Abstract
The endocannabinoid system (ES) is a cell-signalling system widely distributed in biological tissues that includes endogenous ligands, receptors, and biosynthetic and hydrolysing machineries. The impairment of the ES has been associated to several pathological conditions like behavioural, neurological, or metabolic disorders and infertility, suggesting that the modulation of this system may be critical for the maintenance of health status and disease treatment. Lifestyle and environmental factors can exert long-term effects on gene expression without any change in the nucleotide sequence of DNA, affecting health maintenance and influencing both disease load and resistance. This potentially reversible “epigenetic” modulation of gene expression occurs through the chemical modification of DNA and histone protein tails or the specific production of regulatory non-coding RNA (ncRNA). Recent findings demonstrate the epigenetic modulation of the ES in biological tissues; in the same way, endocannabinoids, phytocannabinoids, and cannabinoid receptor agonists and antagonists induce widespread or gene-specific epigenetic changes with the possibility of trans-generational epigenetic inheritance in the offspring explained by the transmission of deregulated epigenetic marks in the gametes. Therefore, this review provides an update on the epigenetics of the ES, with particular attention on the emerging role in reproduction and fertility.
1. Introduction
The endocannabinoid system (ES) is a complex cell-signalling system identified in the early 1990s following studies on the phytocannabinoid Δ9-tetrahydrocannabinol (Δ9THC), the main psychoactive constituent of the marijuana plant Cannabis sativa [1]. It is widely distributed in biological tissues and is involved in many physiological activities such as pain control, motor functions, thermogenesis, sleep/wake cycle, learning and memory, synaptic plasticity, emotional (mood) regulation, stress response, food intake, inflammatory response, lipid and glucose metabolism, heart function, successful gametogenesis, and reproduction, amongst others [2]. The impairment of ES activity has been linked to several pathological conditions, from behavioural, neurological, and metabolic disorders to infertility and cancer, emphasizing the relevance of the pharmacological modulation of this system for the preservation of health status and the treatment of diseases [2,3]. Indeed, the high expression of the ES in brain areas playing a key role in conditioning processes such as drug-seeking behaviour, smoking, and alcohol addiction, emphasises that the ES is widely sensible to environmental epigenetic cues [4,5,6].
In this respect, epigenetics can be defined as the overall biological processes changing gene expression without any change in the nucleotide DNA sequence [7]. This occurs through the chemical modification of DNA and histone protein tails or through the specific production of regulatory non-coding RNA (ncRNA) [8]. The epigenetic signature is first defined in the embryo during development and cell differentiation, and is remodelled during the life course as a direct consequence of lifestyle and environment with impact on health or disease status [9,10]. Therefore, nutritional status, diet, alcohol addiction, physical activity, stress, and exposure to pollutants, pesticides, or endocrine disruptors, among other aspects, can epigenetically affect gene expression. Epigenetic marks can be delivered among tissues within exosomes, extracellular vesicles, or microvesicles, suggesting the existence of new communication routes in which the products of specific cell types may affect gene expression in different target tissues [11,12,13]. However, recent evidence revealed the transfer of epigenetic marks from gametes to the embryo [14,15] with three possibilities of epigenetic inheritance: (1) cross-generational effects or intergenerational inheritance, when the F1 generation is affected as a consequence of in utero or paternal exposure to environmental cues; (2) multigenerational inheritance when F1 and F2 generations are affected; and (3) trans-generational effects when more than three generations stably present the phenotype caused by epigenetic changes [16].
Therefore, the aim of this review is to provide an update on the epigenetic modulation of the ES and the possible ES-dependent epigenetic effects on gene expression, focusing on the emerging role of the ES in male reproduction and fertility.
2. ES in Summary
In 1964 Gaoni and Mechoulam identified Δ9THC as the main biologically active constituent of marijuana plant [17] giving the start to a number of studies aimed at identifying its biological targets and the related mechanisms. Hence, in the 1990s the identification of an endogenous ES composed of ligands, receptors, biosynthetic and hydrolysing enzymes, and possible membrane transporters led to the design and synthesis of high-affinity molecules to differentially modulate this endogenous signalling system [1,2,18].
The main endogenous cannabinoids (“endocannabinoids”), anandamide (AEA) and 2-arachydonoylglicerol (2AG), are derivatives of the n-6 poly unsaturated fatty acid (PUFA) arachidonic acid (ARA), and share some of the effects of Δ9THC [1]. They bind the main endocannabinoid receptors, the central type 1 and the peripheral type 2 cannabinoid receptors (CB1 and CB2), which are membrane G-coupled receptors. AEA has also intracellular binding sites for the cationic channel type 1 vanilloid receptor (TRPV1) and for the nuclear peroxisome proliferator-activated receptor γ (PPARγ), whereas 2-AG binds to specific γ-aminobutyric acid (GABA) receptor A subtypes in neuronal cells [1,2,18]
Apart from endocannabinoid biosynthesis, which is mediated by the N-acyl-phosphatidylethanolamine-specific phospholipase D (NAPE-PLD) and the sn-1-diacylglycerol lipases (DAGLα and DAGLβ) for AEA and 2-AG, respectively, the key feature of endocannabinoid signalling is the modulation of endocannabinoid tone. In this respect, the fatty acid amide hydrolases (FAAH1 and FAAH2) preferentially hydrolyse AEA (and 2-AG to a lesser extent), while the monoacylglycerol lipase (MAGL) is particularly active in hydrolysing 2-AG [19].
On the basis of unexpected evidence for intracellular reservoirs and transporters of endocannabinoids, the classical “dogma” that endocannabinoids—and in particular AEA—are synthesized and released on demand via hydrolysis of cell membrane phospholipid precursors has been revisited as recently reviewed [20]. AEA can be stored in lipid droplets (adiposomes) in association with FAAH1 and FAAH2, and intracellular AEA transporters have been found in different cell types. These transporters shuttle AEA in several cell districts including the nucleus for binding to PPARs (i.e., the fatty acid binding protein 5 (FABP5)) [21], the endoplasmic reticulum for FAAH-dependent degradation, adiposomes for accumulation or degradation or oxidation, the mitochondrion for oxidation, and lysosomes for degradation ([20] and references therein). Among others, intracellular AEA transporters include the potentially sterol carrier protein 2 [22] and the FAAH-like AEA transporter (FLAT-1), a cytosolic variant of FAAH1 that lacks amidase activity but bounds AEA, facilitating its translocation [23].
Currently, the ES represents a key signalling pathway involved in the modulation of most biological functions [1,2,18]. As a consequence, the pharmacological intervention of the ES components may represent a promising strategy for the management and the treatment of diseases, whereas the interference in ES signalling following phytocannabinoid abuse or the impairment of the system may represent a threat for the maintenance of health status.
3. Epigenetic Mechanisms: A Brief Overview
The environmental-dependent modulation of gene expression usually occurs at transcriptional, post-transcriptional, and translational levels. In general, the main epigenetic mechanisms involve the chemical modification of DNA and histone tails with consequences on chromatin architecture and accessibility to transcriptional factors and the production of specific regulatory ncRNA [8].
The main chemical modification of DNA is the covalent transfer of the methyl group (-CH3) to cytosine located within cytosine–phosphate–guanine (CpG) islets in the gene promoter region, thus forming 5-methylcytosine (5mC). This modification changes the chromatin structure from an opened (transcriptionally active) to closed (transcriptionally inactive) state [24]. DNA methylation is typically erased during zygote formation to be newly established in the developing embryo in order to address proper embryo development and to drive gene imprinting, the process causing genes to be expressed from a parent of origin-specific manner [25]. Different DNA methyltransferases (DNMTs), the enzymes involved in this epigenetic modification, are classically responsible for de novo and maintaining methylation, but cooperative activity has also been reported and reviewed [26]. Classically, de novo methylation is established by DNMT3A and DNMT3B in participation with DNMT3L, a DNMT devoid of catalytic activity, but capable of assisting de novo methylation, increasing the ability of DNMTs to bind to the methyl group donor, S-adenosyl-L-methionine (SAM). Once established, DNA methylation status is maintained by DNMT1. Conversely, the Ten–eleven translocation methylcytosine dioxygenases (TET1-3) catalyse the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) [27].
The tight or loose interaction of DNA with basic histone and non-histone proteins deeply affects chromatin structure with consequences for gene expression. The tightly folded part of the chromatin, heterochromatin, is usually transcriptionally inactive, whereas, the loosely folded part of the chromatin, euchromatin, is the site of DNA transcription. At present, nine post-translational modifications occurring at histone protein tails have been identified. The most well-studied are: acetylation (ac), mono- (m1), di- (m2), and tri-methylation (m3) at lysine (K) residues (i.e., the transcription-activating acetylation of histone H3 at lysine 9 or 27 (H3K9ac and H3K27ac, respectively) and the acetylation of histone H4 at lysine 16 (H4K16ac), the transcription activating H3K4me1, H3K4me3, H3K36me3, H3K79me2, and the repressive H3K27me3 and H3K9me3), phosphorylation, and ubiquitination. Crotonylation, citrullination, ADP-ribosylation, proline (P) isomerisation (i.e., H3P30 and H3P38), and O-linked β-d-N-acetylglucosaminylation (O-GlcNAcylation) are less well known [28,29,30].
NcRNAs virtually influence every aspect of gene expression, thus representing emerging epigenetic marks detectable in both tissues and biological fluids with upcoming relevance in the regulation of biological functions, impact on health and disease status and possible employment for the prognosis, diagnosis and treatment of diseases [31,32]. They include microRNA (miRNA), transfer RNA (tRNA) fragments (tiRNA and tRF), long non-coding RNA (lncRNA), P-element induced wimpy testis (PIWI)-interacting RNAs (piRNAs), short interfering RNA (siRNA), and circular RNA (circRNA) [32,33].
MiRNAs (20–22 nt long) are endogenous small ncRNA classically involved in RNA interference and their exogenous counterparts are siRNAs. In most cases, they bind the 3’ untranslated region of target mRNA inhibiting their translation into protein and inducing their degradation; however in the nucleus miRNAs may target mRNA co-transcriptionally, recruiting chromatin-modifying enzymes and inducing epigenetic regulation via DNA methylation or histone tails modifications [34].
Transfer RNAs (tRNAs) produce several fragments involved in repression of translation. Some of them, called 5’- (3’-) tRNA halves (tiRNA, 30–40 nt long), are stress-induced and are produced in humans by the endonuclease angiogenin that cleaves within the anticodon loops of mature tRNAs. Another group (17–26 nt long), usually referred as tRFs, is produced by the processing at the 5’- or 3’-end of mature or precursor tRNAs [35,36].
