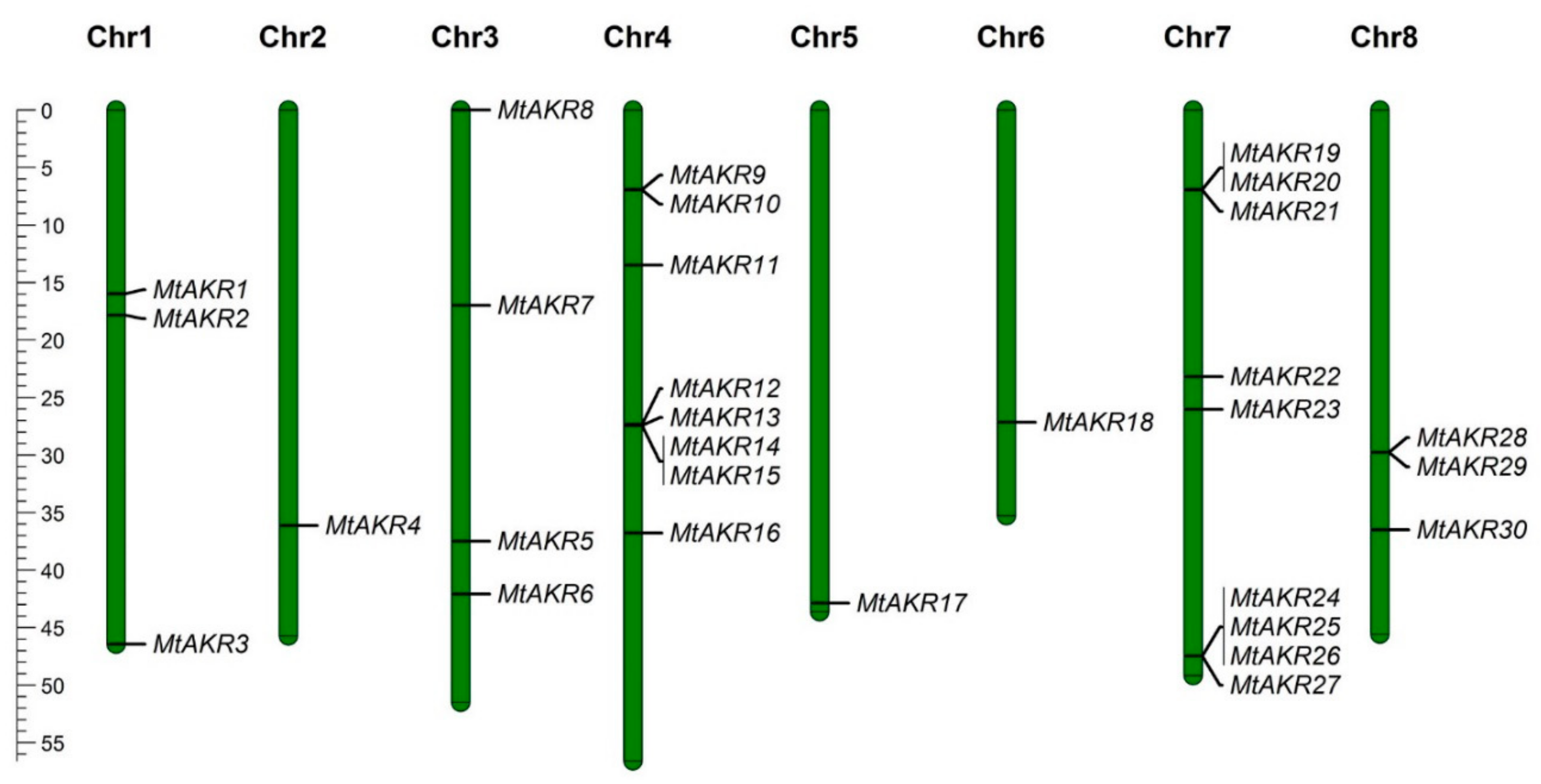
2.2. The Distribution of MtAKR Genes on the Chromosome
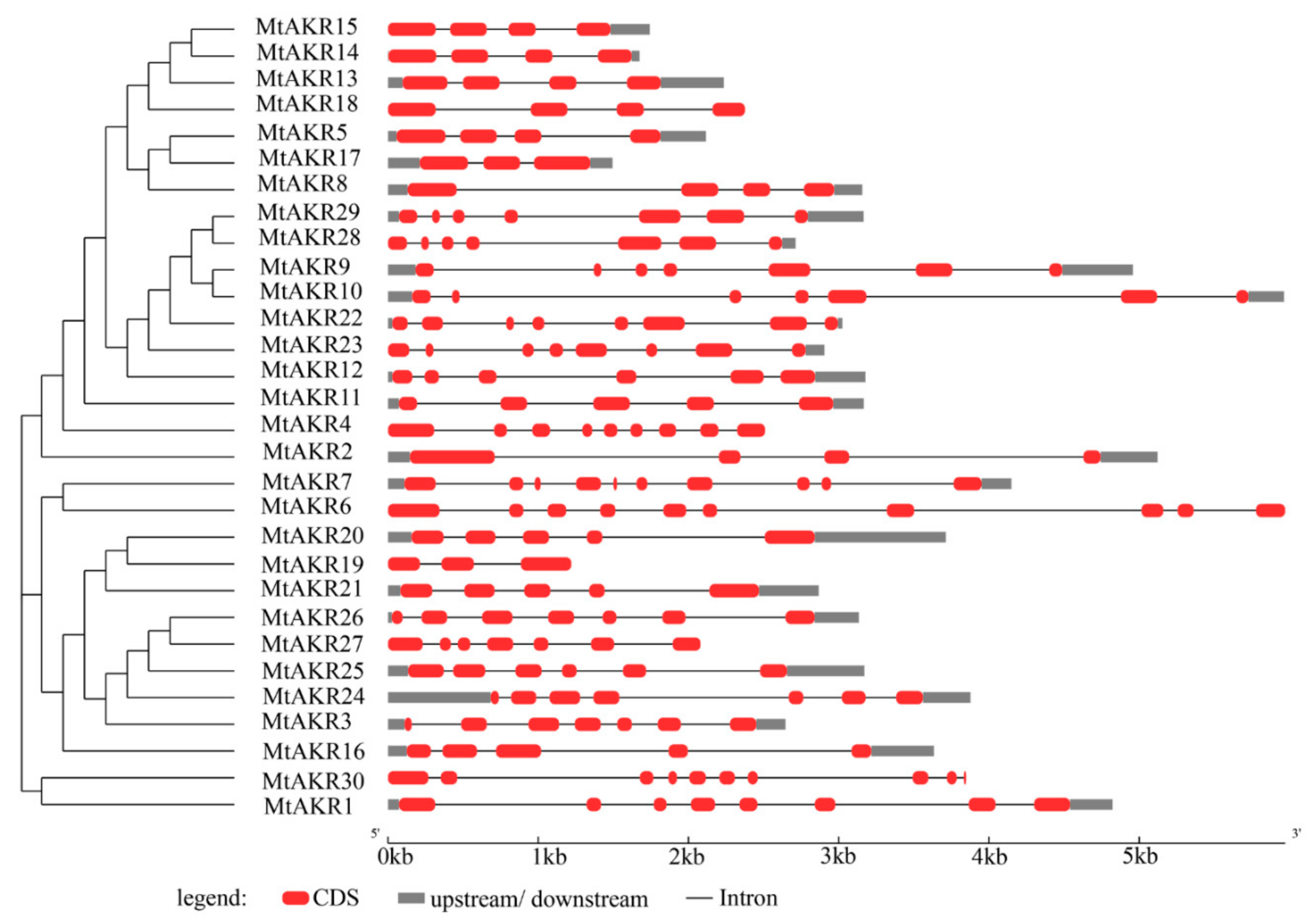
2.3. Structural Analysis of the MtAKR Genes
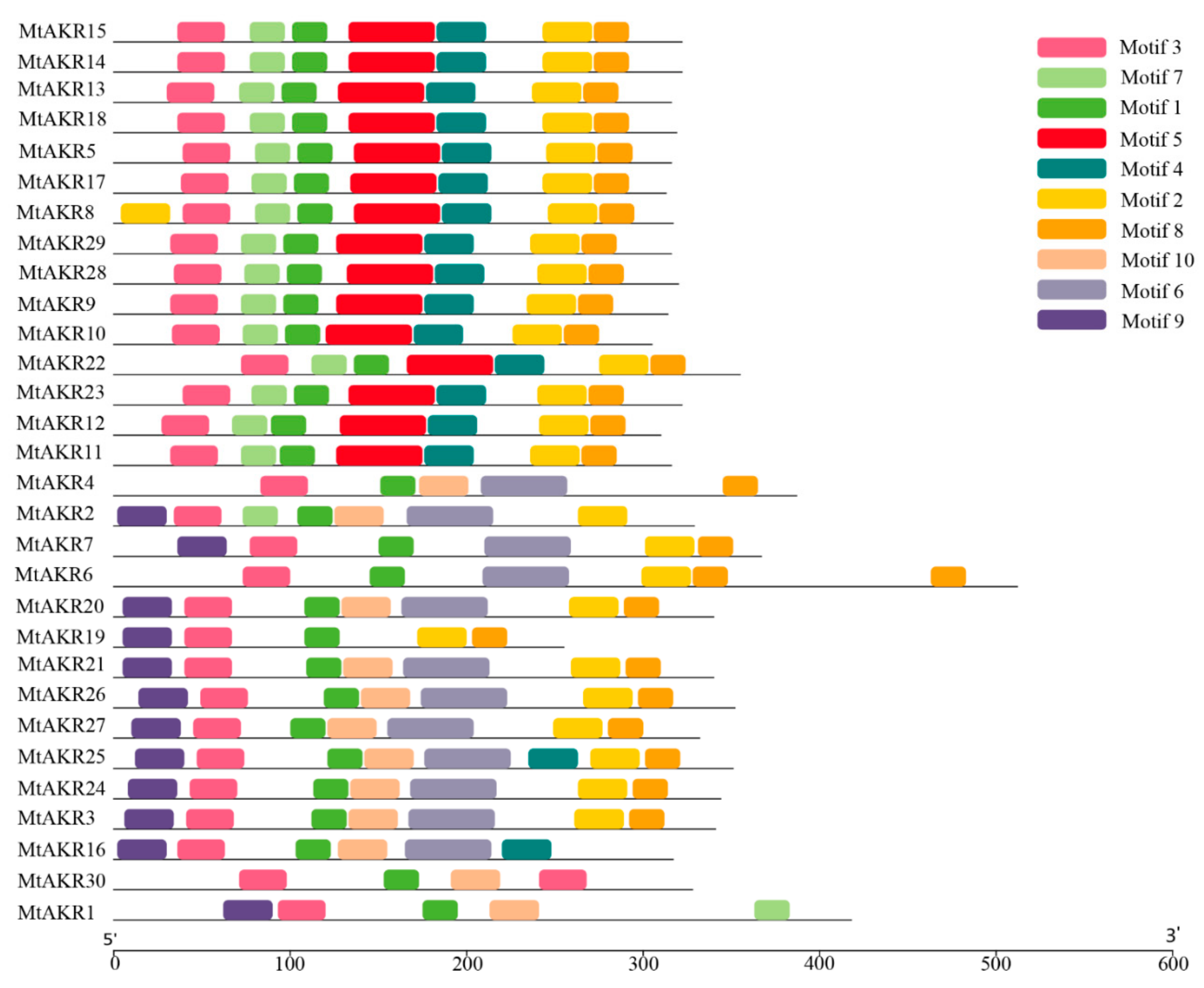
2.4. Conserved Motif in MtAKR Proteins
2.5. Cis-Acting Regulatory Elements in the Promoter of MtAKR Genes
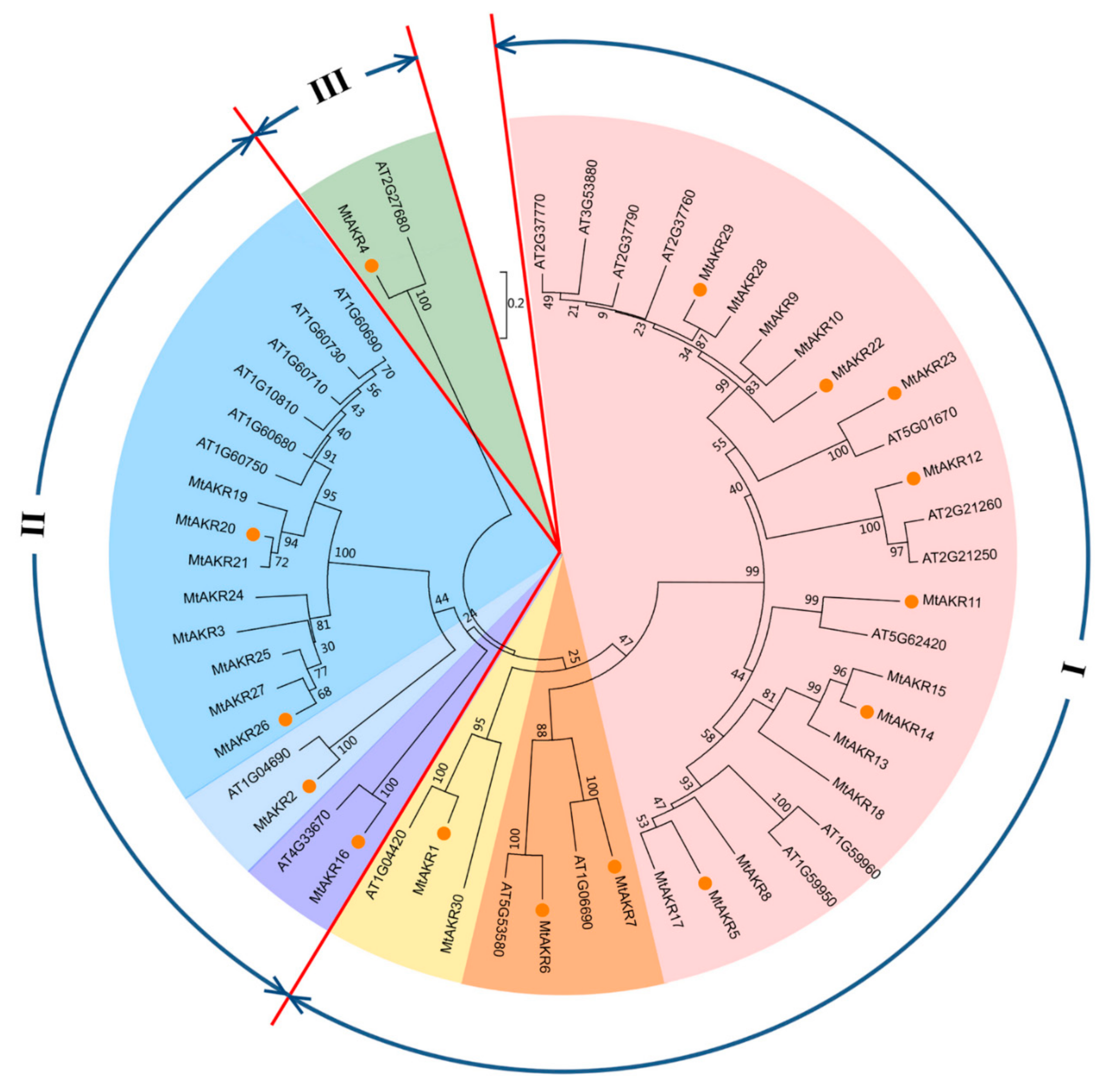
2.6. Phylogenetic Analysis of AKRs in M. Truncatula and A. Thaliana
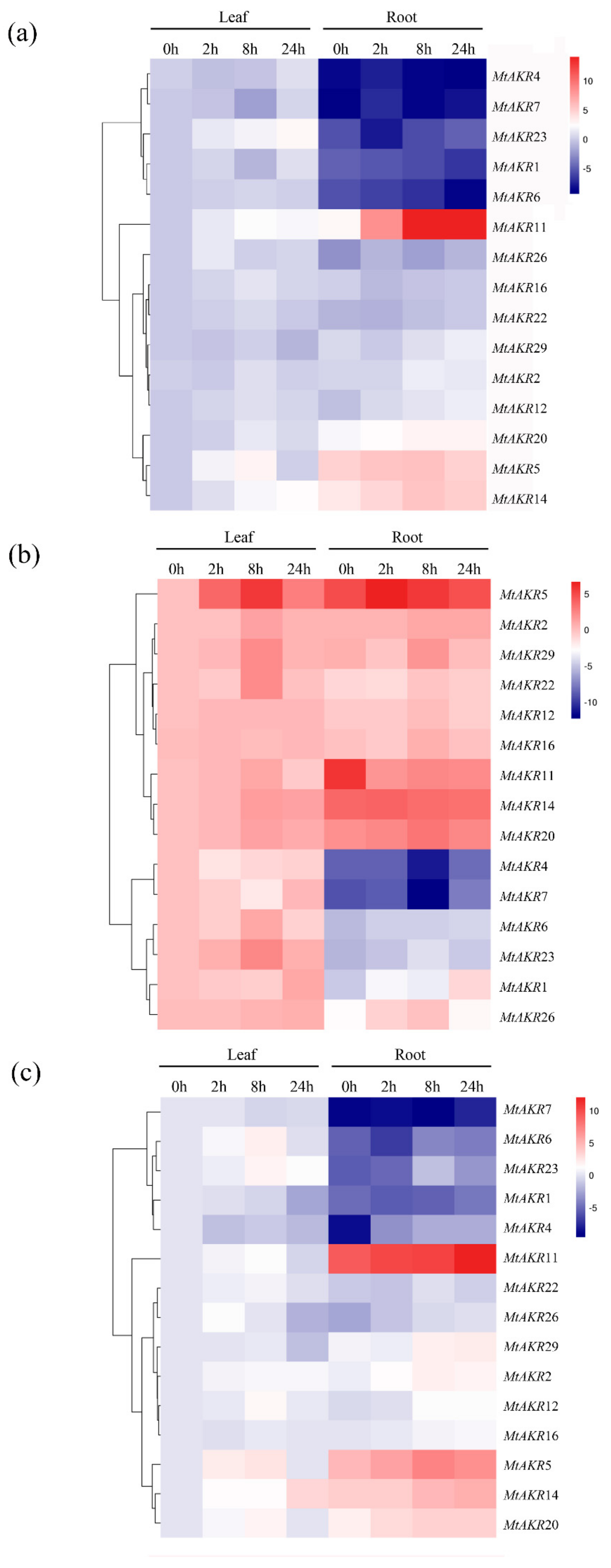
2.7. Expression Profiles of the MtAKR Genes in M. truncatula Leaves and Roots under ABA, PEG, and NaCl Treatment
3. Discussion
3.1. Characterization and Analysis of the MtAKR Gene Family
3.2. The Phylogenetic Analysis of Close Relationship between A. thaliana and M. truncatula AKR Genes and MtAKRs Expression Patterns
4. Materials and Methods
4.1. Identification, Chromosomal Localization, and Preliminary Analysis of the AKR Genes in Medicago truncatula Genome
4.2. Analysis of the Protein Features of the Medicago Truncatula AKRs
4.3. Structural Analysis of the AKRs
4.4. Motif Composition Analysis of MtAKR Proteins
4.5. Prediction of Cis-Acting Elements in MtAKRs
4.6. Phylogenetic of AKRs in Medicago truncatula and Arabidopsis thaliana
4.7. Plant Growth Condition and Treatments
4.8. RNA Preparation, cDNA Synthesis, and Expression Analysis by qRT-PCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Bartosz, G. Oxidative stress in plants. Acta Physiol. Plant. 1997, 19, 47–64. [Google Scholar] [CrossRef]
- Zeid, I.M.; Shedeed, Z.A. Response of alfalfa to putrescine treatment under drought stress. Biol. Plant. 2006, 50, 635–640. [Google Scholar] [CrossRef]
- Esmaeili, N.; Yang, X.; Cai, Y.; Sun, L.; Zhu, X.; Shen, G.; Payton, P.; Fang, W.; Zhang, H. Co-overexpression of AVP1 and OsSIZ1 in Arabidopsis substantially enhances plant tolerance to drought, salt, and heat stresses. Sci. Rep. 2019, 9, 7642. [Google Scholar] [CrossRef]
