Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
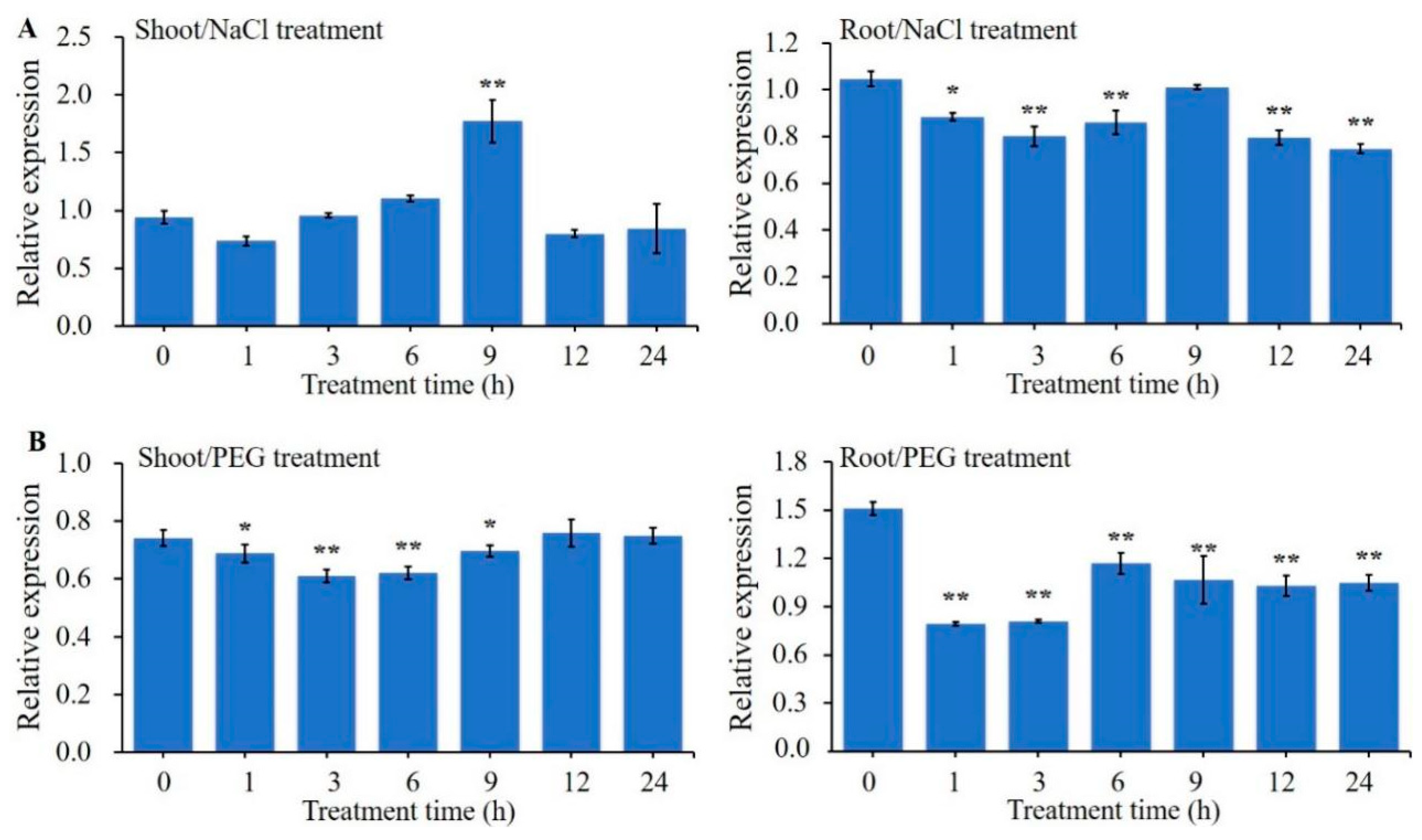
2.1. Expression Analysis of ZmBES1/BZR1-5 Gene
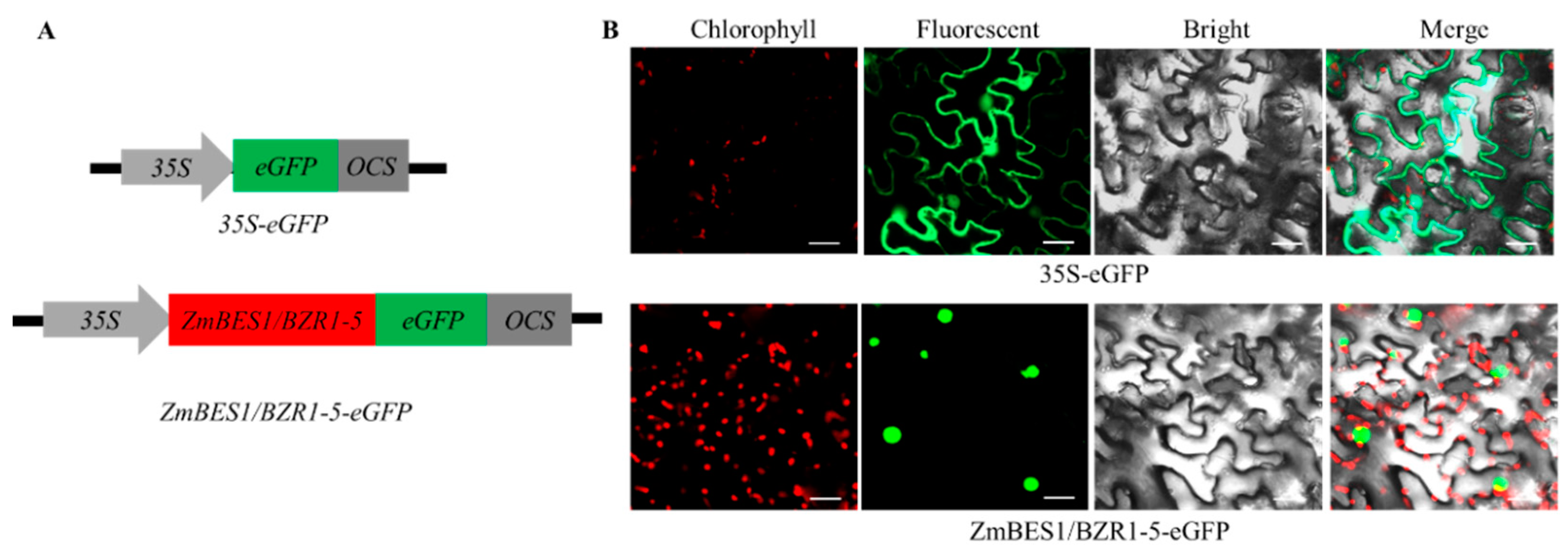
2.2. Subcellular Localization of ZmBES1/BZR1-5
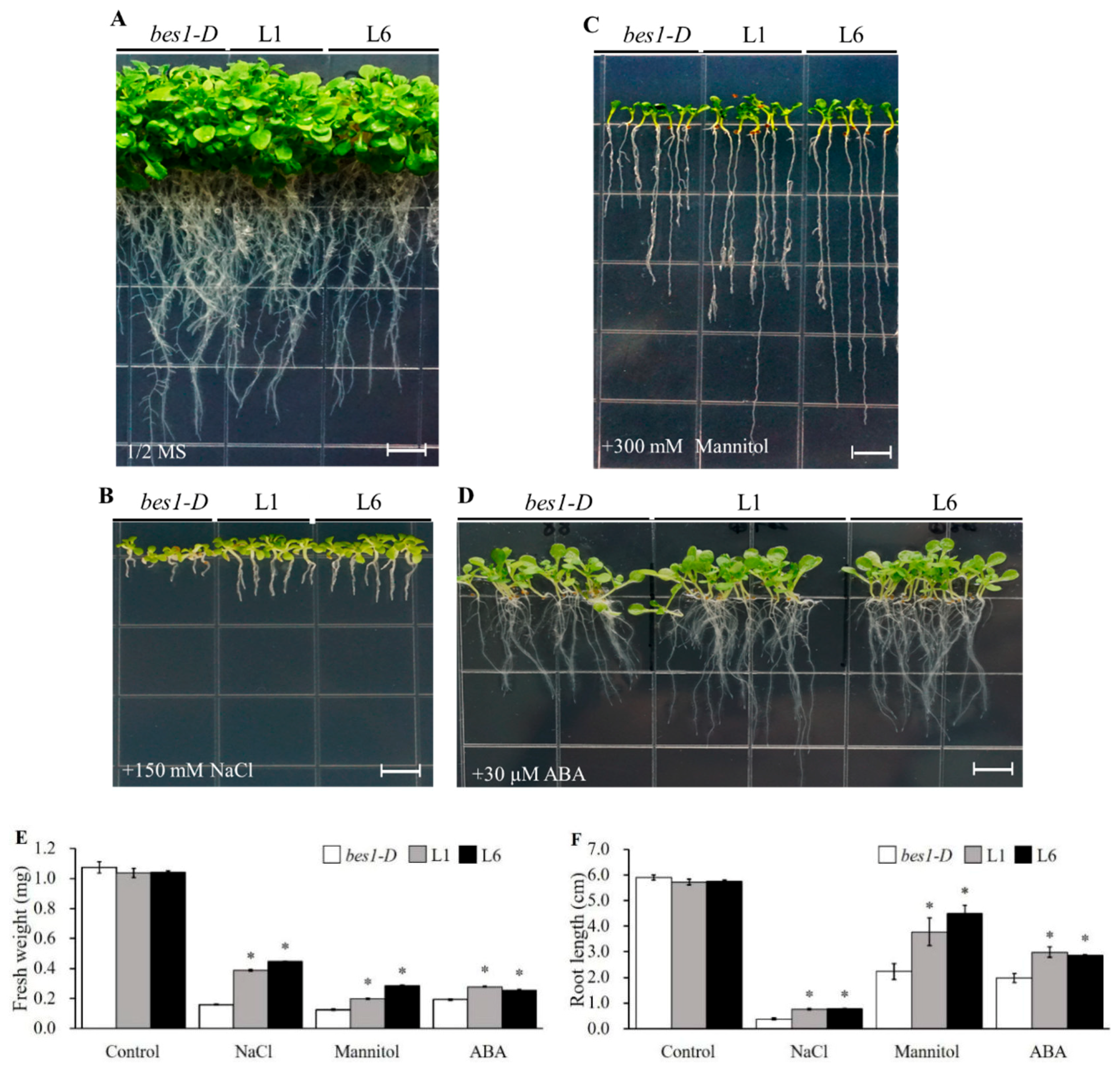
2.3. ZmBES1/BZR1-5 Facilitates Growth Promotion, and Reduces ABA Sensitivity in Arabidopsis
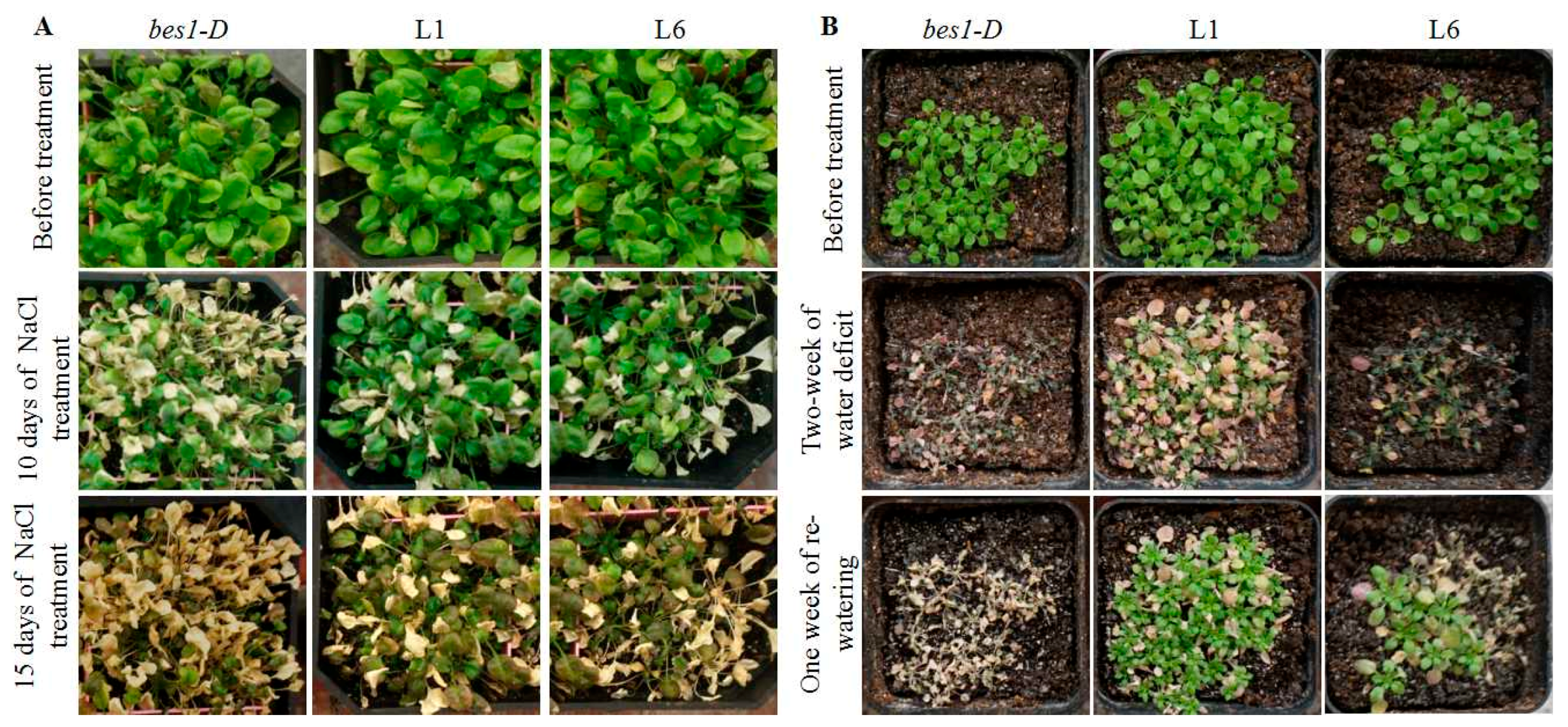
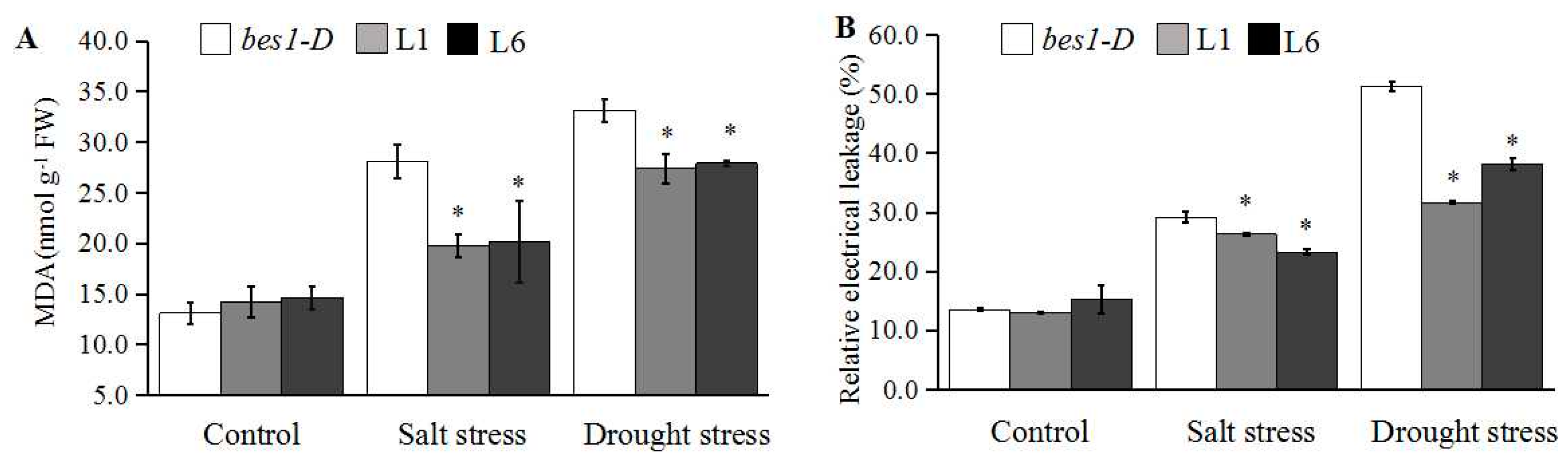
2.4. ZmBES1/BZR1-5 Enhances Salt and Drought Tolerance in Arabidopsis
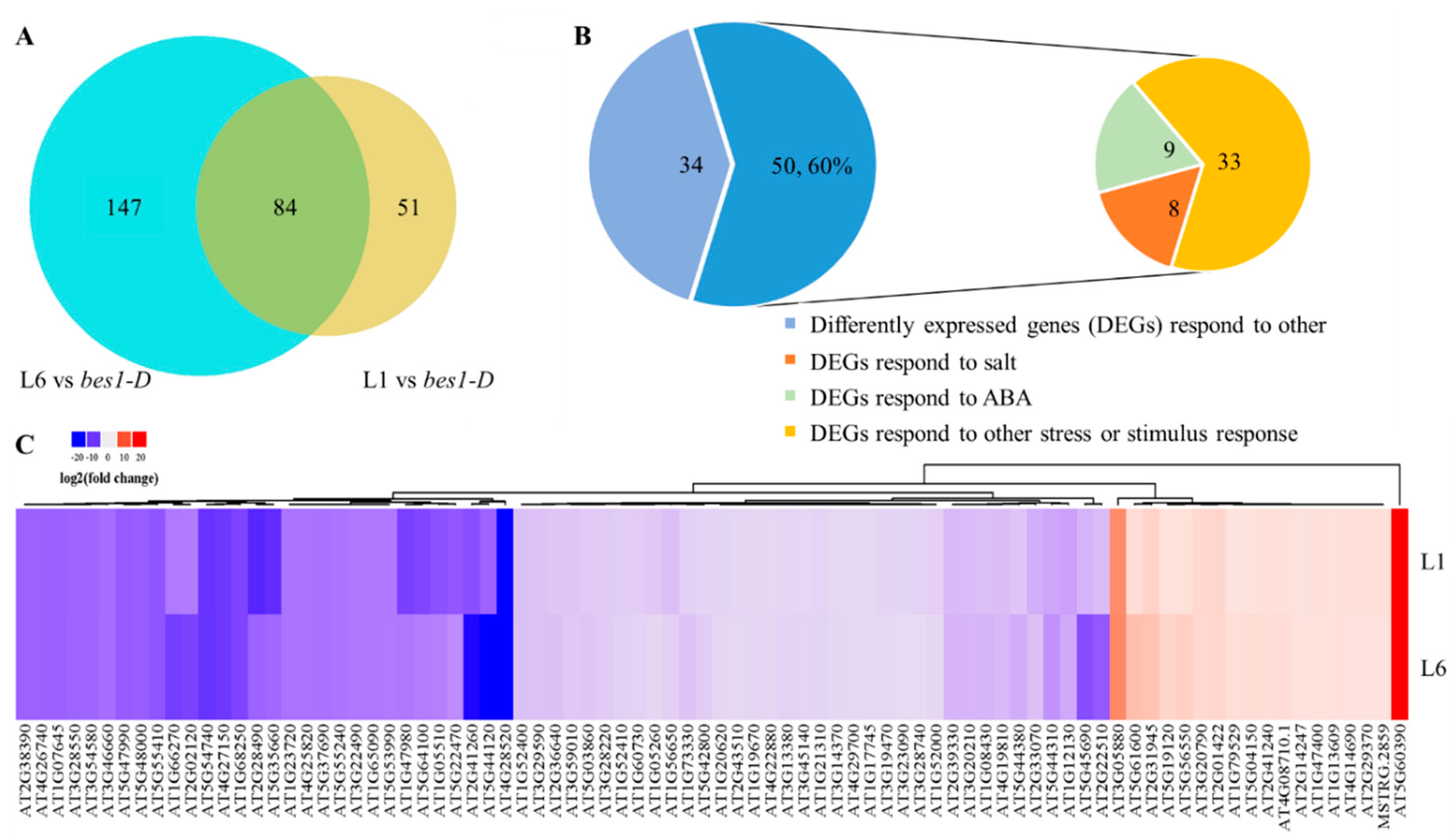
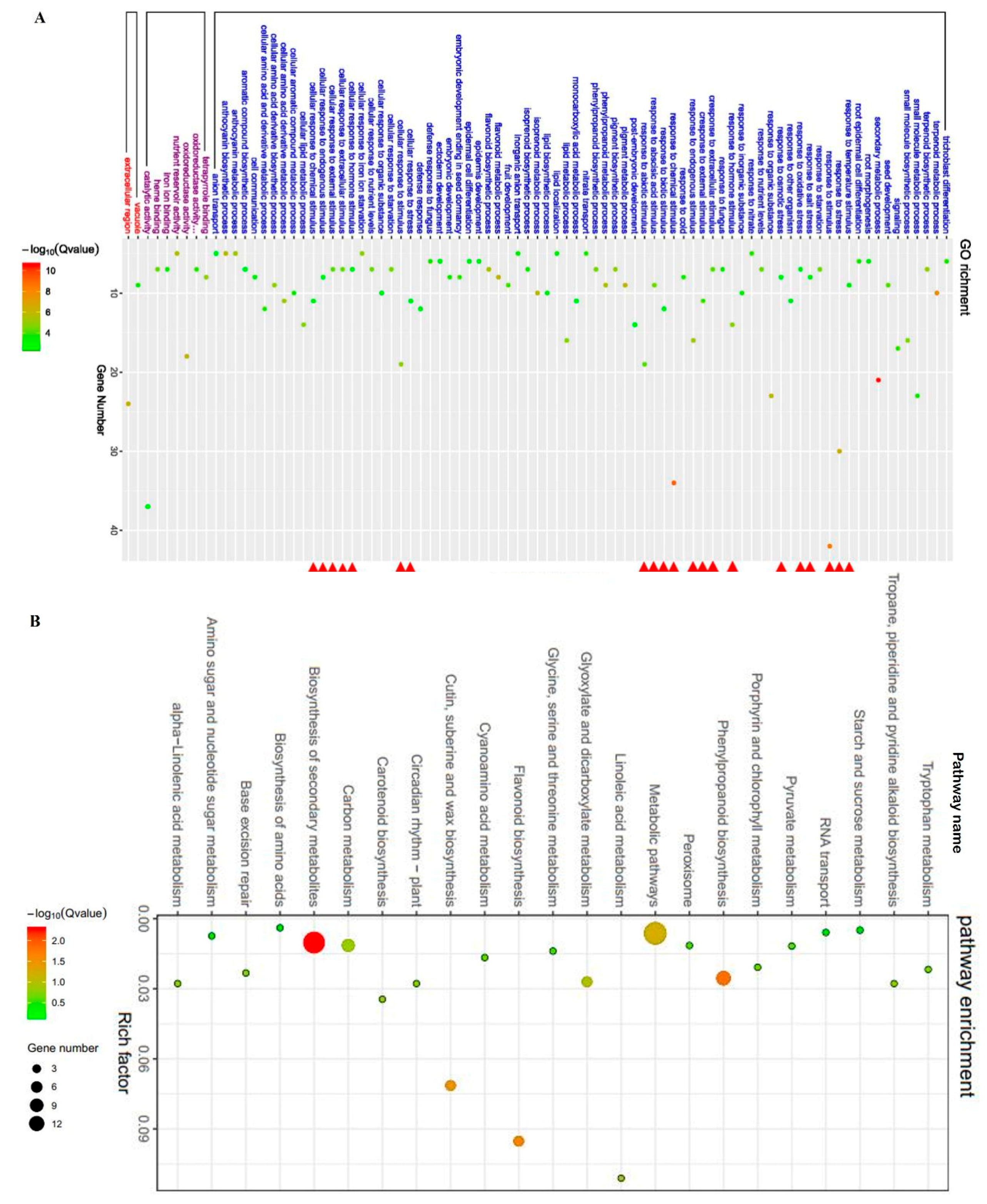
2.5. ZmBES1/BZR1-5 Regulates the Expression of Stress-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Material Preparation and Expression Analysis
4.3. Subcellular Localization of ZmBES1/BZR1-5 in Tobacco
4.4. Transformation of Arabidopsis
4.5. Determination of Root Length and Fresh Weight
4.6. Salt and Drought Treatment
4.7. Determination of MDA Content and REL
4.8. RNA Extraction and RNA Sequencing
4.9. Yeast One Hybrid Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maggio, A.; Bressan, R.A.; Zhao, Y.; Park, J.; Yun, D.J. It’s hard to avoid avoidance: Uncoupling the evolutionary connection between plant growth, productivity and stress “tolerance”. Int. J. Mol. Sci. 2018, 19, 3671. [Google Scholar] [CrossRef] [PubMed]
