Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients
Abstract
1. Introduction
2. Results
2.1. Clinical Assessment
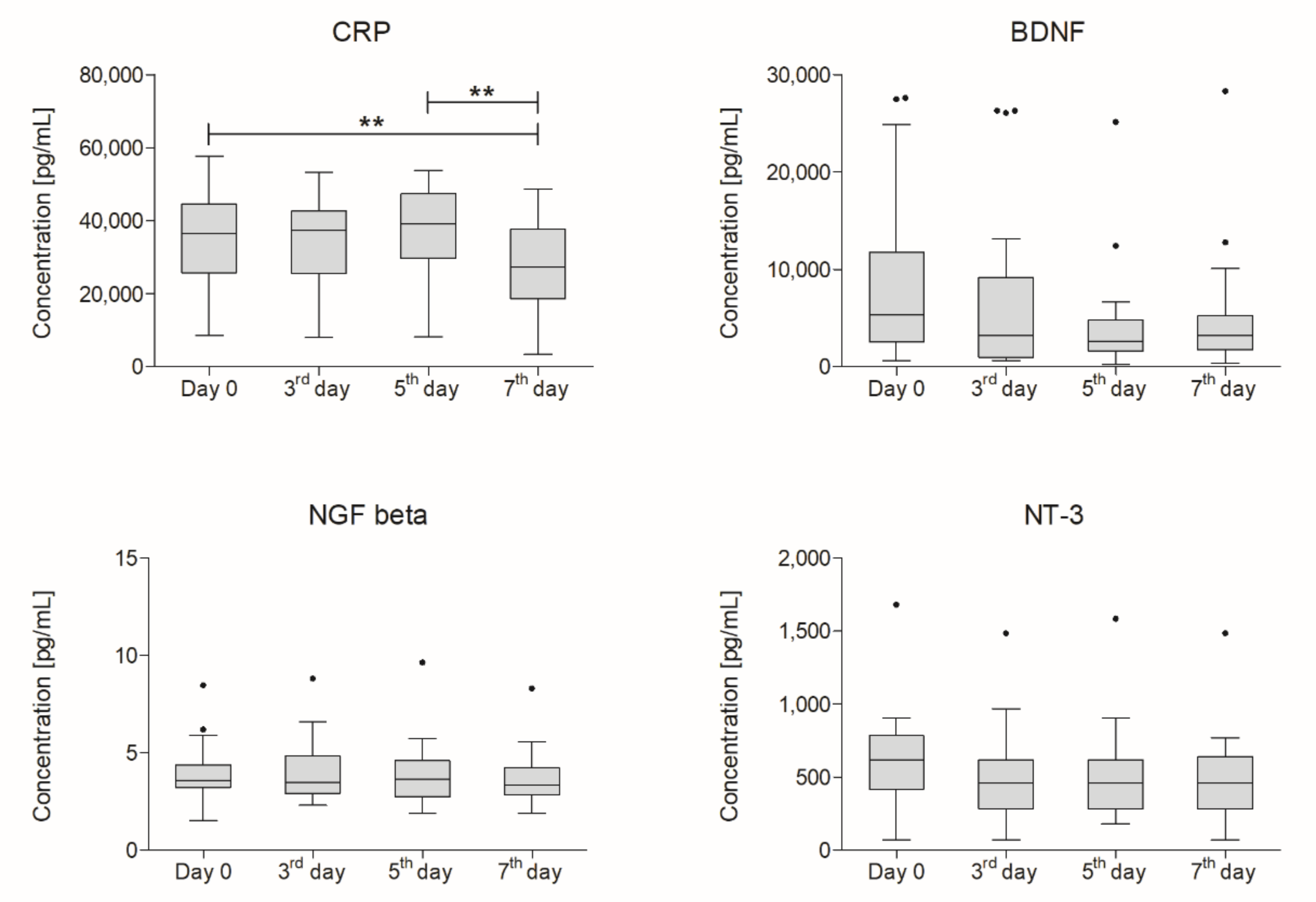
2.2. Concentration of CRP and Selected NTs
2.3. Correlation Between Functional Outcome and Concentration of CRP and NTs
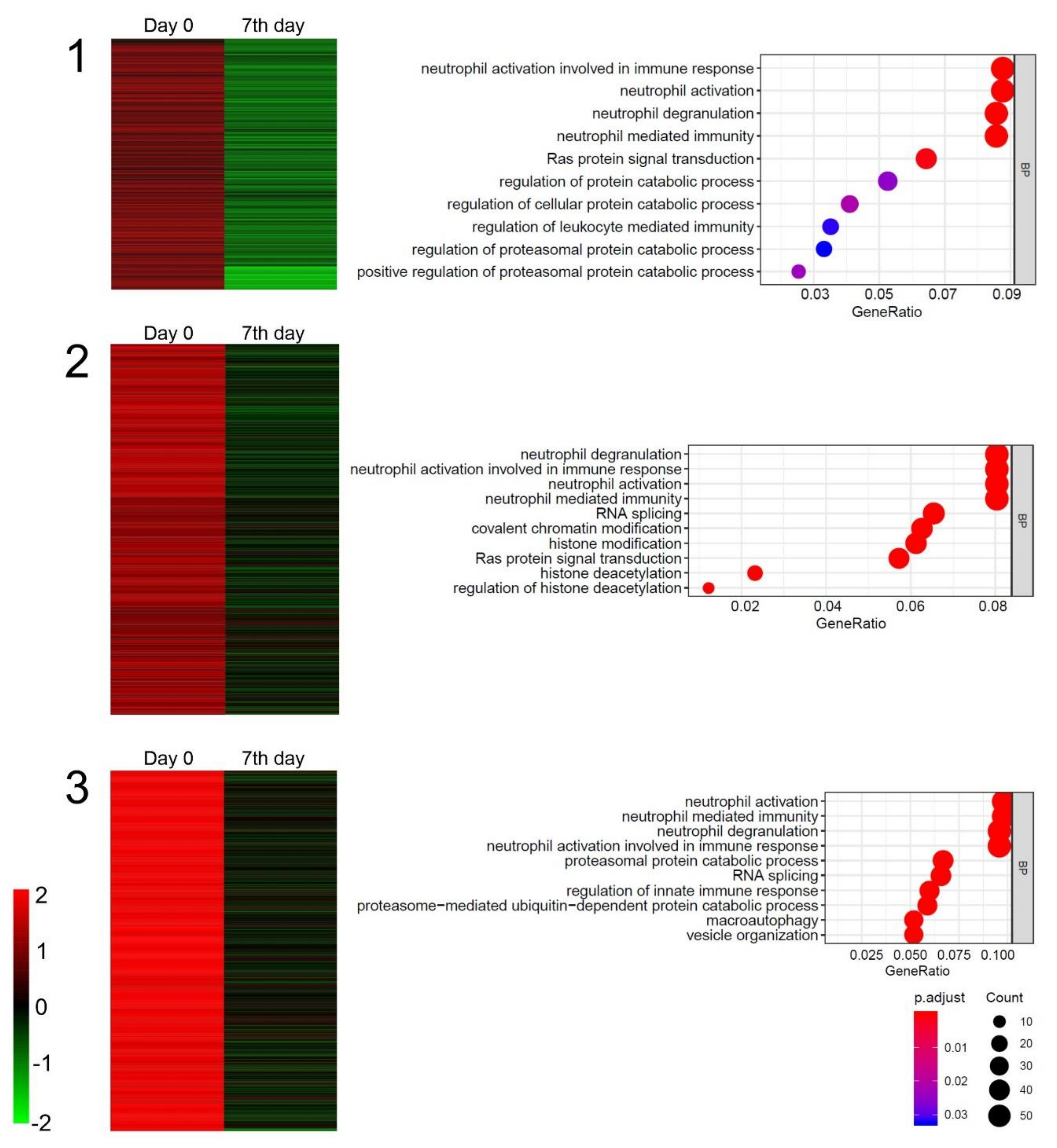
2.4. Gene Microarray Analysis
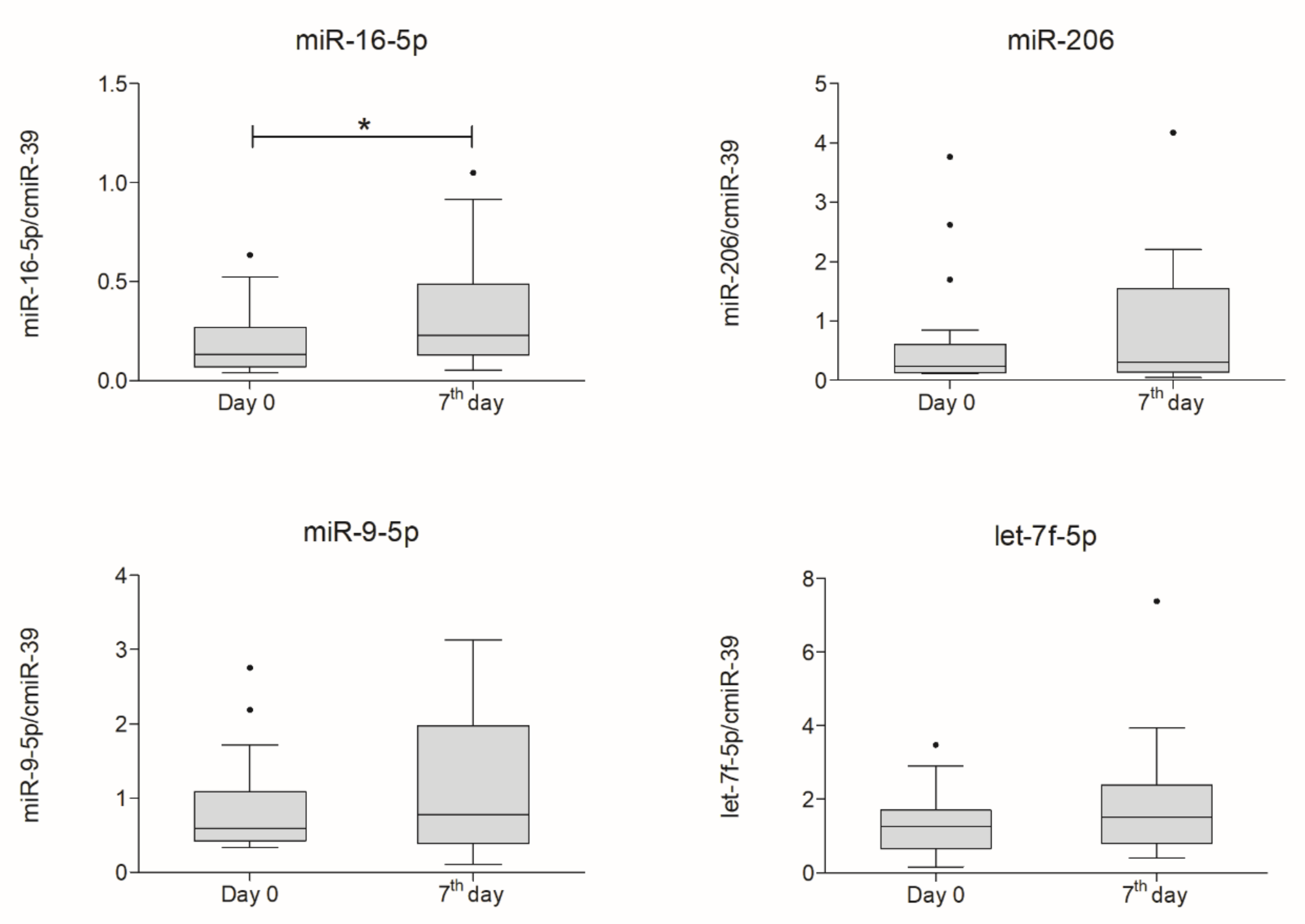
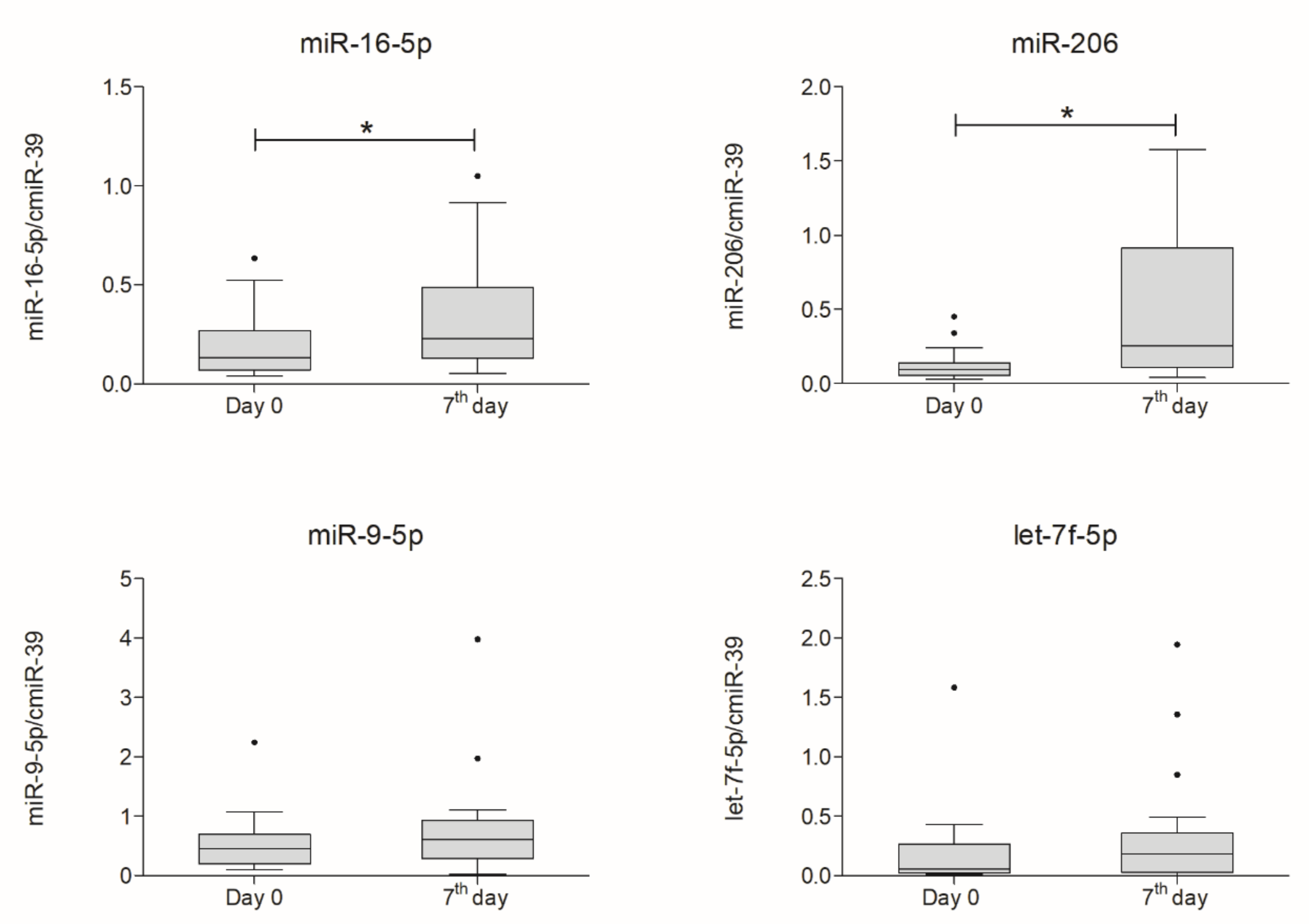
2.5. miRNA Expression Analysis
3. Discussion
Potential Study Limitations
4. Materials and Methods
4.1. Patients
4.2. Clinical Assessment
4.3. Bone Marrow Collection and Lin− Cells Isolation
4.4. Intrathecal Injection of Lin− Cell Suspension
4.5. Sample Collection
4.6. Luminex Multiplex Assay
4.7. Gene Chip Microarray
4.8. miRNA Expression Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic lateral sclerosis |
| ALS-FRSr | Amyotrophic Lateral Sclerosis functional rating scale revised |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| CRP | C-reactive protein |
| CSF | Cerebrospinal fluid |
| CXCR4 | Chemokine receptor type 4 |
| DEGs | Differentially expressed genes |
| FDA | Food and Drug Administration |
| GO | Gene ontology |
| Lin− | Lineage-negative |
| MNs | Motor neurons |
| NGF beta | Nerve growth factor beta |
| NT-3 | Neurotrophin-3 |
| NTs | Neurotrophins |
| PBS | Phosphate-buffered saline |
| sALS | Sporadic amyotrophic lateral sclerosis |
| SCs’ | Stem cells |
| SOD1 | Superoxide dismutase 1 |
| SPCs | Stem and progenitor cells |
| Tx | Transplantation |
References
- Niedermeyer, S.; Murn, M.; Choi, P.J. Respiratory Failure in Amyotrophic Lateral Sclerosis. Chest 2018, 155, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Rowland, L.P.; Shneider, N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001, 344, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Al-Chalabi, A.; Hardiman, O. The epidemiology of ALS: A conspiracy of genes, environment and time. Nat. Rev. Neurol. 2013, 9, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Kurland, L.T.; Mulder, D.W. Epidemiologic investigations of amyotrophic lateral sclerosis. 2. Familial aggregations indicative of dominant inheritance. II. Neurology 1955, 5, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Peters, O.M.; Ghasemi, M.; Brown, R.H., Jr. Emerging mechanisms of molecular pathology in ALS. J. Clin. Investig. 2015, 125, 1767–1779. [Google Scholar] [CrossRef]
- Logroscino, G.; Traynor, B.J.; Hardiman, O.; Chiò, A.; Mitchell, D.; Swingler, R.J.; Millul, A.; Benn, E.; Beghi, E. Incidence of Amyotrophic Lateral Sclerosis in Europe. J. Neurol. Neurosurg. Psychiatry 2010, 81, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, N.; Facciponte, D.; Bradley, W.; Stommel, E. Cytokine expression levels in ALS: A potential link between inflammation and BMAA-triggered protein misfolding. Cytokine Growth Factor Rev. 2017, 37, 81–88. [Google Scholar] [CrossRef]