- Zorb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and crop yield. Plant Biol. (Stuttg.) 2019, 21 (Suppl. 1), 31–38. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Esmaeili, N.; O’Connell, M.; Zhang, J. Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crop. Prod. 2019, 130, 118–129. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef]
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef]
- Sengupta, D.; Naik, D.; Reddy, A.R. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. J. Plant Physiol. 2015, 179, 40–55. [Google Scholar] [CrossRef]
- Jin, Y.; Penning, T.M. Aldo-keto reductases and bioactivation/detoxication. Annu. Rev. Pharm. Toxicol. 2007, 47, 263–292. [Google Scholar] [CrossRef]
- Huo, J.; Bing, D.U.; Sun, S.; Shaozhen, H.E.; Zhao, N.; Liu, Q.; Zhai, H. A novel aldo-keto reductase gene, IbAKR, from sweet potato confers higher tolerance to cadmium stress in tobacco. Front. Agric. Sci. Eng. 2018, 5, 206–213. [Google Scholar] [CrossRef]
- Shimakawa, G.; Kohara, A.; Miyake, C. Medium-chain dehydrogenase/reductase and aldo-keto reductase scavenge reactive carbonyls in Synechocystis sp. PCC 6803. FEBS Lett. 2018, 592, 1010–1019. [Google Scholar] [CrossRef]
- Kanayama, Y.; Mizutani, R.; Yaguchi, S.; Hojo, A.; Ikeda, H.; Nishiyama, M.; Kanahama, K. Characterization of an uncharacterized aldo-keto reductase gene from peach and its role in abiotic stress tolerance. Phytochemistry 2014, 104, 30–36. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, P.; Yusuf, M.A.; Upadhyaya, C.P.; Roy, S.D.; Hohn, T.; Sarin, N.B. The Xerophyta viscosa aldose reductase (ALDRXV4) confers enhanced drought and salinity tolerance to transgenic tobacco plants by scavenging methylglyoxal and reducing the membrane damage. Mol. Biotechnol. 2013, 54, 292–303. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef]
- Young, N.D.; Frédéric, D.; Oldroyd, G.E.D.; Rene, G.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Jér?Me, G.; Heiko, S. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef]
- Bonnin, I.; Prosperi, J.M.; Olivieri, I. Comparison of quantitative genetic parameters between two natural populations of a selfing plant species, Medicago truncatula Gaertn. Appl. Genet. 1997, 94, 641–651. [Google Scholar] [CrossRef]