- Keyster, M.; Klein, A.; Ludidi, N. Caspase-like enzymatic activity and the ascorbate-glutathione cycle participate in salt stress tolerance of maize conferred by exogenously applied nitric oxide. Plant. Signal. Behav. 2012, 7, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Zong, N.; Li, X.J.; Wang, L.; Wang, Y.; Wen, H.T.; Li, L.; Zhang, X.; Fan, Y.L.; Zhao, J. Maize ABP2 enhances tolerance to drought and salt stress in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 15–29. [Google Scholar] [CrossRef]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Cheng, H. Abp9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant. Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Amtmann, A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2009, 2, 3–12. [Google Scholar] [CrossRef]
- Cabot, C.; Sibole, J.V.; Barcelo, J.; Poschenrieder, C. Lessons from crop plants struggling with salinity. Plant Sci. 2014, 226, 2–13. [Google Scholar] [CrossRef]
- Soma, F.; Mogami, J.; Yoshida, T.; Abekura, M.; Takahashi, F.; Kidokoro, S.; Mizoi, J.; Shinozaki, K.; Shinozaki, Y.K. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 2017, 3, 16204. [Google Scholar] [CrossRef]
- Joo, J.; Lee, Y.H.; Song, S.I. OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 2018, 249, 1521–1533. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, D.; Wang, P.; Ma, X.; Shan, L. Regulation of Arabidopsis brassinosteroid receptor bri1 endocytosis and degradation by plant u-box pub12/pub13-mediated ubiquitination. Proc. Nat. Acad. Sci. USA 2018, 115, E1906–E1915. [Google Scholar] [CrossRef]
- Nolan, T.; Chen, J.; Yin, Y. Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J. Crop seed size: BR matters. Mol. Plant 2017, 10, 668–669. [Google Scholar] [CrossRef] [PubMed]
- Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J.; Kinoshita, T.; Cano-Delgado, A.; Yoshida, S. Binding of brassinosteroids to the extracellular domain of plant receptor kinase bri1. Nature 2005, 433, 167–171. [Google Scholar]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase bri1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Martinez, C.; Espinosa-Ruiz, A.; Lucas, M.; Bernardo-Garcia, S.; Franco-Zorrilla, J.M.; Prat, S. PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 2018, 37, e99552. [Google Scholar] [CrossRef]
- Ibanez, C.; Delker, C.; Martinez, C.; Burstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids dominate hormonal regulation of plant thermosmorphogenesis via BZR1. Curr. Biol. 2018, 28, 303–310. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.L.