- Forsberg, K.; Jonsson, P.A.; Andersen, P.M.; Bergemalm, D.; Graffmo, K.S.; Hultdin, M.; Jacobsson, J.; Rosquist, R.; Marklund, S.L.; Brannstrom, T. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE 2010, 5, e11552. [Google Scholar] [CrossRef]
- Scotter, E.L.; Chen, H.J.; Shaw, C.E. TDP-43 Proteinopathy and ALS: Insights into Disease Mechanisms and Therapeutic Targets. Neurotherapeutics 2015, 12, 352–363. [Google Scholar] [CrossRef]
- Stoica, R.; De Vos, K.J.; Paillusson, S.; Mueller, S.; Sancho, R.M.; Lau, K.F.; Vizcay-Barrena, G.; Lin, W.L.; Xu, Y.F.; Lewis, J.; et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat. Commun. 2014, 5, 3996. [Google Scholar] [CrossRef]
- Calvo, A.; Canosa, A.; Bertuzzo, D.; Cugnasco, P.; Solero, L.; Clerico, M.; De Mercanti, S.; Bersano, E.; Cammarosano, S.; Ilardi, A.; et al. Influence of cigarette smoking on ALS outcome: A population-based study. J. Neurol. Neurosurg. Psychiatry 2016, 87, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.S.; Wosiski-Kuhn, M.; Gillespie, R.; Caress, J.; Milligan, C. Inflammation, Immunity, and amyotrophic lateral sclerosis: I. Etiology and pathology. Muscle Nerve 2019, 59, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Almer, G.; Guegan, C.; Teismann, P.; Naini, A.; Rosoklija, G.; Hays, A.P.; Chen, C.; Przedborski, S. Increased expression of the pro-inflammatory enzyme cyclooxygenase-2 in amyotrophic lateral sclerosis. Ann. Neurol. 2001, 49, 176–185. [Google Scholar] [CrossRef]
- Murdock, B.J.; Zhou, T.; Kashlan, S.R.; Little, R.J.; Goutman, S.A.; Feldman, E.L. Correlation of Peripheral Immunity With Rapid Amyotrophic Lateral Sclerosis Progression. JAMA Neurol. 2017, 74, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Doble, A. The pharmacology and mechanism of action of riluzole. Neurology 1996, 47, S233–S241. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.P. Edaravone (Radicava): A Novel Neuroprotective Agent for the Treatment of Amyotrophic Lateral Sclerosis. Pharmacol. Ther. 2018, 43, 25–28. [Google Scholar]
- Miller, R.G.; Appel, S.H. Introduction to supplement: The current status of treatment for ALS. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 1–4. [Google Scholar] [CrossRef]
- Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017, 16, 505–512. [CrossRef]
- Paganoni, S.; Macklin, E.A.; Lee, A.; Murphy, A.; Chang, J.; Zipf, A.; Cudkowicz, M.; Atassi, N. Diagnostic timelines and delays in diagnosing amyotrophic lateral sclerosis (ALS). Amyotroph. Lateral. Scler. Front. Degener. 2014, 15, 453–456. [Google Scholar] [CrossRef]
