The Clustering of mApoE Anti-Amyloidogenic Peptide on Nanoparticle Surface Does Not Alter Its Performance in Controlling Beta-Amyloid Aggregation
Abstract
1. Introduction
2. Result
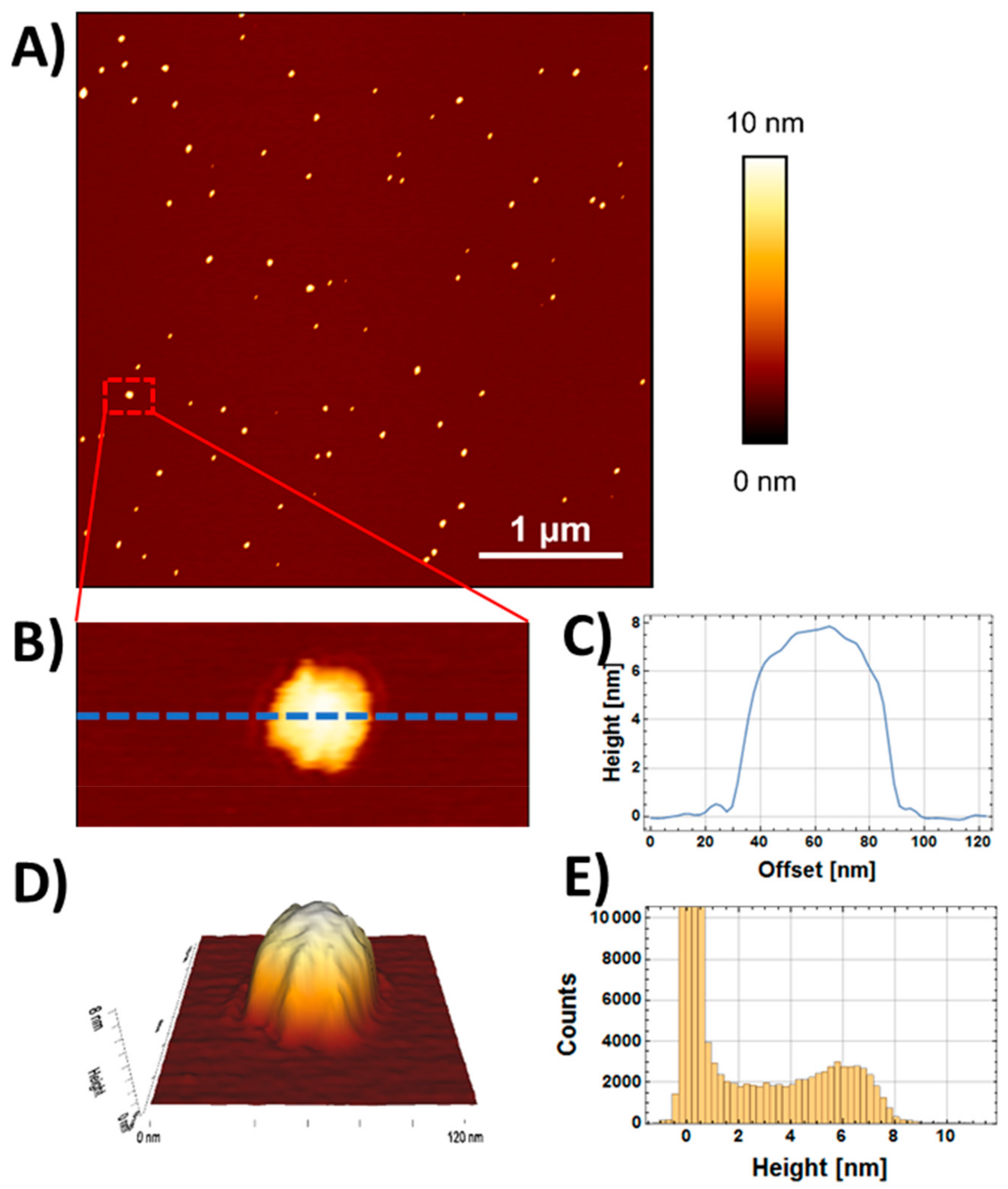