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Feng, W.Q.; Sun, F.A.; Zhang, Y.Y.; Qu, J.T.; Liu, B.L.; Lu, F.Z.; Yang, L.; Fu, F.L.; Li, W.C. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Li, Q.F.; Lu, J.; Yu, J.W.; Zhang, C.Q.; He, J.X.; Liu, Q.Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multi signal-regulated plant growth. BBA Gene Regul. Mech. 2018, 1861, 561–571. [Google Scholar]
- Tong, H.; Chu, C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018, 23, 1016–1028. [Google Scholar] [CrossRef]
- Lv, B.; Tian, H.; Zhang, F.; Liu, J.; Lu, S.; Bai, M.; Li, C.; Ding, Z. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018, 14, e1007144. [Google Scholar] [CrossRef]
- Zhao, X.; Li, R.D.; Gong, Z.Z.; Wang, X.F.; Mao, T.L. BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol. 2018, 221, 908–918. [Google Scholar] [CrossRef]
- He, G.; Liu, J.; Dong, H.; Sun, J. The blue light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol. Plant 2019, 12, 689–703. [Google Scholar] [CrossRef]
- Liang, T.; Mei, S.; Shi, C.; Shi, C.; Yang, Y.; Peng, Y.; Ma, L.; Wang, F.; Li, X.; Huang, X.; et al. UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 2018, 44, 512–523. [Google Scholar] [CrossRef]