- Lunn, J.S.; Sakowski, S.A.; Feldman, E.L. Stem cell therapies for amyotrophic lateral sclerosis: Recent advances and prospects for the future. Stem Cells 2014, 32, 1099–1109. [Google Scholar] [CrossRef]
- Forraz, N.; Pettengell, R.; McGuckin, C.P. Characterization of a lineage-negative stem-progenitor cell population optimized for ex vivo expansion and enriched for LTC-IC. Stem Cells 2004, 22, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Paczkowska, E.; Kaczynska, K.; Pius-Sadowska, E.; Roginska, D.; Kawa, M.; Ustianowski, P.; Safranow, K.; Celewicz, Z.; Machalinski, B. Humoral activity of cord blood-derived stem/progenitor cells: Implications for stem cell-based adjuvant therapy of neurodegenerative disorders. PLoS ONE 2013, 8, e83833. [Google Scholar] [CrossRef] [PubMed]
- Sobus, A.; Baumert, B.; Litwinska, Z.; Golab-Janowska, M.; Stepniewski, J.; Kotowski, M.; Pius-Sadowska, E.; Kawa, M.P.; Grodecka-Szwajkiewicz, D.; Peregud-Pogorzelski, J.; et al. Safety and Feasibility of Lin-Cells Administration to ALS Patients: A Novel View on Humoral Factors and miRNA Profiles. Int. J. Mol. Sci. 2018, 19, 1312. [Google Scholar] [CrossRef] [PubMed]
- Di Terlizzi, R.; Platt, S. The function, composition and analysis of cerebrospinal fluid in companion animals: Part I-function and composition. Vet. J. 2006, 172, 422–431. [Google Scholar] [CrossRef]
- McGuckin, C.P.; Forraz, N.; Allouard, Q.; Pettengell, R. Umbilical cord blood stem cells can expand hematopoietic and neuroglial progenitors in vitro. Exp. Cell Res. 2004, 295, 350–359. [Google Scholar] [CrossRef]
- Bibel, M.; Barde, Y.A. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000, 14, 2919–2937. [Google Scholar] [CrossRef]
- Keefe, K.M.; Sheikh, I.S.; Smith, G.M. Targeting Neurotrophins to Specific Populations of Neurons: NGF, BDNF, and NT-3 and Their Relevance for Treatment of Spinal Cord Injury. Int. J. Mol. Sci. 2017, 18, 548. [Google Scholar] [CrossRef]
- Ikeda, K.; Klinkosz, B.; Greene, T.; Cedarbaum, J.M.; Wong, V.; Lindsay, R.M.; Mitsumoto, H. Effects of brain-derived neurotrophic factor on motor dysfunction in wobbler mouse motor neuron disease. Ann. Neurol. 1995, 37, 505–511. [Google Scholar] [CrossRef]
- Haase, G.; Kennel, P.; Pettmann, B.; Vigne, E.; Akli, S.; Revah, F.; Schmalbruch, H.; Kahn, A. Gene therapy of murine motor neuron disease using adenoviral vectors for neurotrophic factors. Nat. Med. 1997, 3, 429–436. [Google Scholar] [CrossRef]
- A controlled trial of recombinant methionyl human BDNF in ALS: The BDNF Study Group (Phase III). Neurology 1999, 52, 1427–1433. [CrossRef]