- Li, L.; Cheng, H.; Gai, J.; Yu, D. Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 2007, 226, 109–123. [Google Scholar] [CrossRef]
- Bianco, C.; Defez, R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 2009, 60, 3097–3107. [Google Scholar] [CrossRef]
- Lopez, M.; Herrera-Cervera, J.A.; Iribarne, C.; Tejera, N.A.; Lluch, C. Growth and nitrogen fixation in Lotus japonicus and Medicago truncatula under NaCl stress: Nodule carbon metabolism. J. Plant Physiol. 2008, 165, 641–650. [Google Scholar] [CrossRef]
- Filippou, P.; Antoniou, C.; Fotopoulos, V. Effect of drought and rewatering on the cellular status and antioxidant response of Medicago truncatula plants. Plant Signal Behav. 2011, 6, 270–277. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, R.; Xu, Y.; Wang, M.; Zhang, J.; Bai, M.; Han, C.; Xiang, F.; Wang, Z.Y.; Mysore, K.S.; et al. AGLF provides C-function in floral organ identity through transcriptional regulation of AGAMOUS in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2019, 116, 5176–5181. [Google Scholar] [CrossRef]
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Briere, C.; Moreau, S.; Carrere, S.; Blein, T.; Jardinaud, M.F.; Latrasse, D.; Zouine, M.; et al. Whole-genome landscape of Medicago truncatula symbiotic genes. Nat. Plants 2018, 4, 1017–1025. [Google Scholar] [CrossRef]
- Aubert, G.; Morin, J.; Jacquin, F.; Loridon, K.; Quillet, M.C.; Petit, A.; Rameau, C.; Lejeune-Hénaut, I.; Huguet, T.; Burstin, J. Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Appl. Genet. 2006, 112, 1024–1041. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, H.; Zhao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 2013, 32, 1543–1555. [Google Scholar] [CrossRef]
- Khan, Z.; Karamahmutoglu, H.; Elitas, M.; Yuce, M.; Budak, H. Through the looking glass: Real-Time Imaging in Brachypodium Roots and Osmotic Stress Analysis. Plants 2019, 8, 14. [Google Scholar] [CrossRef]