LncRNAs are bidirectional, antisense, intronic, intergenic, or overlapping transcripts capable of modulating the transcription of neighbouring protein-coding genes with remarkable tissue specificity. They also remodel chromatin and genome architecture or stabilize RNA through the recruitment of chromatin-modifying enzymes or directly acting as cis/trans scaffolding factors [37].
Originally characterized in germ cells, piRNAs (26–31 nt long) target heterochromatic regions through the formation of a PIWI–piRNA complex which usually is associated with the repressive histone/lysine methylation marks, but may also recruit different chromatin-modifying enzymes or facilitate transcription [33].
Lastly, circRNAs, a novel class of ncRNAs, are the result of back-splicing and usually are characterized by a covalently closed continuous loop without 5′ or 3′ polarities structure. Highly stable and widely expressed in mammalian cells, including spermatozoa [38], they usually modulate gene expression by acting as miRNA sponges [39].
Once established at embryo stage to define cell fate through the restriction of developmental potential [8], during the course of life genome activity is dynamically modulated under exogenous influence with gene activation or silencing during the life. Therefore, the continuous interaction between the internal and external environment addresses physiological development and health maintenance influencing both disease load and resistance [40]. Hence, there is hypothesis of an epigenetic “clock” phenomenon, a potential tracker of biological age, in which the aging-dependent genome-wide DNA hypomethylation leads to genome instability and occurrence of disease [41]. As a consequence, various epigenetic writers, readers, and erasers like maintenance and de novo DNMTs, TET proteins, histone acetyltransferases (HATs), deacetylases (HDACs), methyltransferases (HMT), and demethylases (amino oxidase homolog lysine demethylase 1 (KDM1) and JmjC domain-containing histone demethylases), or the ncRNA biosynthetic pathways have been identified in living organisms [32] and their activity is strongly related to the preservation of health status. Epigenetic modifications are linked to changes in development and behaviour, cancer, aging-related diseases, infertility, cardiovascular, neurological and metabolic disorders, or drug addiction, among others [42]. Hence, the development of drugs targeting epigenetic machinery represents the first step for the possible employment of personalized epigenetic therapy in the treatment of diseases [43].
4. The Epigenetics of ES
Recent evidence has revealed that ES undergoes epigenetic modulation by alcohol, diet, stress, smoking, exercise, or drugs [44,45,46,47,48,49,50,51,52,53,54,55]. The main targets appear to be the genes encoding for cannabinoid receptors, especially CNR1 which encodes for CB1, and the hydrolysing enzyme FAAH, with subsequent alteration of endocannabinoid signalling or tone. The detected epigenetic mechanisms involve changes in DNA methylation (both global and gene-specific), histone tail modifications such as acetylation, deacetylation, or methylation, and the production of specific miRNAs in different brain regions, peripheral tissues, and cell lines. Of note, epigenetic changes in the ES have been detected in several pathological situations such as Alzheimer’s disease, glioblastoma, and colorectal cancer (CRC), and the ES is the target of several ncRNAs [56,57,58,59,60,61,62,63,64], (details in Table 1).

Table 1.
The epigenetic modulation of ES by non-canonical ligands and during the disease state.
Nevertheless, phytocannabinoids, endocannabinoids, and endocannabinoid receptor agonist/antagonists all affect epigenetic mechanisms in cell lines, animal models, and humans as well [65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96] (details in Table 2), with a long-term impact on health status and the possibility of transmission to the offspring through gametes, leading to trans-generational epigenetic inheritance (Figure 1). Thus, the ES may represent a potential epigenetic target for the assessment of health/disease status, the treatment of disease, and the development of possible epigenetic therapies.

Table 2.
Epigenetic changes induced by phytocannabinoids, endocannabinoids, and ES agonists/antagonists.
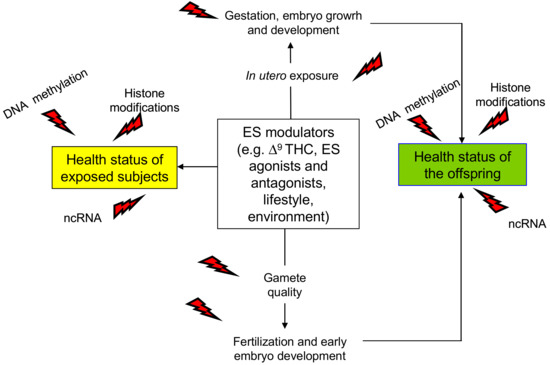
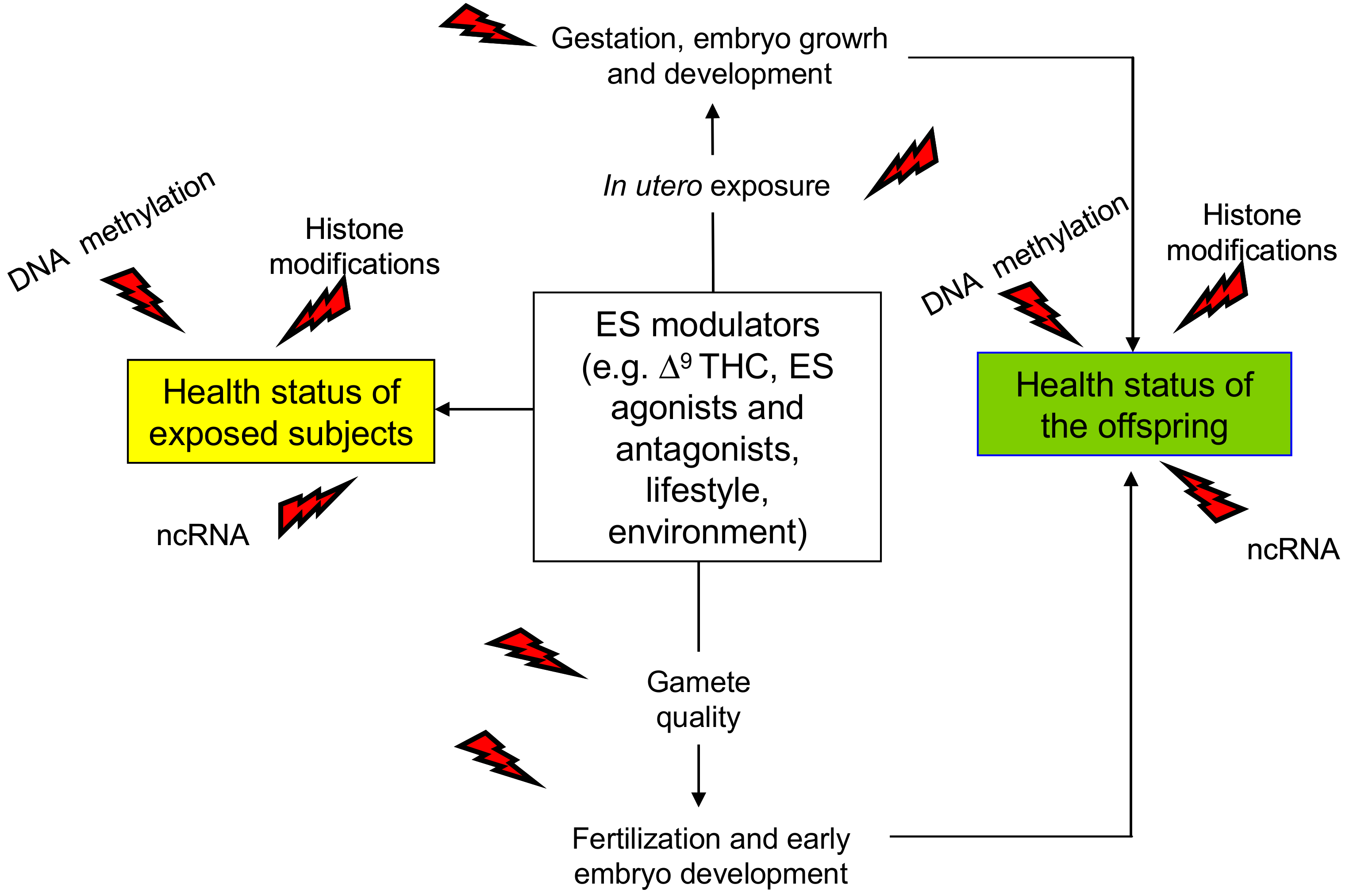
Figure 1.
Schematic representation of the main outcomes of endocannabinoid system (ES) epigenetic modulation. Direct effects are in yellow box; trans-generational effects occurring via gametes or following in utero exposure are in green box. ncRNA: non-coding RNA; Δ9THC: Δ9-tetrahydrocannabinol, red flash of lightning indicates the epigenetic changes.
4.1. Effects on Peripheral Tissues, Brain Functions, and Disease State
Peripherally, the activity of ES may be epigenetically modulated by diet. In fact, extra-virgin olive oil (EVOO), a typical lipid source in the Mediterranean diet which is rich in phenolic compounds, epigenetically modulates the expression rate of CNR1 in vivo and in vitro [51]. Thus, dietary EVOO administration reduces the methylation status of rat CNR1 promoter and the expression of miR23A and miR-301a—two modulators of CB1 in the pathogenesis of colorectal cancer, thus inducing the selective expression of CB1 in rat colon. Accordingly, in Caco-2 cells CB1 is less expressed than normal colon mucosa due to the hypermethylation of DNA at CNR1 promoter; in vitro EVOO, its phenolic extract (OPE), and authentic hydroxytyrosol (HT) upregulate CB1 expression with mechanisms for OPE and HT involving the reduction of CNR1 methylation at promoter level and leading to inhibition of cell proliferation [51].
In the brain, ES is involved in the homeostatic regulation of food intake, through the interplay with peripheral nutrient-sensors and the orexigenic and anorexigenic peptides produced within the arcuate nucleus of the hypothalamus, the brain region capable of capturing and integrating environmental cues with outcomes on feeding behaviour and reproduction [97]. To date, endocannabinoids act as orexigenic factors, stimulating food intake and fat deposition [97]; consistently, rimonabant (SR141716, Acomplia), the selective CB1 antagonist used in clinical trials for the treatment of obesity, reduces body weight, but due to severe psychiatric side effects, its use in patients has been discontinued [98].