- Price, R.D.; Milne, S.A.; Sharkey, J.; Matsuoka, N. Advances in small molecules promoting neurotrophic function. Pharmacol. Ther. 2007, 115, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Ryberg, H.; An, J.; Darko, S.; Lustgarten, J.L.; Jaffa, M.; Gopalakrishnan, V.; Lacomis, D.; Cudkowicz, M.E.; Bowser, R. Discovery and Verification of Amyotrophic Lateral Sclerosis Biomarkers by Proteomics. Muscle Nerve 2010, 42, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Yasojima, K.; Schwab, C.; McGeer, E.G.; McGeer, P.L. Human neurons generate C-reactive protein and amyloid P: Upregulation in Alzheimer’s disease. Brain Res. 2000, 887, 80–89. [Google Scholar] [CrossRef]
- McGeer, P.L.; McGeer, E.G.; Yasojima, K. Alzheimer disease and neuroinflammation. J. Neural. Transm. Suppl. 2000, 59, 53–57. [Google Scholar] [CrossRef]
- Lunnon, K.; Teeling, J.L.; Tutt, A.L.; Cragg, M.S.; Glennie, M.J.; Perry, V.H. Systemic inflammation modulates Fc receptor expression on microglia during chronic neurodegeneration. J. Immunol. 2011, 186, 7215–7224. [Google Scholar] [CrossRef]
- Murdock, B.J.; Bender, D.E.; Kashlan, S.R.; Figueroa-Romero, C.; Backus, C.; Callaghan, B.C.; Goutman, S.A.; Feldman, E.L. Increased ratio of circulating neutrophils to monocytes in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e242. [Google Scholar] [CrossRef]
- Ricci, C.; Marzocchi, C.; Battistini, S. MicroRNAs as Biomarkers in Amyotrophic Lateral Sclerosis. Cells 2018, 7, 219. [Google Scholar] [CrossRef]
- Toivonen, J.M.; Manzano, R.; Olivan, S.; Zaragoza, P.; Garcia-Redondo, A.; Osta, R. MicroRNA-206: A potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS ONE 2014, 9, e89065. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534. [Google Scholar] [CrossRef]
- Majer, A.; Medina, S.J.; Niu, Y.; Abrenica, B.; Manguiat, K.J.; Frost, K.L.; Philipson, C.S.; Sorensen, D.L.; Booth, S.A. Early mechanisms of pathobiology are revealed by transcriptional temporal dynamics in hippocampal CA1 neurons of prion infected mice. PLoS Pathog. 2012, 8, e1003002. [Google Scholar] [CrossRef]
- De Andrade, H.M.; de Albuquerque, M.; Avansini, S.H.; Rocha, C.d.S.; Dogini, D.B.; Nucci, A.; Carvalho, B.; Lopes-Cendes, I.; Franca, M.C., Jr. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 2016, 368, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Delaloy, C.; Liu, L.; Lee, J.A.; Su, H.; Shen, F.; Yang, G.Y.; Young, W.L.; Ivey, K.N.; Gao, F.B. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 2010, 6, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, I.; Hama, Y.; Matsushima, M.; Hirotani, M.; Kano, T.; Hohzen, H.; Yabe, I.; Utsumi, J.; Sasaki, H. Identification of plasma microRNAs as a biomarker of sporadic Amyotrophic Lateral Sclerosis. Mol. Brain 2015, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.; Zhang, B.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Gordon, P.H. Amyotrophic lateral sclerosis: Pathophysiology, diagnosis and management. CNS Drugs 2011, 25, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hennigan, A.; O’Callaghan, R.M.; Kelly, Á.M. Neurotrophins and their receptors: Roles in plasticity, neurodegeneration and neuroprotection. Biochem. Soc. Trans. 2007, 35, 424–427. [Google Scholar] [CrossRef]