- Galvez, S.; Merida-Garcia, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramirez-Gonzalez, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef]
- Akpınar, B.A.; Lucas, S.J.; Budak, H. Genomics approaches for crop improvement against abiotic stress. Sci. World J. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome. Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Jez, J.M.; Flynn, T.G.; Penning, T.M. A new nomenclature for the aldo-keto reductase superfamily. Biochem. Pharmacol. 1997, 54, 639–647. [Google Scholar] [CrossRef]
- Simpson, P.J.; Tantitadapitak, C.; Reed, A.M.; Mather, O.C.; Bunce, C.M.; White, S.A.; Ride, J.P. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J. Mol. Biol. 2009, 392, 465–480. [Google Scholar] [CrossRef]
- Paven, M.C.M.; Viau, L.; Hamon, A.; Vandecasteele, C.; Pellizzaro, A.; Bourdin, C.; Laffont, C.; Lapied, B.; Lepetit, M.; Frugier, F. Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 2012, 62, 5595. [Google Scholar] [CrossRef]
- Cook, D.R. Medicago truncatula—A model in the making! Commentary. Curr. Opin. Plant. Biol. 1999, 2, 301–304. [Google Scholar] [CrossRef]
- Barker, D.G.; Bianchi, S.; Blondon, F.; Dattée, Y.; Duc, G.; Essad, S.; Flament, P.; Gallusci, P.; Génier, G.; Guy, P. Medicago truncatula, a model plant for studying the molecular genetics of theRhizobium-legume symbiosis. Plant Mol. Biol. Rep. 1990, 8, 40–49. [Google Scholar] [CrossRef]
- Mun, J.H.; Kim, D.J.; Choi, H.K.; Gish, J.; Debelle, F.; Mudge, J.; Denny, R.; Endre, G.; Saurat, O.; Dudez, A.M.; et al. Distribution of microsatellites in the genome of Medicago truncatula: A resource of genetic markers that integrate genetic and physical maps. Genetics 2006, 172, 2541–2555. [Google Scholar] [CrossRef]
- Hyndman, D.; Bauman, D.R.; Heredia, V.V.; Penning, T.M. The aldo-keto reductase superfamily homepage. Chem. Biol. Interact. 2003, 143, 621–631. [Google Scholar] [CrossRef]
- Rojas, B.; Wurman, J.; Zamudio, M.S.; Donoso, A.; Cabedo, P.; Díaz, F.; Stange, C.; Handford, M. AtA6PR1 and AtA6PR2 encode putative aldose 6-phosphate reductases that are cytosolically localized and respond differentially to cold and salt stress in Arabidopsis thaliana. J. Plant Biochem. Biotechnol. 2018. [Google Scholar] [CrossRef]