Endocannabinoid are n-6 PUFA derivatives and an ideal ratio of 5 (n-6): 1 (n-3) has been suggested in order to preserve brain functions [97]. The evaluation of the plasma n6:n3 fatty acid ratio is therefore a possible risk factor to metabolic disease and might indicate an over activation of endocannabinoid signalling. Leptin, an anorexigenic peptide produced by white adipose tissue, inhibits hypothalamic ES [99], and Ob/Ob mice, lacking leptin, over-activate the hypothalamic endocannabinoid signalling [99] and are affected by infertility due to hypogonadotropic hypogonadism [100]. Consistently, leptin resistance has been associated to the over-activation of endocannabinoid signalling, with alterations in food intake and obesity development by a molecular mechanism involving the activation of CB1 [99]. Recently, sex-specific epigenetic changes related to leptin and endocannabinoid signalling have been reported in the hypothalamus of newborn rats following maternal high-fat diet HFD [52]. Prior obesity development, maternal HFD selectively induces the expression of CB1 in the hypothalamus of males, and of CB2 in females, with the former involved in the control of food intake and the latter mainly exerting a neuromodulatory role. Following maternal HFD, the hypothalamic expression of the transcriptional factor STAT3—a signalling intermediate in the leptin-dependent downregulation of the central ES—is down regulated in all newborns, but Almeida and co-workers reported sex-specific mechanisms in the leptin/ES interplay. In fact, while hypoleptinaemia occurred in newborn male rats only, in female rats only a decreased phosphorylation of STAT3 was observed. Thus, two complementary mechanisms impair leptin signalling, leading to the over expression of CB1. Furthermore, in male offspring maternal HFD causes CNR1 overexpression, with mechanisms involving chromatin remodelling at CNR1 promoter region by means of increased histone acetylation rate and increased binding of androgen receptor at CNR1 promoter as a consequence [52].
At present, the FAAH gene is the only component of ES epigenetically modulated in the hypothalamus by binge-eating, a recurrent process potentially influencing the development of eating disorders. An epigenetic mechanism consisting in the reduction of H3K4ac, without any change in DNA methylation and H3K27met3 at FAAH promoter region have been reported [45].
Interestingly, a highly conserved regulatory sequence in CNR1 intron 2 is responsible for the differential transcriptional activation of CB1 in brain regions like the hippocampus and hypothalamus, with effects on the sex-specific anxiety-related behavioural profile, ethanol intake, and hypothermic response following CB1 agonism, but without any significant changes in feeding patterns [101].
Lastly, Jiang and co-workers proposed a central adiponectin-dependent mechanism to promote the peripheral bone formation through the epigenetic regulation of the hypothalamic expression of CB1, requiring HDAC5 binding to the transcription start site 2 (TSS2) region of the CNR1 gene in embryonic mouse hypothalamus cell line N1 [83].
4.2. Effects on Male Reproduction and Embryo Development
Virtually all steps of reproduction are affected by one or more elements of the ES. In fact, this signalling system is deeply involved in the central and local control of reproduction in both sexes, with functions related to the modulation of the hypothalamus–pituitary–gonad (HPG) axis, germ cell development, successful gametogenesis, production of high-quality gametes, fertilization, embryo implantation and growth, pregnancy, and delivery [18,102,103,104,105,106,107,108,109]. Centrally, ES regulates the hypothalamic release of gonadotropin releasing hormone (GnRH) which in turn mediates the discharge of pituitary gonadotropins (follicle stimulating hormone (FSH) and luteinizing hormone (LH)), the hormones responsible for sex steroid biosynthesis in the gonads [105,106,107,108]. Nevertheless, the full ES has been characterized in mammalian and non-mammalian vertebrates. Endocannabinoids are produced and hydrolysed within the gonads and reproductive tissues, and are released in reproductive fluids, whereas somatic cells in the gonads, germ cells, and gametes in both sexes have the ability to respond to endocannabinoids [102,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124]. Lastly, experimental evidence revealed the need for a suitable gradient of endocannabinoids in reproductive tracts to modulate key steps in reproduction such as the acquisition of sperm motility in the epididymis, acrosome reaction, successful embryo implantation, and delivery, among others [102,105,106,125].
Reproduction is a process highly sensitive to environmental factors like diet, stress, or endocrine disruptor exposure among others [32,126]. Thus its epigenetic modulation has been reported and reviewed elsewhere [32,126,127,128]. At present, there is a knowledge gap on the possible epigenetic regulation of ES in the modulation of GnRH pulse and reproductive hormones, but, as reported in Section 4.1, the epigenetic modulation of hypothalamic ES has been recently reported in relationship to diet and nutritional status, conditions notably affecting reproductive ability [129]. On the contrary, the epigenetic modulation of ES has been reported in male gonads, with emerging roles in spermatozoa and consequences for fertility and embryo development (Figure 2). Therefore, we will deeply analyse the epigenetic modulation of the ES in the testis.
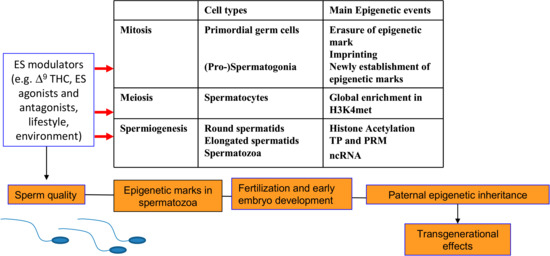
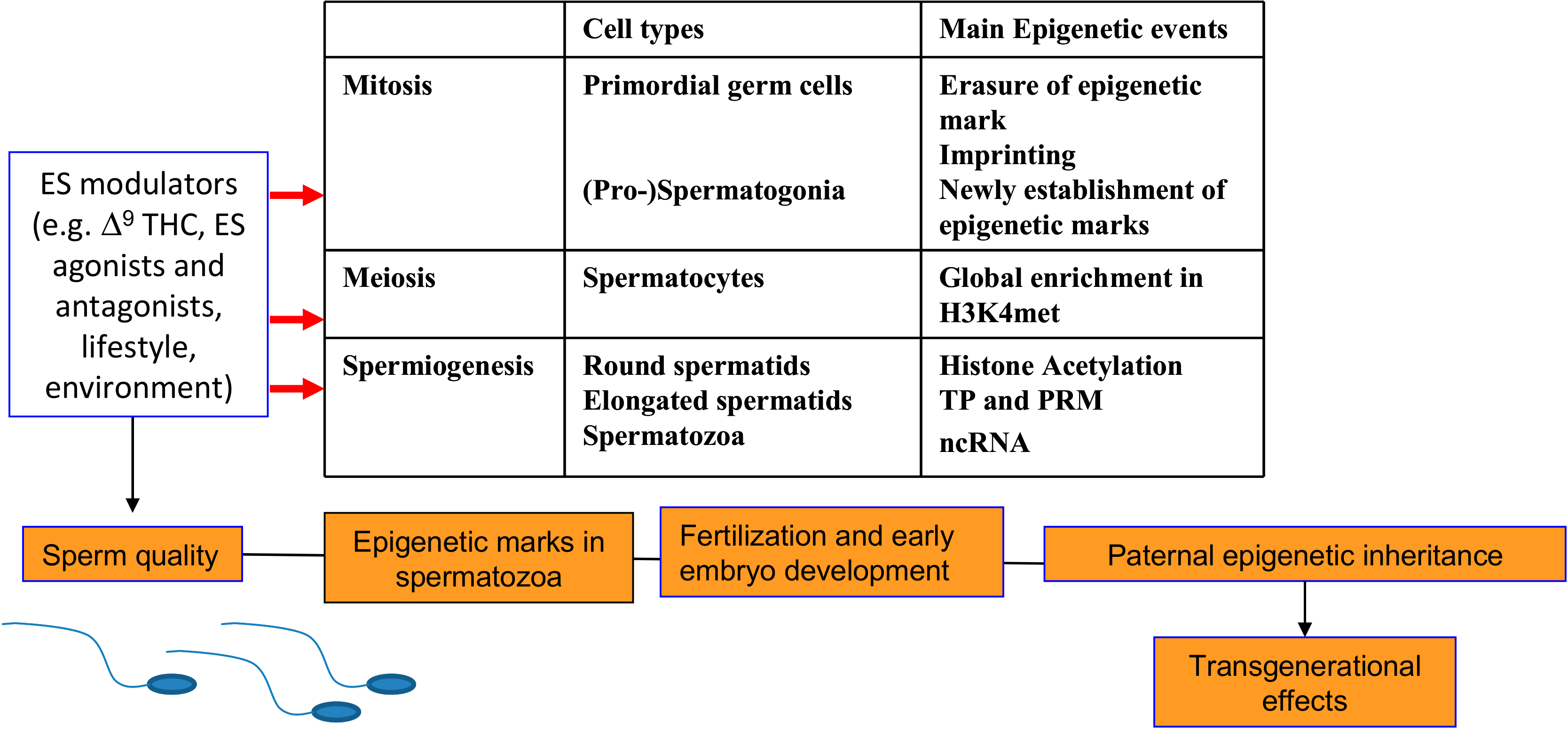
Figure 2.
A summary of the main epigenetic changes occurring during the spermatogenesis and the effects of ES modulation. PRM: protamine; TP: transition protein; ncRNA: non-coding RNA.
The first evidence of the epigenetic modulation of ES in the gonad concerns the expression of FAAH1 in Sertoli cells, the nurse cells within the testis whose survival depends on AEA tone, FSH, and oestradiol activity [130]. The FAAH1 gene promoter contains an oestrogen-responsive element (ERE) and is notably expressed under oestradiol control in primary mouse Sertoli cells [131]. However, the oestradiol-dependent transcriptional activation of FAAH1 requires not only the binding of ERβ to proximal ERE sequences (ERE2/3), but also the involvement of the histone demethylase LSD1, and decreased methylation of both DNA at the CpG site and H3K9 in the proximal promoter region [132].
The ES deeply affects the development and the activity of Leydig cells [111], but at present there is no evidence of the possible epigenetic modulation of ES in Leydig cells.