- Sakane, T.; Pardridge, W.M. Carboxyl-directed pegylation of brain-derived neurotrophic factor markedly reduces systemic clearance with minimal loss of biologic activity. Pharm Res. 1997, 14, 1085–1091. [Google Scholar] [CrossRef]
- Tria, M.A.; Fusco, M.; Vantini, G.; Mariot, R. Pharmacokinetics of nerve growth factor (NGF) following different routes of administration to adult rats. Exp. Neurol. 1994, 127, 178–183. [Google Scholar] [CrossRef]
- Ikeda, O.; Murakami, M.; Ino, H.; Yamazaki, M.; Koda, M.; Nakayama, C.; Moriya, H. Effects of brain-derived neurotrophic factor (BDNF) on compression-induced spinal cord injury: BDNF attenuates down-regulation of superoxide dismutase expression and promotes up-regulation of myelin basic protein expression. J. Neuropathol. Exp. Neurol. 2002, 61, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Bemelmans, A.P.; Husson, I.; Jaquet, M.; Mallet, J.; Kosofsky, B.E.; Gressens, P. Lentiviral-mediated gene transfer of brain-derived neurotrophic factor is neuroprotective in a mouse model of neonatal excitotoxic challenge. J. Neurosci. Res. 2006, 83, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Shruthi, S.; Sumitha, R.; Varghese, A.M.; Ashok, S.; Chandrasekhar Sagar, B.K.; Sathyaprabha, T.N.; Nalini, A.; Kramer, B.W.; Raju, T.R.; Vijayalakshmi, K.; et al. Brain-Derived Neurotrophic Factor Facilitates Functional Recovery from ALS-Cerebral Spinal Fluid-Induced Neurodegenerative Changes in the NSC-34 Motor Neuron Cell Line. Neurodegener. Dis. 2017, 17, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, L.; Bonafede, R.; Scambi, I.; Parrella, E.; Pizzi, M.; Mariotti, R. Acetylation state of RelA modulated by epigenetic drugs prolongs survival and induces a neuroprotective effect on ALS murine model. Sci. Rep. 2018, 8, 12875. [Google Scholar] [CrossRef]
- Tremolizzo, L.; Pellegrini, A.; Conti, E.; Arosio, A.; Gerardi, F.; Lunetta, C.; Magni, P.; Appollonio, I.; Ferrarese, C. BDNF Serum Levels with Respect to Multidimensional Assessment in Amyotrophic Lateral Sclerosis. Neurodegener. Dis. 2016, 16, 192–198. [Google Scholar] [CrossRef]
- Prins, B.P.; Abbasi, A.; Wong, A.; Vaez, A.; Nolte, I.; Franceschini, N.; Stuart, P.E.; Guterriez Achury, J.; Mistry, V.; Bradfield, J.P.; et al. Investigating the Causal Relationship of C-Reactive Protein with 32 Complex Somatic and Psychiatric Outcomes: A Large-Scale Cross-Consortium Mendelian Randomization Study. PLoS Med. 2016, 13, e1001976. [Google Scholar] [CrossRef]