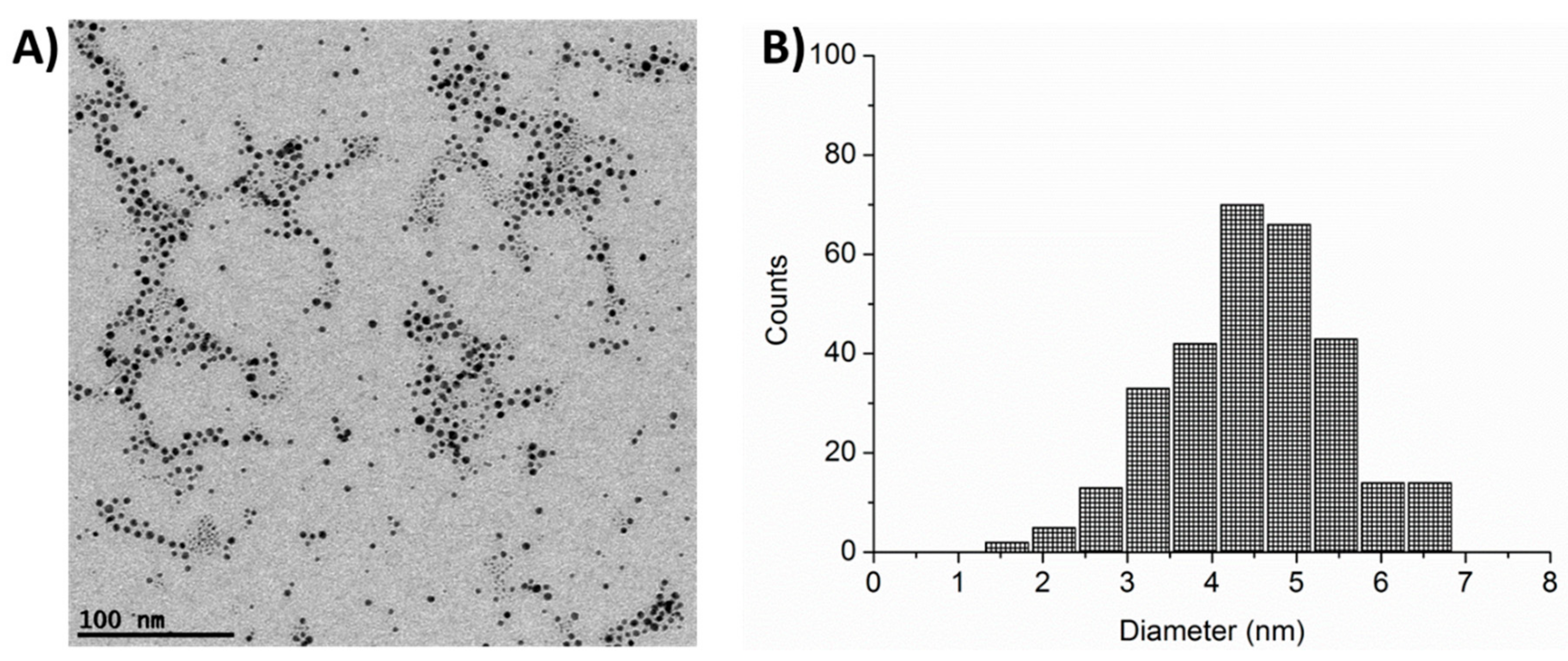
2.1. AuNP Characterization
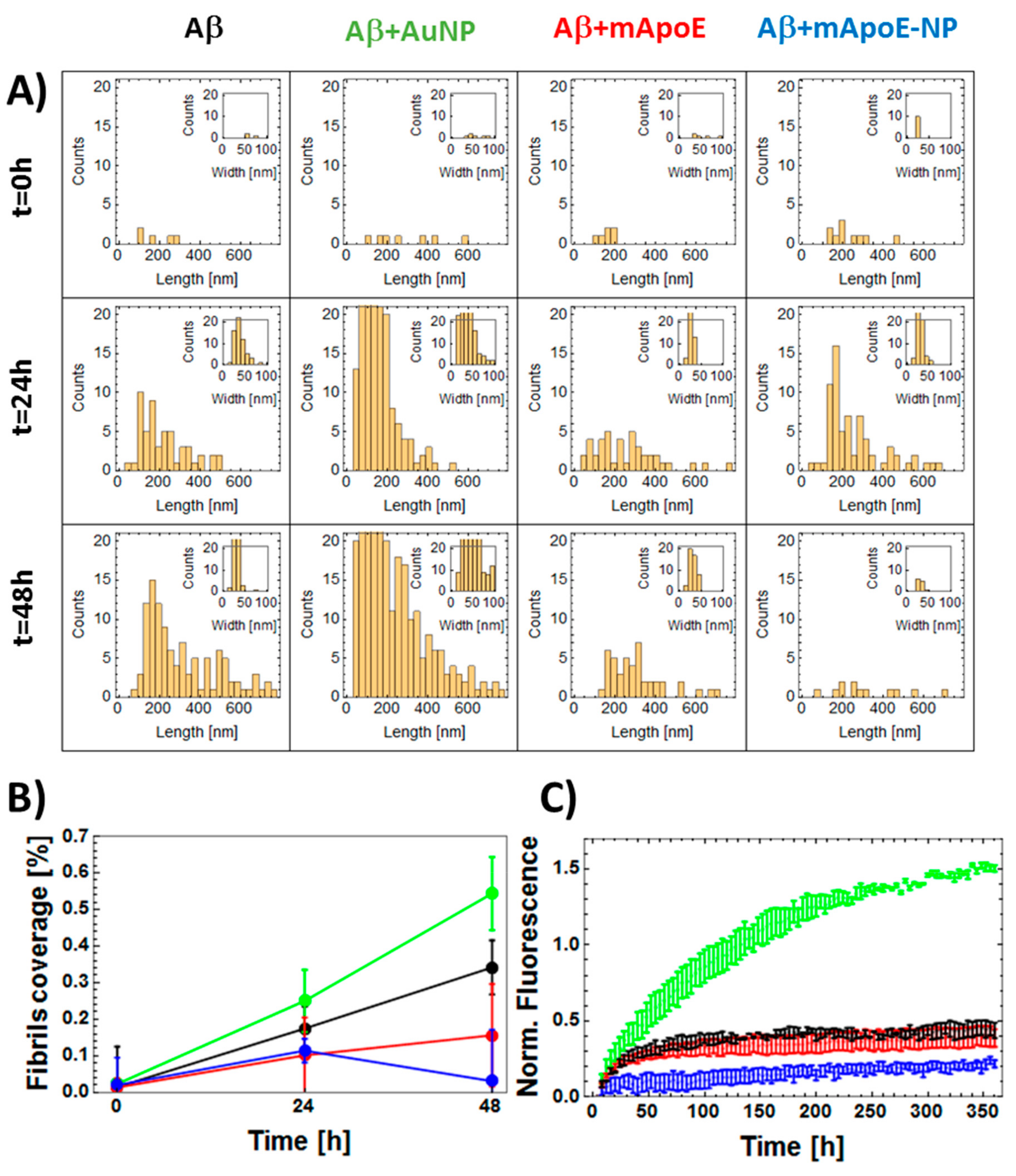