In spite of the lack of data on the possible epigenetic modulation of testicular somatic cells, new insights concern the possible ES-dependent modulation of spermatogenesis through epigenetic mechanisms. Germ cell development is deeply modulated by epigenetic mechanisms which first erase the epigenetic signature (DNA methylation and histone tail remodelling) in primordial germ cells (PGCs) from specifications to migration and proliferation, and subsequently resettle DNA methylation status with de novo development of epigenetic marks and gene imprinting in pro-spermatogonia [9,10]. During post-natal testis development, additional reprogramming of epigenetic marks occurs in two particular time frames: at the entry in meiosis—a process requiring CB2 activity [112,122]—and in post meiotic stages, notably under CB1 control [117,120]. Lastly, during the transit in male and female reproductive tracts, spermatozoa need ES activity for the acquisition of motility and capacitation, respectively [118,125], and this occurs through a deep remodelling process which includes, among others, the exchange/acquisition of such epigenetic marks as ncRNA through epidydymosomes, prostasomes, or oviductosomes [133,134,135,136] that integrates the marks already available in spermatozoa [38,137,138].
In detail, during the spermatogenesis in rodents, CB2 exerts a pivotal role in meiosis entry [112,122] and its hyper- or hypostimulation disrupts the temporal dynamics of the spermatogenesis with possible epigenetic mechanisms [92]. In fact, JWH-133, a CB2 specific agonist, stimulates the expression of the meiotic genes c-Kit and Stra8 by increasing and decreasing the levels of H3K4m3 and H3K9m2, respectively, in genomic regions flanking the transcription start sites. Interestingly, the global increase in H3K4m3 occurs through the JWH-133-dependent transcriptional activation of Prdm9, the gene encoding for a zinc finger protein with HMT activity that catalyses H3K4me3 during the meiotic prophase. As a consequence, prolonged exposure to JWH-133 or administration of the specific CB2 antagonist AM630 accelerates or delays spermatogenesis onset in immature mice, pointing out the importance of correct endocannabinoid signalling for proper spermatogenesis and the deleterious effect of exogenous cannabinoids on male fertility [92].
In this respect, the modulation or the interference in the endogenous ES may affect gamete quality or impact the epigenetic mark of gametes, both critical for pregnancy and embryo development, with the (remote, but real) possibility of trans-generational inheritance in the offspring.
Spermiogenesis is the process leading to the formation of spermatozoa and is characterized by round spermatid elongation, acrosome and tail formation, nuclear shaping, and DNA packaging with transcriptional silencing as a consequence. Chromatin remodelling and DNA packaging are therefore the main nuclear events in spermiogenesis, consisting of a double-step process that requires histone replacement, first by transition proteins (TP2 and TP1) and then by protamines (PRM1 and PRM2), a class of small basic proteins [139]. The cooperation between HATs, HDACs, molecular chaperones, ubiquitination, and DNA repair systems drives the shift from a nucleosomal-based to a mainly protamine-based chromatin configuration [139]. In this respect, data from CB1-/- mice revealed the requirement of ES signalling for proper chromatin remodelling during spermiogenesis [117], and production of high-quality spermatozoa [140]. In fact, the genetic ablation of the CNR1 negatively affects the chromatin packaging, by affecting the content of TP2 mRNA and reducing histone displacement, with consequences on chromatin condensation and DNA integrity in the spermatozoa [117], which exhibit nuclear size elongation [140]. Such a mechanism is reversed by oestradiol administration, a treatment promoting histone displacement and chromatin condensation rescue in epididymal sperm collected from knock down animals [120].
In line with the results reported above, recent data revealed that the chronic administration of JWH-133 reduces sperm count in mouse and affects the epigenome of spermatozoa. Interestingly, the sperm from JWH-133 treated mice maintains the ability to fertilize eggs from untreated females, but impairment of embryo growth and defects in placental size have been reported, suggesting a possible interference in paternal inheritance through epigenetic mechanisms [93]. Accordingly defects in DNA methylation/hydroxymethylation at paternally expressed imprinted genes (i.e., Peg10 and Plagl1) have been reported in the sperm of JWH-133 treated animals and are maintained in placental tissue following fertilization. Thus, CB2 signalling may be critical for the integrity of the epigenome in the sperm, with the possibility of paternal epigenetic inheritance in the embryo, a process in which spermatozoa act as vectors for the delivery of epigenetic marks into the developing embryo. Consistently, two isoforms of CircNAPE-PLD (CircNAPE-PLD1 and CircNAPE-PLD2) are expressed in human and murine spermatozoa, and CircNAPE-PLD1 physically interacts with oocyte miRNAs involved in the progression of cell cycle [38]. Therefore, a new role of ES in the zygote to regulate the first stages of embryo development, through epigenetic paternal inheritance aimed at miRNA decoy, has emerged.
The above observations point out the possible risk for epigenome integrity of spermatozoa following marijuana use. Spermatozoa contain a complete ES devoted to the control of sperm physiology; acrosome reaction, acquisition of motility, spermatozoa–oocyte interaction all require the physiological activity of the endogenous ES [105,106]. Classically marijuana smokers exhibit a large set of reproductive failures, from imbalanced hormonal milieu and poor sperm quality to impairment of menstrual cycle, poor oocyte retrieval rate, low pregnancy rate, pre-term delivery, and prematurity with low fetal birth weight [102]. As a consequence, a recent study from the group of Murphy has been focused on the possible epigenetic effects of marijuana smoking and Δ9THC on reproductive health status. In humans and rats, Δ9THC exposure lowers sperm concentration and alters DNA sperm methylome with substantial shifts in both hypo- and hyper-DNA methylation, with the latter predominating [80]. In particular, 10.3% of differentially methylated CpG sites (409/3979) significantly correlate with sperm count and 183 individual CpG sites representing 177 named genes have methylation levels significantly correlated with measured Δ9THC levels [80]. Altered CpG sites associated with genes involved in the Hippo signalling pathway and in pathways in cancer are common in both cannabis users and Δ9THC-exposed rats [80]. Interestingly, Δ9THC target genes in rat sperm substantially overlap with genes having altered methylation rate in the brain of rat offspring born to parents both exposed to Δ9THC during adolescence [79].
Consistently with the above observation, Δ9THC exposure-dependent changes in the DNA methylation of rat sperm [80] do not significantly impact the clinical health of the offspring (e.g., litter size, sex ratio, pup birth weight, survival, and growth) but cause long-lasting neurobehavioral effects in the offspring with impairment in attentional performance [78]. In addition, cannabis use in humans causes in the sperm the hypomethylation at 9 CpG sites located in intron 7 of the autism candidate gene Discs-Large Associated Protein 2 (DLGAP2), a gene involved in synapse organization and neuronal signalling [80,81]. Similarly, Δ9THC exposure in adult rats differently methylated DLGAP2 gene in spermatozoa [81].
Lastly, IBN Lahmar Andaloussi et al. recently reported the behavioural and epigenetic effects of stress in male rats whose fathers were exposed to cannabinoids during adolescence [95] in the presence or absence of the synthetic CB1 agonist WIN55.212-2. Interestingly, stress exposure induced a significant anxiogenic-like effect but did not affect the episodic-like memory in the offspring of WIN55.212-2-exposed fathers only, with significant increases in global DNA methylation and DNMT1 and DNMTa3 transcription in the prefrontal cortex [95]. Thus, these results suggest that chronic exposure to cannabinoids during adolescence may lead to a trans-generational transfer of stress susceptibility to the offspring through the transfer of epigenetic marks in gametes.
Taken together, the use of cannabis for recreational use may represent a serious risk for both the fertility of marijuana smokers and the health of the offspring.
5. Conclusions
The ES is an almost ubiquitous cell signalling system regulating several processes inside cells that are not yet completely understood. From its discovery in 1990s, it was clear that this system could be modulated by both extrinsic (cannabis and its derivatives) and intrinsic (the endogenous ligands) signals. Subsequent studies pointed out that the ES is much more complex than was thought since it is able to cross-talk with many other transduction cell signalling pathways, therefore regulating key biological processes such as cell proliferation and differentiation, synaptic plasticity, gametogenesis, and fertility. From the data herein reported, it emerges that epigenetic modifications of the ES by means of DNA methylation, histone acetylation/deacetylation at the CNR1, and FAAH genes encoding the CB1 receptor and FAAH hydrolysing enzyme may play a relevant role both in physiological processes regulating (male) fertility and reproduction as well as in disease pathogenesis and progression including cancer. Interestingly, it has been documented that external epigenetic cues such as alcohol induce DNA methylation changes in the mouse model of foetal alcohol spectrum disorder, and the lack of a functional CNR1 gene protects against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation [49]. This suggests that the ES itself may act as an epigenetic signal regulating gene expression. On the other hand, the altered DNA methylation of both GPR55 and CB1 encoding genes, resulting in increased expression of GPR55 and reduced levels of the CB1 in CRC patients [58], also supports the hypothesis that the ES receptors could behave either as tumour promoting or tumour suppressor genes depending on the kind of epigenetics changes they undergo. The ES has been recognized as a strong modulator in the central and local control of reproduction in both sexes. It is involved in the regulation of HPG axis, successful gametogenesis, fertilization, and embryo implantation and development. Therefore, it is conceivable that environmental factors, by epigenetically affecting the ES, could induce adverse effects on reproductive system functions per se or alternatively, change gene expression profile with a transgenerational inheritance in the offspring. Within this context, the emerging literature tags the ES signalling as critical for the integrity of the epigenome in the sperm suggesting the possibility of a paternal epigenetic inheritance in the embryo through a process in which spermatozoa act as vectors for delivering epigenetic marks into the developing embryo, and in our opinion this is a very interesting issue that has to be further studied. To date, most studies have been conducted in experimental models other than humans and the few studies in humans involve a limited number of subjects and do not extend to the offspring. However, the overlapping DNA methylation profiles detected in both human and rat sperm suggest that data in experimental models may be the basis for further investigations and confirmation in human subjects. Similarly, the use of phytocannabinoids or synthetic cannabinoids for therapeutic purposes point out the possible trans-generational transmission of epigenetic marks, revealing the need for particular caution and attention in the field. Finally, epigenetic modifiers of the ES could be also a promising tool to treat eating disorders or manage pathological conditions involving alterations of the ES system. However, the significance of ES epigenetics is still an open question needing deeper investigation to better characterize the real consequences of the epigenetic changes of this intriguing cell signalling system.
Author Contributions
Conceptualization, R.M. (Rosaria Meccariello) and R.P.; writing—original draft preparation, R.M. (Rosaria Meccariello), A.S., S.D., R.M. (Rossella Morrone), A.V.; writing—review and editing, A.S., A.V., S.F.; supervision, R.M. (Rosaria Meccariello) and R.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Miur (Prin 2017 grant number 20175MT5EM, to R.M., A.V., R.P.)