- Keizman, D.; Rogowski, O.; Berliner, S.; Ish-Shalom, M.; Maimon, N.; Nefussy, B.; Artamonov, I.; Drory, V.E. Low-grade systemic inflammation in patients with amyotrophic lateral sclerosis. Acta Neurol. Scand. 2009, 119, 383–389. [Google Scholar] [CrossRef]
- Sutedja, N.A.; van der Schouw, Y.T.; Fischer, K.; Sizoo, E.M.; Huisman, M.H.; Veldink, J.H.; Van den Berg, L.H. Beneficial vascular risk profile is associated with amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2011, 82, 638–642. [Google Scholar] [CrossRef]
- Nagel, G.; Peter, R.S.; Rosenbohm, A.; Koenig, W.; Dupuis, L.; Rothenbacher, D.; Ludolph, A.C. Adipokines, C-reactive protein and Amyotrophic Lateral Sclerosis-results from a population-based ALS registry in Germany. Sci. Rep. 2017, 7, 4374. [Google Scholar] [CrossRef]
- Lunetta, C.; Lizio, A.; Maestri, E.; Sansone, V.A.; Mora, G.; Miller, R.G.; Appel, S.H.; Chio, A. Serum C-Reactive Protein as a Prognostic Biomarker in Amyotrophic Lateral Sclerosis. JAMA Neurol. 2017, 74, 660–667. [Google Scholar] [CrossRef]
- Lu, C.H.; Allen, K.; Oei, F.; Leoni, E.; Kuhle, J.; Tree, T.; Fratta, P.; Sharma, N.; Sidle, K.; Howard, R.; et al. Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e244. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Trias, E.; King, P.H.; Si, Y.; Kwon, Y.; Varela, V.; Ibarburu, S.; Kovacs, M.; Moura, I.C.; Beckman, J.S.; Hermine, O.; et al. Mast cells and neutrophils mediate peripheral motor pathway degeneration in ALS. Jci Insight 2018, 3. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, W.; Du, Y.; Guo, Q.; Mao, Y.; Zhou, X.; Hua, D. Serum miR-16 as a potential biomarker for human cancer diagnosis: Results from a large-scale population. J. Cancer Res. Clin. Oncol. 2019, 145, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Grasedieck, S.; Scholer, N.; Bommer, M.; Niess, J.H.; Tumani, H.; Rouhi, A.; Bloehdorn, J.; Liebisch, P.; Mertens, D.; Dohner, H.; et al. Impact of serum storage conditions on microRNA stability. Leukemia Engl. 2012, 26, 2414–2416. [Google Scholar] [CrossRef]
- Waller, R.; Wyles, M.; Heath, P.R.; Kazoka, M.; Wollff, H.; Shaw, P.J.; Kirby, J. Small RNA Sequencing of Sporadic Amyotrophic Lateral Sclerosis Cerebrospinal Fluid Reveals Differentially Expressed miRNAs Related to Neural and Glial Activity. Front. Neurosci. 2017, 11, 731. [Google Scholar] [CrossRef]
- Liguori, M.; Nuzziello, N.; Introna, A.; Consiglio, A.; Licciulli, F.; D’Errico, E.; Scarafino, A.; Distaso, E.; Simone, I.L. Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2018, 11, 288. [Google Scholar] [CrossRef]
- Sharma, M.; Juvvuna, P.K.; Kukreti, H.; McFarlane, C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. 2014, 5, 239. [Google Scholar] [CrossRef]