- Turoczy, Z.; Kis, P.; Torok, K.; Cserhati, M.; Lendvai, A.; Dudits, D.; Horvath, G.V. Overproduction of a rice aldo-keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol. Biol. 2011, 75, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.S.; Shin, K.H.; Kim, S.; Nam, K.H.; Lee, M.S.; Chun, J.Y.; Cheon, C.I. Overexpression of GmAKR1, a stress-induced aldo/keto reductase from soybean, retards nodule development. Mol. Cells 2009, 27, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Yue, R.; Sun, T.; Zhang, L.; Xu, L.; Tie, S.; Wang, H.; Yang, Y. Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front Plant Sci. 2015, 6, 73. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Vasconcelos, A.C.; Berkowitz, G.A. Evidence that plant K+ channel proteins have two different types of subunits. Plant Physiol. 1995, 109, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Ashley, M.K.; Grant, M.; Grabov, A. Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 2006, 57, 425–436. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Arisz, S.A.; Dekker, H.L.; Kramer, G.; de Koster, C.G.; Haring, M.A.; Munnik, T.; Testerink, C. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 2013, 450, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.Y.; Xue, H.W. Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 2018, 60, 851–863. [Google Scholar] [CrossRef]
- Oberschall, A.; Deak, M.K.; Sass, L.; Vass, I.; Kovacs, I.; Feher, A. A novel aldose/aldehyde reductase protects transgenic plants against lipidperoxidation under chemical and drought stresses. Plant J. 2000, 24, 437–446. [Google Scholar] [CrossRef]
- Vemanna, R.S.; Vennapusa, A.R.; Easwaran, M.; Chandrashekar, B.K.; Rao, H.; Ghanti, K.; Sudhakar, C.; Mysore, K.S.; Makarla, U. Aldo-keto reductase enzymes detoxify glyphosate and improve herbicide resistance in plants. Plant Biotechnol. J. 2017, 15, 794–804. [Google Scholar] [CrossRef]