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACEA | arachidonyl-2′-chloroethylamine |
| ACPA | arachidonylcyclopropylamide |
| AEA | anandamide |
| 2AG | 2-arachydonoylglicerol |
| APN | adiponectin |
| ARA | arachidonic acid |
| CB1 | type 1 cannabinoid receptor |
| CB2 | type 2 cannabinoid receptor |
| CHF | chronic heart failure |
| circRNA | circular RNA |
| CNR1 | gene encoding for CB1 |
| CpG | cytosine–phosphate-guanine |
| CRC | Colon Rectal Cancer |
| DAGL | sn-1-diacylglycerol lipase |
| Dex | dexmedetomidine |
| DIO | diet-induced obesity |
| DLGAP2 | discs large associated protein 2 |
| DNMT | DNA methyltransferases |
| Drd2 | gene encoding for dopamine receptor D2 |
| Δ9THC | Δ9-tetrahydrocannabinol |
| ER | oestrogen receptor |
| ERE | oestrogen responsive element |
| ES | endocannabinoid system |
| EVOO | extra-virgin olive oil |
| FAAH1 | fatty acid amide hydrolase 1 |
| FAAH2 | fatty acid amide hydrolase 2 |
| FABP5 | fatty acid binding protein 5 |
| FLAT-1 | FAAH-like AEA transporter |
| FSH | follicle stimulating hormone |
| GABA | γ-aminobutyric acid |
| GlcNAcylation | O-linked β-d-N-acetylglucosaminylation |
| GnRH | gonadotropin releasing hormone |
| HAT | histone acetyltransferases |
| HDAC | histone deacetylases |
| HFD | high-fat diet |
| 5hmC | 5-hydroxymethylcytosine |
| HMT | histone methyltransferases |
| HPG | hypothalamus-pituitary-gonad |
| HT | hydroxytyrosol |
| KDM1 | amino oxidase homolog lysine demethylase 1 |
| LH | luteinizing hormone |
| lncRNA | long non-coding RNA |
| LOAD | late-onset Alzheimer’s disease |
| MAGL | monoacylglycerol lipase |
| 5mC | 5-methylcytosine |
| miRNA | microRNA |
| NAPE-PLD | N-acyl-phosphatidylethanolamine-specific phospholipase D |
| ncRNA | non coding RNA |
| OPE | olive oil phenolic extract |
| PBMC | peripheral blood mononuclear cells |
| Penk | gene encoding for Proenkephalin |
| PGC | primordial germ cell |
| P-LGG | paediatric low-grade gliomas |
| piRNA | PIWI-interacting RNA |
| piwi | P-element induced wimpy testis |
| PPARγ | peroxisome proliferator-activated receptor γ |
| PRM | protamine |
| PUFA | poly unsaturated fatty acids |
| SAM | S-adenosyl-L-methionine |
| siRNA | short interfering RNA |
| TET | Ten-eleven translocation methylcytosine dioxygenases |
| tiRNA and tRF | tRNA fragments |
| TP | transition protein |
| TRPV1 | cationic channel type 1 vanilloid receptor |
| TSS | transcription start site |
References
- Lu, H.-C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatr. 2016, 79, 516–525. [Google Scholar] [CrossRef]
- Meccariello, R.; Chianese, R. (Eds.) Cannabinoids in Health and Disease; IntechOpen: Rijeka, Croatia, 2016; pp. 1–248. ISBN 978-953-51-2429-0. [Google Scholar] [CrossRef]
- Laezza, C.; Pagano, C.; Navarra, G.; Pastorino, O.; Proto, M.C.; Fiore, D.; Piscopo, C.; Gazzerro, P.; Bifulco, M. The Endocannabinoid System: A Target for Cancer Treatment. Int. J. Mol. Sci. 2020, 21, 747. [Google Scholar] [CrossRef]
- Serrano, A.; Parsons, L.H. Endocannabinoid influence in drug reinforcement, dependence and addiction-related behaviors. Pharmacol. Ther. 2011, 132, 215–241. [Google Scholar] [CrossRef] [PubMed]
- Basavarajappa, B.S.; Joshi, V.; Shivakumar, M.; Subbanna, S. Distinct functions of endogenous cannabinoid system in alcohol abuse disorders. Br. J. Pharmacol. 2019, 176, 3085–3109. [Google Scholar] [CrossRef] [PubMed]
- Hayase, T. Epigenetic mechanisms associated with addiction-related behavioural effects of nicotine and/or cocaine: Implication of the endocannabinoid system. Behav. Pharmacol. 2017, 28, 493–511. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 2009, 66, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Seisenberger, S.; Peat, J.R.; Hore, T.A.; Santos, F.; Dean, W.; Reik, W. Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20110330. [Google Scholar] [CrossRef]
- Hogg, K.; Western, P.S. Refurbishing the germline epigenome: Out with the old, in with the new. Semin. Cell. Dev. Biol. 2015, 45, 104–113. [Google Scholar] [CrossRef]
- Bakhshandeh, B.; Kamaleddin, M.A.; Aalishah, K.A. Comprehensive review on exosomes and microvesicles as epigenetic factors. Curr. Stem Cell Res. Ther. 2017, 12, 31–36. [Google Scholar] [CrossRef]
- Qian, Z.; Shen, Q.; Yang, X.; Qiu, Y.; Zhang, W. The role of extracellular vesicles: An epigenetic view of the cancer microenvironment. BioMed Res. Int. 2015, 2015, 649161. [Google Scholar] [CrossRef] [PubMed]
- Motti, M.L.; D’Angelo, S.; Meccariello, R. MicroRNAs, cancer and diet: Facts and new exciting perspectives. Curr. Mol. Pharmacol. 2018, 11, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Daxinger, L.; Whitelaw, E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 2012, 13, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Derghal, A.; Djelloul, M.; Trouslard, J.; Mounien, L. An Emerging Role of micro-RNA in the Effect of the Endocrine Disruptors. Front. Neurosci. 2016, 10, 318. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Baruah, K.; Vanrompay, D.; Bossier, P. Can epigenetics translate environmental cues into phenotypes? Sci. Total Environ. 2019, 647, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Gaoni, Y.; Mechoulam, R. Isolation, structure and partial synthesis of an active constituent of hashish. J. Am. Chem. Soc. 1964, 86, 1646–1647. [Google Scholar] [CrossRef]
- Cacciola, G.; Chianese, R.; Chioccarelli, T.; Ciaramella, V.; Fasano, S.; Pierantoni, R.; Meccariello, R.; Cobellis, G. Cannabinoids and Reproduction: A Lasting and Intriguing History. Pharmaceuticals 2010, 3, 3275–3323. [Google Scholar] [CrossRef]
- Di Marzo, V. Endocannabinoids: Synthesis and degradation. Rev. Physiol. Biochem. Pharmacol. 2008, 160, 1–24. [Google Scholar] [CrossRef]
- Maccarrone, M. Metabolism of the Endocannabinoid Anandamide: Open Questions after 25 Years. Front. Mol. Neurosci. 2017, 10, 166. [Google Scholar] [CrossRef]
- Kaczocha, M.; Vivieca, S.; Sun, J.; Glaser, S.T.; Deutsch, D.G. Fatty acid-binding proteins transport N-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J. Biol. Chem. 2012, 287, 3415–3424. [Google Scholar] [CrossRef]
- Liedhegner, E.S.; Vogt, C.D.; Sem, D.S.; Cunningham, C.W.; Hillard, C.J. Sterol carrier protein-2: Binding protein for endocannabinoids. Mol. Neurobiol. 2014, 50, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bottegoni, G.; Sasso, O.; Bertorelli, R.; Rocchia, W.; Masetti, M.; Guijarro, A.; Lodola, A.; Armirotti, A.; Garau, G.; et al. A catalytically silent FAAH-1 variant drives anandamide transport in neurons. Nat. Neurosci. 2011, 15, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Li, Y.; Robertson, K.D. DNA Methylation Superior or Subordinate in the Epigenetic Hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef]
- Javaid, N.; Choi, S. Acetylation- and Methylation-Related Epigenetic Proteins in the Context of Their Targets. Genes 2017, 8, 196. [Google Scholar] [CrossRef]
- Wu, D.; Cai, Y.; Jin, J. Potential coordination role between O-GlcNAcylation and epigenetics. Protein Cell 2017, 8, 713–723. [Google Scholar] [CrossRef]
- Xu, Y.M.; Du, J.Y.; Lau, A.T. Posttranslational modifications of human histone H3: An update. Proteomics 2014, 14, 2047–2060. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef]
- Chianese, R.; Troisi, J.; Richards, S.; Scafuro, M.; Fasano, S.; Guida, M.; Pierantoni, R.; Meccariello, R. Bisphenol A in Reproduction: Epigenetic Effects. Curr. Med. Chem. 2018, 25, 748–770. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.A.; Webb, W.M.; Lubin, F.D. Regulatory RNAs and control of epigenetic mechanisms: Expectations for cognition and cognitive dysfunction. Epigenomics 2016, 8, 135–151. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar]
- Saikia, M.; Hatzoglou, M. The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health. J. Biol. Chem. 2015, 290, 29761–29768. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Barbagallo, D.; Chioccarelli, T.; Manfrevola, F.; Cobellis, G.; Di Pietro, C.; Brex, D.; Battaglia, R.; Fasano, S.; Ferraro, B.; et al. CircNAPEPLD is expressed in human and murine spermatozoa and physically interacts with oocyte miRNAs. RNA Biol. 2019, 16, 1237–1248. [Google Scholar] [CrossRef]
- Rong, D.; Sun, H.; Li, Z.; Liu, S.; Dong, C.; Fu, K.; Tang, W.; Cao, H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017, 8, 73271–73281. [Google Scholar] [CrossRef] [PubMed]