- Tasca, E.; Pegoraro, V.; Merico, A.; Angelini, C. Circulating microRNAs as biomarkers of muscle differentiation and atrophy in ALS. Clin. Neuropathol. 2016, 35, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.; Goodall, E.F.; Milo, M.; Cooper-Knock, J.; Da Costa, M.; Hobson, E.; Kazoka, M.; Wollff, H.; Heath, P.R.; Shaw, P.J.; et al. Serum miRNAs miR-206, 143–3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 2017, 55, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Nakao, H.; Kiyonari, H.; Abe, T.; Aizawa, S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011, 31, 3407–3422. [Google Scholar] [CrossRef] [PubMed]
- Dajas-Bailador, F.; Bonev, B.; Garcez, P.; Stanley, P.; Guillemot, F.; Papalopulu, N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012, 15, 697–699. [Google Scholar] [CrossRef]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.W.; Wills, A.M.; Wirguin, I.; et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron. Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Kaufmann, P.; Levy, G.; Thompson, J.L.; Delbene, M.L.; Battista, V.; Gordon, P.H.; Rowland, L.P.; Levin, B.; Mitsumoto, H. The ALSFRSr predicts survival time in an ALS clinic population. Neurology 2005, 64, 38–43. [Google Scholar] [CrossRef]
- Oda, E.; Ohashi, Y.; Tashiro, K.; Mizuno, Y.; Kowa, H.; Yanagisawa, N. Reliability and factorial structure of a rating scale for amyotrophic lateral sclerosis. No Shinkei 1996, 48, 999–1007. [Google Scholar]
- Cymbaluk-Ploska, A.; Chudecka-Glaz, A.; Pius-Sadowska, E.; Sompolska-Rzechula, A.; Chudecka, K.; Bulsa, M.; Machalinski, B.; Menkiszak, J. Clinical Relevance of NGAL/MMP-9 Pathway in Patients with Endometrial Cancer. Dis. Markers 2017, 2017, 6589262. [Google Scholar] [CrossRef]
- Wu, F.-X. Genetic weighted k-means algorithm for clustering large-scale gene expression data. BMC Bioinform. 2008, 9, 1–10. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef] [PubMed]
- Fresno, C.; Fernandez, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef] [PubMed]
- Jazwa, A.; Kasper, L.; Bak, M.; Sobczak, M.; Szade, K.; Jozkowicz, A.; Sladek, K.; Dulak, J. Differential inflammatory microRNA and cytokine expression in pulmonary sarcoidosis. Arch. Immunol. Exp. 2015, 63, 139–146. [Google Scholar] [CrossRef] [PubMed]


| Score Fluctuations | Norris Scale | ALS-FRSr |
|---|---|---|
| Stable (score in day 28 the same as in day 0 or ± 1 point) | n = 9 | n = 9 |
| Increase (by at least 2 points) | n = 3 (mean = +11 ± 8.66) | n = 3 (mean = +4 ± 1) |
| Decrease (by at least 2 points) | n = 4 (mean = −6.75 ± 5.5) | n = 4 (mean = −4.5 ± 2.08) |
| CSF Concentrations | ALS-FRSr day 28 vs. 0 (Valid Calculations n = 16) | Norris day 28 vs. 0 (Valid Calculations n = 16) | |||