- Wu, J.; Wang, W.; Xu, P.; Pan, J.; Zhang, T.; Li, Y.; Li, G.; Yang, H.; Lian, H. PhyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol. 2019, 60, 353–366. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ye, K.; Shi, Y.; Cheng, J.; Zhang, X.; Yang, S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 2017, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signaling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Kang, S.; Yang, F.; Li, L.; Chen, H.; Chen, S.; Zhang, J. The Arabidopsis transcription factor BES1 is a direct substrate of MPK6 and regulates immunity. Plant Physiol. 2015, 167, 1076–1086. [Google Scholar] [CrossRef]
- Singh, A.P.; Fridman, Y.; Friedlander-Shani, L.; Tarkowska, D.; Strnad, M.; Savaldi-Goldstein, S. Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol. 2014, 166, 678–688. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.J.; Wang, K.X.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Zhou, J. BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation tolerance in tomato. Plant Physiol. 2019, 179, 671–685. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Cagirici, H.B.; Alptekin, B.; Budak, H. RNA sequencing and co-expressed long non-coding RNA in modern and wild wheats. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Gálvez, S.; Mérida-García, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramírez-González, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef]
- Mitsuya, S.; Taniguchi, M.; Miyake, H.; Takabe, T. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta 2005, 222, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Long, R.; Zhang, F.; Li, Z.; Li, M.; Cong, L.; Kang, J.; Zhang, T.; Zhao, Z.; Sun, Y.; Yang, Q. Isolation and functional characterization of salt-stress induced RCI2-like genes from Medicago sativa and Medicago truncatula. J. Plant Res. 2015, 128, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Kang, J.G.; Nou, I.S. Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 2015, 92, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Guo, G.; Yan, H.; Qiu, Z.; Liu, Q.; Zeng, B. Characterization of Brassinazole resistant (BZR) gene family and stress induced expression in Eucalyptus grandis. Physiol. Mol. Biol. Plants 2018, 24, 821–831. [Google Scholar] [CrossRef]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W.; et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef]
- Salazar-Henao, J.E.; Lehner, R.; Betegón-Putze, I.; Vilarrasa-Blasi, J.; Caño-Delgado, A.I. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4951–4961. [Google Scholar] [CrossRef]