- Kanherkar, R.R.; Bhatia-Dey, N.; Csoka, A.B. Epigenetics across the human lifespan. Front. Cell. Dev. Biol. 2014, 2, 49. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Feinberg, A.P. Phenotypic plasticity and the epigenetics of human disease. Nature 2007, 447, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.D.; Issa, J.J. The promise of epigenetic therapy: Reprogramming the cancer epigenome. Curr. Opin. Genet. Dev. 2017, 42, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hutchison, K.E.; Bryan, A.D.; Filbey, F.M.; Calhoun, V.D.; Claus, E.D.; Lin, D.; Sui, J.; Du, Y.; Liu, J. Opposite Epigenetic Associations With Alcohol Use and Exercise Intervention. Front. Psychiatry 2018, 9, 594. [Google Scholar] [CrossRef]
- Pucci, M.; Micioni Di Bonaventura, M.V.; Zaplatic, E.; Bellia, F.; Maccarrone, M.; Cifani, C.; D’Addario, C. Transcriptional regulation of the endocannabinoid system in a rat model of binge-eating behavior reveals a selective modulation of the hypothalamic fatty acid amide hydrolase gene. Int. J. Eat Disord. 2019, 52, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Rotter, A.; Bayerlein, K.; Hansbauer, M.; Weiland, J.; Sperling, W.; Kornhuber, J.; Biermann, T. CB1 and CB2 receptor expression and promoter methylation in patients with cannabis dependence. Eur. Addict. Res. 2013, 19, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lomazzo, E.; König, F.; Abassi, L.; Jelinek, R.; Lutz, B. Chronic stress leads to epigenetic dysregulation in the neuropeptide-Y and cannabinoid CB1 receptor genes in the mouse cingulate cortex. Neuropharmacology 2017, 113 Pt A, 301–313. [Google Scholar] [CrossRef]
- Subbanna, S.; Nagre, N.N.; Umapathy, N.S.; Pace, B.S.; Basavarajappa, B.S. Ethanol exposure induces neonatal neurodegeneration by enhancing CB1R Exon1 histone H4K8 acetylation and up-regulating CB1R function causing neurobehavioral abnormalities in adult mice. Int. J. Neuropsychopharmacol. 2014, 18, pyu028. [Google Scholar] [CrossRef] [PubMed]
- Nagre, N.N.; Subbanna, S.; Shivakumar, M.; Psychoyos, D.; Basavarajappa, B.S. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. J. Neurochem. 2015, 132, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Correa, F.; De Laurentiis, A.; Franchi, A.M. Ethanol downregulates N-acyl phosphatidylethanolamine-phospholipase D expression in BV2 microglial cells via epigenetic mechanisms. Eur. J. Pharmacol. 2016, 786, 224–233. [Google Scholar] [CrossRef]
- Di Francesco, A.; Falconi, A.; Di Germanio, C.; Micioni Di Bonaventura, M.V.; Costa, A.; Caramuta, S.; Del Carlo, M.; Compagnone, D.; Dainese, E.; Cifani, C.; et al. Extravirgin olive oil up-regulates CB1 tumor suppressor gene in human colon cancer cells and in rat colon via epigenetic mechanisms. J. Nutr. Biochem. 2015, 26, 250–258. [Google Scholar] [CrossRef]
- Almeida, M.M.; Dias-Rocha, C.P.; Reis-Gomes, C.F.; Wang, H.; Atella, G.C.; Cordeiro, A.; Pazos-Moura, C.C.; Joss-Moore, L.; Trevenzoli, I.H. Maternal high-fat diet impairs leptin signaling and up-regulates type-1 cannabinoid receptor with sex-specific epigenetic changes in the hypothalamus of newborn rats. Psychoneuroendocrinology 2019, 103, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, W.; Wang, W.; Wang, Z.; Pu, Y.; Chen, H.; Wang, F.; Qian, J. Involvement of miR-665 in protection effect of dexmedetomidine against Oxidative Stress Injury in myocardial cells via CB2 and CK1. Biomed. Pharmacother. 2019, 115, 108894. [Google Scholar] [CrossRef] [PubMed]
- Börner, C.; Martella, E.; Höllt, V.; Kraus, J. Regulation of opioid and cannabinoid receptor genes in human neuroblastoma and T cells by theepigenetic modifiers trichostatin A and 5-aza-2′-deoxycytidine. Neuroimmunomodulation 2012, 19, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Proto, M.C.; Gazzerro, P.; Di Croce, L.; Santoro, A.; Malfitano, A.M.; Pisanti, S.; Laezza, C.; Bifulco, M. Interaction of endocannabinoid system and steroid hormones in the control of colon cancer cell growth. J. Cell. Physiol. 2012, 227, 250–258. [Google Scholar] [CrossRef]
- D’Addario, C.; Di Francesco, A.; Arosio, B.; Gussago, C.; Dell’Osso, B.; Bari, M.; Galimberti, D.; Scarpini, E.; Altamura, A.C.; Mari, D.; et al. Epigenetic regulation of fatty acid amide hydrolase in Alzheimer disease. PLoS ONE 2012, 7, e39186. [Google Scholar] [CrossRef] [PubMed]
- Etcheverry, A.; Aubry, M.; de Tayrac, M.; Vauleon, E.; Boniface, R.; Guenot, F.; Saikali, S.; Hamlat, A.; Riffaud, L.; Menei, P.; et al. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genom. 2010, 11, 701. [Google Scholar] [CrossRef] [PubMed]
- Hasenoehrl, C.; Feuersinger, D.; Sturm, E.M.; Bärnthaler, T.; Heitzer, E.; Graf, R.; Grill, M.; Pichler, M.; Beck, S.; Butcher, L.; et al. G protein-coupled receptor GPR55 promotes colorectal cancer and has opposing effects to cannabinoid receptor 1. Int. J. Cancer 2018, 142, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qian, X.; Zhu, M.; Li, A.; Fang, M.; Zhu, Y.; Zhang, J. miR-1273g-3p promotes proliferation, migration and invasion of LoVo cells via cannabinoid receptor 1 through activation of ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Mol. Med. Rep. 2018, 17, 4619–4626. [Google Scholar] [CrossRef]
- Tung, C.W.; Ho, C.; Hsu, Y.C.; Huang, S.C.; Shih, Y.H.; Lin, C.L. MicroRNA-29a Attenuates Diabetic Glomerular Injury through Modulating Cannabinoid Receptor 1 Signaling. Molecules 2019, 24, 264. [Google Scholar] [CrossRef]
- Möhnle, P.; Schütz, S.V.; Schmidt, M.; Hinske, C.; Hübner, M.; Heyn, J.; Beiras-Fernandez, A.; Kreth, S. MicroRNA-665 is involved in the regulation of the expression of the cardioprotective cannabinoid receptor CB2 in patients with severe heart failure. Biochem. Biophys. Res. Commun. 2014, 451, 516–521. [Google Scholar] [CrossRef]
- Sredni, S.T.; Huang, C.C.; Suzuki, M.; Pundy, T.; Chou, P.; Tomita, T. Spontaneous involution of pediatric low-grade gliomas: High expression of cannabinoid receptor 1 (CNR1) at the time of diagnosis may indicate involvement of the endocannabinoid system. Childs Nerv. Syst. 2016, 32, 2061–2067. [Google Scholar] [CrossRef] [PubMed]
- Chiarlone, A.; Börner, C.; Martín-Gómez, L.; Jiménez-González, A.; García-Concejo, A.; García-Bermejo, M.L.; Lorente, M.; Blázquez, C.; García-Taboada, E.; de Haro, A.; et al. MicroRNA let-7d is a target of cannabinoid CB1 receptor and controls cannabinoid signaling. Neuropharmacology 2016, 108, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Most, D.; Salem, N.A.; Tiwari, G.R.; Blednov, Y.A.; Mayfield, R.D.; Harris, R.A. Silencing synaptic MicroRNA-411 reduces voluntary alcohol consumption in mice. Addict. Biol. 2019, 24, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Ehrlich, S.; Walton, E.; Whitem, T.; Perrone-Bizzozero, N.; Bustillo, J.; Turner, J.A.; Calhoun, V.D. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr. Bull. 2014, 40, 769–776. [Google Scholar] [CrossRef]
- Pucci, M.; Rapino, C.; Di Francesco, A.; Dainese, E.; D’Addario, C.; Maccarrone, M. Epigenetic control of skin differentiation genes by phytocannabinoids. Br. J. Pharmacol. 2013, 170, 581–591. [Google Scholar] [CrossRef]
- Molina, P.E.; Amedee, A.; LeCapitaine, N.J.; Zabaleta, J.; Mohan, M.; Winsauer, P.; Vande Stouwe, C. Cannabinoid neuroimmune modulation of SIV disease. J. Neuroimmune Pharmacol. 2011, 6, 516–527. [Google Scholar] [CrossRef]
- Khare, M.; Taylor, A.H.; Konje, J.C.; Bell, S.C. Delta9-tetrahydrocannabinol inhibits cytotrophoblast cell proliferation and modulates gene transcription. Mol. Hum. Reprod. 2006, 12, 321–333. [Google Scholar] [CrossRef]
- Tomasiewicz, H.C.; Jacobs, M.M.; Wilkinson, M.B.; Wilson, S.P.; Nestler, E.J.; Hurd, Y.L. Proenkephalin mediates the enduring effects of adolescent cannabis exposure associated with adult opiate vulnerability. Biol. Psychiatry 2012, 72, 803–810. [Google Scholar] [CrossRef]
- DiNieri, J.A.; Wang, X.; Szutorisz, H.; Spano, S.M.; Kaur, J.; Casaccia, P.; Dow-Edwards, D.; Hurd, Y.L. Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol. Psychiatry 2011, 70, 763–769. [Google Scholar] [CrossRef]
- Prini, P.; Penna, F.; Sciuccati, E.; Alberio, T.; Rubino, T. Chronic Δ⁸-THC Exposure Differently Affects Histone Modifications in the Adolescent and Adult Rat Brain. Int. J. Mol. Sci. 2017, 18, 2094. [Google Scholar] [CrossRef]
- Prini, P.; Rusconi, F.; Zamberletti, E.; Gabaglio, M.; Penna, F.; Fasano, M.; Battaglioli, E.; Parolaro, D.; Rubino, T. Adolescent THC exposure in female rats leads to cognitive deficits through a mechanism involving chromatin modifications in the prefrontal cortex. J. Psychiatry Neurosci. 2017, 42, 170082. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hegde, V.L.; Rao, R.; Zhang, J.; Nagarkatti, P.S.; Nagarkatti, M. Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J. Biol. Chem. 2014, 289, 18707–18718. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bam, M.; Nagarkatti, P.S.; Nagarkatti, M. RNA-seq Analysis of δ9-Tetrahydrocannabinol-treated T Cells Reveals Altered Gene Expression Profiles That Regulate Immune Response and Cell Proliferation. J. Biol. Chem. 2016, 291, 15460–15472. [Google Scholar] [CrossRef] [PubMed]