- Pan, L.; Yu, Q.; Han, H.; Mao, L.; Nyporko, A.; Fan, L.; Bai, L.; Powles, S.B. Aldo-keto reductase metabolizes glyphosate and confers glyphosate resistance in Echinochloa colona. Plant Physiol. 2019, 181, 1519–1534. [Google Scholar] [CrossRef]
- Lee, S.P.; Chen, T.H. Molecular cloning of abscisic acid-responsive mRNAs expressed during the induction of freezing tolerance in bromegrass (Bromus inermis Leyss) suspension culture. Plant Physiol. 1993, 101, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Engelhardt, K.; Roncarati, R.; Schneider, K.; Rotter, M.; Salamini, F. An ABA and GA modulated gene expressed in the barley embryo encodes an aldose reductase related protein. EMBO J. 1991, 10, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Perin, E.C.; da Silva Messias, R.; Borowski, J.M.; Crizel, R.L.; Schott, I.B.; Carvalho, I.R.; Rombaldi, C.V.; Galli, V. ABA-dependent salt and drought stress improve strawberry fruit quality. Food Chem. 2019, 271, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Tran, C.D.; Chu, H.D.; Nguyen, K.H.; Watanabe, Y.; La, H.V.; Tran, K.D.; Tran, L.-S.P. Genome-Wide Identification of the TCP Transcription Factor Family in Chickpea (Cicer arietinum L.) and Their Transcriptional Responses to Dehydration and Exogenous Abscisic Acid Treatments. J. Plant Growth Regul. 2018, 37, 1286–1299. [Google Scholar] [CrossRef]
- Herrero, S.; González, E.; Gillikin, J.W.; Vélëz, H.; Daub, M.E. Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol. Biol. 2011, 76, 157. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BiorXiv 2018, 289660. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.D.P.; Pierre, R.; Stephane, R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]

| No. | Gene Name | gDNA Length a | cds Length a | Gene ID b | Protein Length c | Mass c (kDa) | PI c | GRAVY c | TargetP d |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MtAKR1 | 4822 | 1254 | MTR1g042730 | 417 | 46.65 | 7.58 | −0.251 | C* |
| 2 | MtAKR2 | 5122 | 987 | MTR1g047250 | 328 | 36.48 | 7.57 | −0.251 | M |