- Martins, S.; Montiel-Jorda, A.; Cayrel, A.; Huguet, S.; Roux, C.P.; Ljung, K.; Vert, G. Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 2017, 8, 309. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef]
- Breusegem, F.V.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2016, 141, 384–390. [Google Scholar] [CrossRef]
- Zhang, S.; Cai, Z.; Wang, X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, H.; Soyk, S.; Simkova, K.; Hostettler, C.; Marafino, J.; Mainiero, S.; Vaughan, C.K.; Monroe, J.D.; Zeemana, S.C. β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 2011, 23, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Simkova, K.; Zurcher, E.; Luginbuhl, L.; Brand, L.H.; Vaughan, C.K.; Wanke, D.; Zeeman, S.C. The enzyme-like domain of Arabidopsis nuclear β-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 2014, 26, 1746–1763. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L.Z. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef]
- Lima, J.M.; Nath, M.; Dokku, P.; Raman, K.V.; Kulkarni, K.P.; Vishwakarma, C.S.P.; Sahoo, U.B.; Mohapatra, S.V.; Amitha, M.V.; Chinnusamy, S.; et al. Physiological, anatomical and transcriptional alterations in a rice mutant leading to enhanced water stress tolerance. AoB Plants 2015, 7, plv023. [Google Scholar] [CrossRef]
- Desfeux, C.; Clough, S.J.; Bent, A.F. Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 2000, 123, 895–904. [Google Scholar] [CrossRef]