- Szutorisz, H.; DiNieri, J.A.; Sweet, E.; Egervari, G.; Michaelides, M.; Carter, J.M.; Ren, Y.; Miller, M.L.; Blitzer, R.D.; Hurd, Y.L. Parental THC Exposure Leads to Compulsive Heroin-Seeking and Altered Striatal Synaptic Plasticity in the Subsequent Generation. Neuropsychopharmacology 2014, 39, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Hegde, V.L.; Tomar, S.; Jackson, A.; Rao, R.; Yang, X.; Singh, U.P.; Singh, N.P.; Nagarkatti, P.S.; Nagarkatti, M. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Delta9-tetrahydrocannabinol in vivo: Regulation of CCAAT/enhancer-binding protein alpha by microRNA-690. J. Biol. Chem. 2013, 288, 36810–36826. [Google Scholar] [CrossRef]
- Chandra, L.C.; Kumar, V.; Torben, W.; Vande Stouwe, C.; Winsauer, P.; Amedee, A.; Molina, P.E.; Mohan, M. Chronic administration of Delta9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J. Virol. 2015, 89, 1168–1181. [Google Scholar] [CrossRef]
- Levin, E.D.; Hawkey, A.B.; Hall, B.J.; Cauley, M.; Slade, S.; Yazdani, E.; Kenou, B.; White, H.; Wells, C.; Rezvani, A.H.; et al. Paternal THC exposure in rats causes long-lasting neurobehavioral effects in the offspring. Neurotoxicol. Teratol. 2019, 74, 106806. [Google Scholar] [CrossRef]
- Watson, C.T.; Szutorisz, H.; Garg, P.; Martin, Q.; Landry, J.A.; Sharp, A.J.; Hurd, Y.L. Genome-Wide DNA Methylation Profiling Reveals Epigenetic Changes in the Rat Nucleus Accumbens Associated With Cross-Generational Effects of Adolescent THC Exposure. Neuropsychopharmacology 2015, 40, 2993–3005. [Google Scholar] [CrossRef]
- Murphy, S.K.; Itchon-Ramos, N.; Visco, Z.; Huang, Z.; Grenier, C.; Schrott, R.; Acharya, K.; Boudreau, M.H.; Price, T.M.; Raburn, D.J.; et al. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 2018, 13, 1208–1221. [Google Scholar] [CrossRef]
- Schrott, R.; Acharya, K.; Itchon-Ramos, N.; Hawkey, A.B.; Pippen, E.; Mitchell, J.T.; Kollins, S.H.; Levin, E.D.; Murphy, S.K. Cannabis use is associated with potentially heritable widespread changes in autism candidate gene DLGAP2 DNA methylation in sperm. Epigenetics 2020, 15, 161–173. [Google Scholar] [CrossRef]
- Ernst, J.; Grabiec, U.; Greither, T.; Fischer, B.; Dehghani, F. The endocannabinoid system in the human granulosa cell line KGN. Mol. Cell. Endocrinol. 2016, 423, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wu, Y.; Valverde, P.; Murray, D.; Tang, J.; Yao, Q.; Han, Q.; Zhang, J.; Zhang, L.; Sui, L.; et al. Central adiponectin induces trabecular bone mass partly through epigenetic downregulation of cannabinoid receptor CB1. J. Cell Physiol. 2019, 234, 7062–7069. [Google Scholar] [CrossRef] [PubMed]
- Hayase, T. Putative Epigenetic Involvement of the Endocannabinoid System in Anxiety- and Depression-Related Behaviors Caused by Nicotine as a Stressor. PLoS ONE 2016, 11, e0158950. [Google Scholar] [CrossRef] [PubMed]
- Paradisi, A.; Pasquariello, N.; Barcaroli, D.; Maccarrone, M. Anandamide regulates keratinocyte differentiation by inducing DNA methylation in a CB1 receptor-dependent manner. J. Biol. Chem. 2008, 283, 6005–6012. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.R.; Nagarkatti, P.; Nagarkatti, M. Anandamide attenuates Th-17 cell-mediated delayed-type hypersensitivity response by triggering IL-10 production and consequent microRNA induction. PLoS ONE 2014, 9, e93954. [Google Scholar] [CrossRef]
- Večeřa, J.; Bártová, E.; Krejčí, J.; Legartová, S.; Komůrková, D.; Rudá-Kučerová, J.; Štark, T.; Dražanová, E.; Kašpárek, T.; Šulcová, A.; et al. HDAC1 and HDAC3 underlie dynamic H3K9 acetylation during embryonic neurogenesis and in schizophrenia-like animals. J. Cell Physiol. 2018, 233, 530–548. [Google Scholar] [CrossRef]
- Miranda, K.; Mehrpouya-Bahrami, P.; Nagarkatti, P.S.; Nagarkatti, M. Cannabinoid Receptor 1 Blockade Attenuates Obesity and Adipose Tissue Type 1 Inflammation Through miR-30e-5p Regulation of Delta-Like-4 in Macrophages and Consequently Downregulation of Th1 Cells. Front. Immunol. 2019, 10, 1049. [Google Scholar] [CrossRef]
- Shamran, H.; Singh, N.P.; Zumbrun, E.E.; Murphy, A.; Taub, D.D.; Mishra, M.K.; Price, R.L.; Chatterjee, S.; Nagarkatti, M.; Nagarkatti, P.S.; et al. Fatty acid amide hydrolase (FAAH) blockade ameliorates experimental colitis by altering microRNA expression and suppressing inflammation. Brain Behav. Immun. 2017, 59, 10–20. [Google Scholar] [CrossRef]
- Hollins, S.L.; Zavitsanou, K.; Walker, F.R.; Cairns, M.J. Alteration of imprinted Dlk1-Dio3 miRNA cluster expression in the entorhinal cortex induced by maternal immune activation and adolescent cannabinoid exposure. Transl. Psychiatry 2014, 4, e452. [Google Scholar] [CrossRef]
- Aguado, T.; Carracedo, A.; Julien, B.; Velasco, G.; Milman, G.; Mechoulam, R.; Alvarez, L.; Guzmán, M.; Galve-Roperh, I. Cannabinoids induce glioma stem-like cell differentiation and inhibit gliomagenesis. J. Biol. Chem. 2007, 282, 6854–6862. [Google Scholar] [CrossRef]
- Di Giacomo, D.; De Domenico, E.; Sette, C.; Geremia, R.; Grimaldi, P. Type 2 cannabinoid receptor contributes to the physiological regulation of spermatogenesis. FASEB J. 2016, 30, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Innocenzi, E.; De Domenico, E.; Ciccarone, F.; Zampieri, M.; Rossi, G.; Cicconi, R.; Bernardini, R.; Mattei, M.; Grimaldi, P. Paternal activation of CB2 cannabinoid receptor impairs placental and embryonic growth via an epigenetic mechanism. Sci. Rep. 2019, 9, 17034. [Google Scholar] [CrossRef] [PubMed]
- Mehrpouya-Bahrami, P.; Miranda, K.; Singh, N.P.; Zumbrun, E.E.; Nagarkatti, M.; Nagarkatti, P.S. Role of microRNA in CB1 antagonist-mediated regulation of adipose tissue macrophage polarization and chemotaxis during diet-induced obesity. J. Biol. Chem. 2019, 294, 7669–7681. [Google Scholar] [CrossRef] [PubMed]
- Ibn Lahmar Andaloussi, Z.; Taghzouti, K.; Abboussi, O. Behavioural and epigenetic effects of paternal exposure to cannabinoids during adolescence on offspring vulnerability to stress. Int. J. Dev. Neurosci. 2019, 72, 48–54. [Google Scholar] [CrossRef]
- Tomas-Roig, J.; Benito, E.; Agis-Balboa, R.C.; Piscitelli, F.; Hoyer-Fender, S.; Di Marzo, V.; Havemann-Reinecke, U. Chronic exposure to cannabinoids during adolescence causes long-lasting behavioral deficits in adult mice. Addict Biol. 2017, 22, 1778–1789. [Google Scholar] [CrossRef]
- Chianese, R.; Coccurello, R.; Viggiano, A.; Scafuro, M.; Fiore, M.; Coppola, G.; Operto, F.F.; Fasano, S.; Layé, S.; Pierantoni, R.; et al. Impact of dietary fat on brain functions. Curr. Neuropharmacol. 2018, 16, 1059–1085. [Google Scholar] [CrossRef]
- Sam, A.H.; Salem, V.; Ghatei, M.A. Rimonabant: From RIO to Ban. J. Obes. 2011, 2011, 432607. [Google Scholar] [CrossRef]
- Di Marzo, V.; Goparaju, S.K.; Wang, L.; Liu, J.; Bátkai, S.; Járai, Z.; Fezza, F.; Miura, G.I.; Palmiter, R.D.; Sugiura, T.; et al. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 2001, 410, 822–825. [Google Scholar] [CrossRef]
- Swerdloff, R.S.; Batt, R.A.; Bray, G.A. Reproductive hormonal function in the genetically obese (ob/ob) mouse. Endocrinology 1976, 98, 1359–1364. [Google Scholar] [CrossRef]
- Hay, E.A.; McEwan, A.; Wilson, D.; Barrett, P.; D’Agostino, G.; Pertwee, R.G.; MacKenzie, A. Disruption of an enhancer associated with addictive behaviour within the cannabinoid receptor-1 gene suggests a possible role in alcohol intake, cannabinoid response and anxiety-related behaviour. Psychoneuroendocrinology 2019, 109, 104407. [Google Scholar] [CrossRef]
- Wang, H.; Dey, S.K.; Maccarrone, M. Jekyll and Hyde: Two faces of cannabinoid signaling in male and female fertility. Endocr. Rev. 2006, 27, 427–448. [Google Scholar] [CrossRef] [PubMed]
- Fasano, S.; Meccariello, R.; Cobellis, G.; Chianese, R.; Cacciola, G.; Chioccarelli, T.; Pierantoni, R. The endocannabinoid system: An ancient signaling involved in the control of male fertility. Annals Ann. N. Y. Acad. Sci. 2009, 1163, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Pierantoni, R.; Cobellis, G.; Meccariello, R.; Cacciola, G.; Chianese, R.; Chioccarelli, T.; Fasano, S. Testicular gonadotropin-releasing hormone activity, progression of spermatogenesis and sperm transport in vertebrates. Ann. N. Y. Acad. Sci. 2009, 1163, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Battista, N.; Meccariello, R.; Cobellis, G.; Fasano, S.; Di Tommaso, M.; Pirazzi, V.; Konje, J.C.; Pierantoni, R.; Maccarrone, M. The role of endocannabinoids in gonadal function and fertility along the evolutionary axis. Mol. Cell. Endocrinol. 2012, 355, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meccariello, R.; Battista, N.; Bradshaw, H.B.; Wang, H. Updates in reproduction coming from the endocannabinoid system. Int. J. Endocrinol. 2014, 2014, 412354. [Google Scholar] [CrossRef]