| 3 | MtAKR3 | 2646 | 1023 | MTR1g102750 | 340 | 38.02 | 5.50 | −0.250 | _ |
| 4 | MtAKR4 | 2510 | 1161 | MTR2g085125 | 386 | 43.44 | 8.15 | −0.300 | M |
| 5 | MtAKR5 | 2116 | 948 | MTR3g083130 | 315 | 35.16 | 5.70 | −0.175 | _ |
| 6 | MtAKR6 | 5971 | 1536 | MTR3g092140 | 511 | 56.96 | 9.20 | −0.109 | C* |
| 7 | MtAKR7 | 4149 | 1101 | MTR3g449790 | 366 | 40.25 | 8.98 | −0.303 | C |
| 8 | MtAKR8 | 3157 | 951 | MTR0374s0050 | 316 | 35.58 | 5.40 | −0.184 | C |
| 9 | MtAKR9 | 4958 | 942 | MTR4g021350 | 313 | 34.77 | 6.10 | −0.301 | _ |
| 10 | MtAKR10 | 5964 | 915 | MTR4g021410 | 304 | 33.97 | 6.00 | −0.290 | _ |
| 11 | MtAKR11 | 3166 | 948 | MTR4g036845 | 315 | 36.00 | 5.40 | −0.419 | M |
| 12 | MtAKR12 | 3177 | 930 | MTR4g072060 | 309 | 34.82 | 5.88 | −0.213 | _ |
| 13 | MtAKR13 | 2235 | 948 | MTR4g072320 | 315 | 35.84 | 5.96 | −0.314 | _ |
| 14 | MtAKR14 | 1674 | 966 | MTR4g072350 | 321 | 36.27 | 6.27 | −0.260 | _ |
| 15 | MtAKR15 | 1742 | 966 | MTR4g072360 | 321 | 36.55 | 6.42 | −0.274 | _ |
| 16 | MtAKR16 | 3633 | 951 | MTR4g092750 | 316 | 34.29 | 5.44 | −0.096 | _ |
| 17 | MtAKR17 | 1494 | 939 | MTR5g097910 | 312 | 34.89 | 5.93 | −0.231 | _ |
| 18 | MtAKR18 | 2376 | 957 | MTR6g073110 | 318 | 35.62 | 7.09 | −0.160 | _ |
| 19 | MtAKR19 | 1220 | 765 | MTR7g021670 | 254 | 28.05 | 5.90 | −0.321 | _ |
| 20 | MtAKR20 | 3713 | 1020 | MTR7g021680 | 339 | 37.40 | 5.77 | −0.285 | _ |
| 21 | MtAKR21 | 2867 | 1020 | MTR7g021850 | 339 | 37.66 | 5.60 | −0.318 | _ |
| 22 | MtAKR22 | 3024 | 1065 | MTR7g063580 | 354 | 39.51 | 6.46 | −0.106 | S* |
| 23 | MtAKR23 | 2904 | 966 | MTR7g070500 | 321 | 36.20 | 6.02 | −0.462 | _ |
| 24 | MtAKR24 | 3877 | 1032 | MTR7g114970 | 343 | 37.95 | 6.02 | −0.181 | _ |
| 25 | MtAKR25 | 3171 | 1053 | MTR7g114980 | 350 | 38.36 | 6.13 | −0.222 | _ |
| 26 | MtAKR26 | 3134 | 1056 | MTR7g114990 | 351 | 38.35 | 5.64 | −0.165 | _ |
| 27 | MtAKR27 | 2080 | 996 | MTR7g115010 | 331 | 36.33 | 5.25 | −0.191 | _ |
| 28 | MtAKR28 | 2713 | 960 | MTR8g070095 | 319 | 35.69 | 5.73 | −0.206 | _ |
| 29 | MtAKR29 | 3165 | 948 | MTR8g070115 | 315 | 34.92 | 6.46 | −0.242 | _ |