- Yu, H.Q.; Zhou, X.Y.; Wang, Y.G.; Zhou, S.F.; Fu, F.L.; Li, W.C. A betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus enhances tolerance of Arabidopsis to high salt and drought stresses. Plant Growth Regul. 2017, 83, 265–276. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with hisat, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, F.; Yu, H.; Qu, J.; Cao, Y.; Ding, L.; Feng, W.; Khalid, M.H.B.; Li, W.; Fu, F. Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 996. https://doi.org/10.3390/ijms21030996
Sun F, Yu H, Qu J, Cao Y, Ding L, Feng W, Khalid MHB, Li W, Fu F. Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2020; 21(3):996. https://doi.org/10.3390/ijms21030996
Chicago/Turabian StyleSun, Fuai, Haoqiang Yu, Jingtao Qu, Yang Cao, Lei Ding, Wenqi Feng, Muhammad Hayder Bin Khalid, Wanchen Li, and Fengling Fu. 2020. "Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis" International Journal of Molecular Sciences 21, no. 3: 996. https://doi.org/10.3390/ijms21030996
APA StyleSun, F., Yu, H., Qu, J., Cao, Y., Ding, L., Feng, W., Khalid, M. H. B., Li, W., & Fu, F. (2020). Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis. International Journal of Molecular Sciences, 21(3), 996. https://doi.org/10.3390/ijms21030996