- Bovolin, P.; Cottone, E.; Pomatto, V.; Fasano, S.; Pierantoni, R.; Cobellis, G.; Meccariello, R. Endocannabinoids are involved in male vertebrate reproduction: Regulatory mechanisms at central and gonadal level. Front. Endocrinol. 2014, 5, 54. [Google Scholar] [CrossRef]
- Cobellis, G.; Meccariello, R.; Chianese, R.; Chioccarelli, T.; Fasano, S.; Pierantoni, R. Effects of neuroendocrine CB1 activity on adult Leydig cells. Front. Endocrinol. 2016, 7, 47. [Google Scholar] [CrossRef]
- Cecconi, S.; Rapino, C.; Di Nisio, V.; Rossi, G.; Maccarrone, M. The (endo)cannabinoid signaling in female reproduction: What are the latest advances? Prog. Lipid Res. 2019, 77, 101019. [Google Scholar] [CrossRef]
- Meccariello, R.; Franzoni, M.F.; Chianese, R.; Cottone, E.; Scarpa, D.; Donna, D.; Cobellis, G.; Guastalla, A.; Pierantoni, R.; Fasano, S. Interplay between the endocannabinoid system and GnRH-I in the forebrain of the anuran amphibian Rana esculenta. Endocrinology 2008, 149, 2149–2158. [Google Scholar] [CrossRef]
- Cacciola, G.; Chioccarelli, T.; Mackie, K.; Meccariello, R.; Ledent, C.; Fasano, S.; Pierantoni, R.; Cobellis, G. Expression of type-1 cannabinoid receptor during rat postnatal testicular development: Possible involvement in adult Leydig cell differentiation. Biol. Reprod. 2008, 79, 758–765. [Google Scholar] [CrossRef]
- Grimaldi, P.; Orlando, P.; Di Siena, S.; Lolicato, F.; Petrosino, S.; Bisogno, T.; Geremia, R.; De Petrocellis, L.; Di Marzo, V. The endocannabinoid system and pivotal role of the CB2 receptor in mouse spermatogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 11131–11136. [Google Scholar] [CrossRef]
- Trabucco, E.; Acone, G.; Marenna, A.; Pierantoni, R.; Cacciola, G.; Chioccarelli, T.; Mackie, K.; Fasano, S.; Colacurci, N.; Meccariello, R.; et al. Endocannabinoid System in First Trimester Placenta: Low FAAH and High CB1 Expression Characterize Spontaneous Miscarriage. Placenta 2009, 30, 516–522. [Google Scholar] [CrossRef]
- Acone, G.; Trabucco, E.; Colacurci, N.; Cobellis, L.; Mackie, K.; Meccariello, R.; Cacciola, G.; Chioccarelli, T.; Fasano, S.; Pierantoni, R.; et al. Low type I cannabinoid receptor levels characterize placental villous in labouring delivery. Placenta 2009, 30, 203–205. [Google Scholar] [CrossRef]
- Aquila, S.; Guido, C.; Laezza, C.; Santoro, A.; Pezzi, V.; Panza, S.; Andò, S.; Bifulco, M. A new role of anandamide in human sperm: Focus on metabolism. J. Cell Physiol. 2009, 221, 147–153. [Google Scholar] [CrossRef]
- Aquila, S.; Guido, C.; Santoro, A.; Perrotta, I.; Laezza, C.; Bifulco, M.; Sebastiano, A. Human sperm anatomy: Ultrastructural localization of the cannabinoid1 receptor and a potential role of anandamide in sperm survival and acrosome reaction. Anat. Rec. 2010, 293, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Chioccarelli, T.; Cacciola, G.; Altucci, L.; Lewis, S.E.; Simon, L.; Ricci, G.; Ledent, C.; Meccariello, R.; Fasano, S.; Pierantoni, R.; et al. Cannabinoid Receptor 1 Influences Chromatin Remodeling in Mouse Spermatids by Affecting Content of Transition Protein 2 mRNA and Histone Displacement. Endocrinology 2010, 151, 5017–5029. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, G.; Battista, N.; Rossi, G.; Di Tommaso, M.; Pucci, M.; Pirazzi, V.; Cecconi, S.; Maccarrone, M. Effect of capacitation on the endocannabinoid system of mouse sperm. Mol. Cell. Endocrinol. 2011, 343, 88–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chianese, R.; Ciaramella, V.; Scarpa, D.; Fasano, S.; Pierantoni, R.; Meccariello, R. Anandamide regulates the expression of GnRH1, GnRH2 and GnRHRs in frog testis. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E475–E487. [Google Scholar] [CrossRef]
- Cacciola, G.; Chioccarelli, T.; Altucci, L.; Ledent, C.; Mason, J.I.; Fasano, S.; Pierantoni, R.; Cobellis, G. Low 17beta-estradiol levels in CNR1 knock-out mice affect spermatid chromatin remodeling by interfering with chromatin reorganization. Biol. Reprod. 2013, 88, 152. [Google Scholar] [CrossRef]
- Ciaramella, V.; Meccariello, R.; Chioccarelli, T.; Sirleto, M.; Fasano, S.; Pierantoni, R.; Chianese, R. Anandamide acts via kisspeptin in the regulation of testicular activity of the frog, Pelophylax esculentus. Mol. Cell. Endocrinol. 2016, 420, 75–84. [Google Scholar] [CrossRef]
- Migliaccio, M.; Ricci, G.; Suglia, A.; Manfrevola, F.; Mackie, K.; Fasano, S.; Pierantoni, R.; Chioccarelli, T.; Cobellis, G. Analysis of Endocannabinoid System in Rat Testis During the First Spermatogenetic Wave. Front. Endocrinol. 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.E.; Rolland, A.D.; Rajpert-De Meyts, E.; Janfelt, C.; Jørgensen, A.; Winge, S.B.; Kristensen, D.M.; Juul, A.; Chalmel, F.; Jégou, B.; et al. Characterisation and localisation of the endocannabinoid system components in the adult human testis. Sci. Rep. 2019, 9, 12866. [Google Scholar] [CrossRef] [PubMed]
- Barchi, M.; Innocenzi, E.; Giannattasio, T.; Dolci, S.; Rossi, P.; Grimaldi, P. Cannabinoid receptor signalling in the development, epigenetics and tumors of male germ cells. Int. J. Mol. Sci. 2019, 21, 25. [Google Scholar] [CrossRef] [PubMed]
- Cobellis, G.; Ricci, G.; Cacciola, G.; Orlando, P.; Petrosino, S.; Cascio, M.G.; Bisogno, T.; De Petrocellis, L.; Chioccarelli, T.; Altucci, L.; et al. A Gradient of 2-Arachidonoylglycerol Regulates Mouse Epididymal Sperm Cell Start-Up. Biol. Reprod. 2010, 82, 451–458. [Google Scholar] [CrossRef]
- Santoro, A.; Chianese, R.; Troisi, J.; Richards, S.; Nori, S.L.; Fasano, S.; Guida, M.; Plunk, E.; Viggiano, A.; Pierantoni, R.; et al. Neuro-toxic and Reproductive Effects of BPA. Curr. Neuropharmacol. 2019, 17, 1109–1132. [Google Scholar] [CrossRef]
- Motti, M.L.; Meccariello, R. Minireview: The epigenetic modulation of KISS1 in cancer and reproduction. Int. J. Environ. Res. Public Health 2019, 16, 2607. [Google Scholar] [CrossRef]
- Chamani, I.J.; Keefe, D.L. Epigenetics and Female Reproductive Aging. Front. Endocrinol. 2019, 10, 473. [Google Scholar] [CrossRef]
- Fernandez-Fernandez, R.; Martini, A.C.; Navarro, V.M.; Castellano, J.M.; Dieguez, C.; Aguilar, E.; Pinilla, L.; Tena-Sempere, M. Novel signals for the integration of energy balance and reproduction. Mol. Cell. Endocrinol. 2006, 254–255, 127–132. [Google Scholar] [CrossRef]
- Rossi, G.; Gasperi, V.; Paro, R.; Barsacchi, D.; Cecconi, S.; Maccarrone, M. Follicle-stimulating hormone activates fatty acid amide hydrolase by protein kinase A and aromatase-dependent pathways in mouse primary Sertoli cells. Endocrinology 2007, 148, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Waleh, N.S.; Cravatt, B.F.; Apte-Deshpande, A.; Terao, A.; Kilduff, T.S. Transcriptional regulation of the mouse fatty acid amide hydrolase gene. Gene 2002, 291, 203–210. [Google Scholar] [CrossRef]
- Grimaldi, P.; Pucci, M.; Di Siena, S.; Di Giacomo, D.; Pirazzi, V.; Geremia, R.; Maccarrone, M. The faah gene is the first direct target of estrogen in the testis: Role of histone demethylase LSD1. Cell. Mol. Life Sci. 2012, 69, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R. Epididymosomes: Role of extracellular microvesicles in sperm maturation. Front. Biosci. (Schol. Ed.) 2016, 8, 106–114. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.W.; Li, H.M.; Qing, X.R.; Huang, D.H.; Li, H.G. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Sci. Rep. 2016, 6, 39080. [Google Scholar] [CrossRef] [PubMed]
- Fereshteh, Z.; Schmidt, S.A.; Al-Dossary, A.A.; Accerbi, M.; Arighi, C.; Cowart, J.; Song, J.L.; Green, P.J.; Choi, K.; Yoo, S.; et al. Murine Oviductosomes (OVS) microRNA profiling during the estrous cycle: Delivery of OVS-borne microRNAs to sperm where miR-34c-5p localizes at the centrosome. Sci. Rep. 2018, 8, 16094. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Rathke, C.; Baarends, W.M.; Awe, S.; Renkawitz-Pohl, R. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 2014, 1839, 155–168. [Google Scholar] [CrossRef]
- Cacciola, G.; Chioccarelli, T.; Altucci, L.; Viggiano, A.; Fasano, S.; Pierantoni, R.; Cobellis, G. Nuclear size as estrogen-responsive chromatin quality parameter of mouse spermatozoa. Gen. Comp. Endocrinol. 2013, 193, 201–209. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).