| 30 | MtAKR30 | 3848 | 984 | MTR8g088160 | 327 | 36.83 | 6.71 | −0.223 | M* |
| Motif Number | Length (Amino Acid) | Best Possible Match | Function |
|---|---|---|---|
| Motif 1 | 21 | CEASLKRLQLDYIDLYYIHWP | Aldo/keto reductase domain |
| Motif 2 | 29 | CTPAQIALRWGYQQGHDVCPKSFNTERMN | unmatched |
| Motif 3 | 28 | ISAIHHAIKAGYRHFDTADIYGNHEENG | Aldo/keto reductase domain |
| Motif 4 | 29 | PAVNQVEMHPSWHQDKLREFCNQKGIHVS | unmatched |
| Motif 5 | 50 | FKAEDMTPFDMKGTWKAMEECYRS GLARAIGVSNFSIKKLQDLLEYARIP | Aldo/keto reductase domain |
| Motif 6 | 50 | IRRAHAVHPITAVQMEWSLWTRDIE EEIIPLCRELGIGIVPYSPLGRGFF | Aldo/keto reductase domain |
| Motif 7 | 21 | NRDDLFITSKLWCTDHHPEDV | unmatched |
| Motif 8 | 21 | QNIGAFDWKLTQEDMRKISQI | unmatched |
| Motif 9 | 29 | PRMKLGTQGMEVSKQGFGCMGMSGFYNPP | unmatched |
| Motif 10 | 29 | DTSVPIEDTMGELKKLVEEGKIKYIGLSE | Aldo/keto reductase domain |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Sun, H.; Zhang, J.; Hou, Y.; Zhang, T.; Kang, J.; Wang, Z.; Yang, Q.; Long, R. Analysis of Aldo–Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula. Int. J. Mol. Sci. 2020, 21, 754. https://doi.org/10.3390/ijms21030754
Yu J, Sun H, Zhang J, Hou Y, Zhang T, Kang J, Wang Z, Yang Q, Long R. Analysis of Aldo–Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula. International Journal of Molecular Sciences. 2020; 21(3):754. https://doi.org/10.3390/ijms21030754
Chicago/Turabian StyleYu, Jie, Hao Sun, Jiaju Zhang, Yiyao Hou, Tiejun Zhang, Junmei Kang, Zhen Wang, Qingchuan Yang, and Ruicai Long. 2020. "Analysis of Aldo–Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula" International Journal of Molecular Sciences 21, no. 3: 754. https://doi.org/10.3390/ijms21030754
APA StyleYu, J., Sun, H., Zhang, J., Hou, Y., Zhang, T., Kang, J., Wang, Z., Yang, Q., & Long, R. (2020). Analysis of Aldo–Keto Reductase Gene Family and Their Responses to Salt, Drought, and Abscisic Acid Stresses in Medicago truncatula. International Journal of Molecular Sciences, 21(3), 754. https://doi.org/10.3390/ijms21030754