|---|---|---|---|---|---|
| rs | p Value | rs | p Value | ||
| CRP | day 7 vs. 0 | −0.631 | 0.009 | −0.483 | 0.080 |
| BDNF | day 7 vs. 0 | 0.534 | 0.033 | 0.360 | 0.207 |
| NGF beta | day 7 vs. 0 | 0.098 | 0.719 | −0.288 | 0.318 |
| NT-3 | day 7 vs. 0 | 0.046 | 0.866 | 0.099 | 0.737 |
| Parameter | ALS Patients (n = 18) | |
|---|---|---|
| Age (mean ± SD, years) | 53.11 ± 11.14 | |
| Sex (male/female) | 9/9 | |
| Disease onset (bulbar/spinal) | 2/16 | |
| Age at disease onset (mean ± SD, years) | 50.00 ± 12.95 | |
| Symptom duration (mean ± SD, months) | 37.39 ± 34.93 | |
| Bone marrow volume (mean ± SD, mL) | 109.78 ± 21.37 | |
| Number of mononuclear cells (mean ± SD) | 200 x 106 ± 95.8 | |
| Number of administered Lin− cells (mean ± SD) | 5.49 x 106 ± 4.40 | |
| ALS-FRSr score (mean ± SD) | Before Lin− Tx | 29.17 ± 5.92 |
| 3 days after Lin− Tx | 29.17 ± 5.92 | |
| 5 days after Lin− Tx | 29.17 ± 5.92 | |
| 7 days after Lin− Tx | 29.17 ± 5.92 | |
| 28 days after Lin− Tx | 29.75 ± 6.44 | |
| Norris scale score (mean ± SD) | Before Lin− Tx | 80.38 ± 17.49 |
| 3 days after Lin− Tx | 82.83 ± 18.28 | |
| 5 days after Lin− Tx | 82.89 ± 18.34 | |
| 7 days after Lin− Tx | 82.89 ± 18.34 | |
| 28 days after Lin− Tx | 86.63 ± 17.30 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumert, B.; Sobuś, A.; Gołąb-Janowska, M.; Ulańczyk, Z.; Paczkowska, E.; Łuczkowska, K.; Zawiślak, A.; Milczarek, S.; Osękowska, B.; Meller, A.; et al. Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients. Int. J. Mol. Sci. 2020, 21, 1070. https://doi.org/10.3390/ijms21031070
Baumert B, Sobuś A, Gołąb-Janowska M, Ulańczyk Z, Paczkowska E, Łuczkowska K, Zawiślak A, Milczarek S, Osękowska B, Meller A, et al. Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients. International Journal of Molecular Sciences. 2020; 21(3):1070. https://doi.org/10.3390/ijms21031070
Chicago/Turabian StyleBaumert, Bartłomiej, Anna Sobuś, Monika Gołąb-Janowska, Zofia Ulańczyk, Edyta Paczkowska, Karolina Łuczkowska, Alicja Zawiślak, Sławomir Milczarek, Bogumiła Osękowska, Agnieszka Meller, and et al. 2020. "Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients" International Journal of Molecular Sciences 21, no. 3: 1070. https://doi.org/10.3390/ijms21031070
APA StyleBaumert, B., Sobuś, A., Gołąb-Janowska, M., Ulańczyk, Z., Paczkowska, E., Łuczkowska, K., Zawiślak, A., Milczarek, S., Osękowska, B., Meller, A., Machowska-Sempruch, K., Wełnicka, A., Safranow, K., Nowacki, P., & Machaliński, B. (2020). Local and Systemic Humoral Response to Autologous Lineage-Negative Cells Intrathecal Administration in ALS Patients. International Journal of Molecular Sciences, 21(3), 1070. https://doi.org/10.3390/ijms21031070