2.2. Effect of mApoE-Functionalized AuNP on Aβ Peptide Aggregation
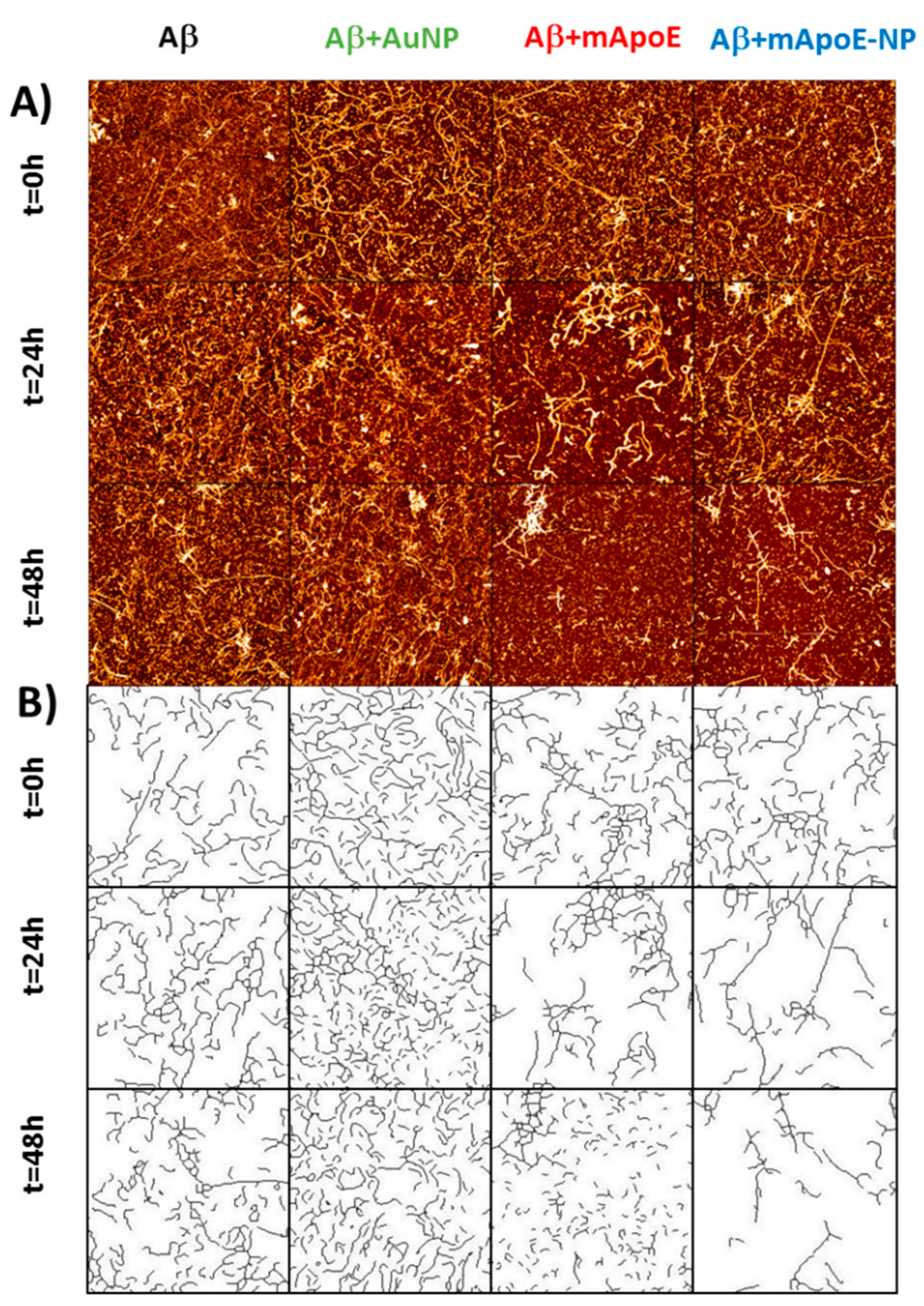
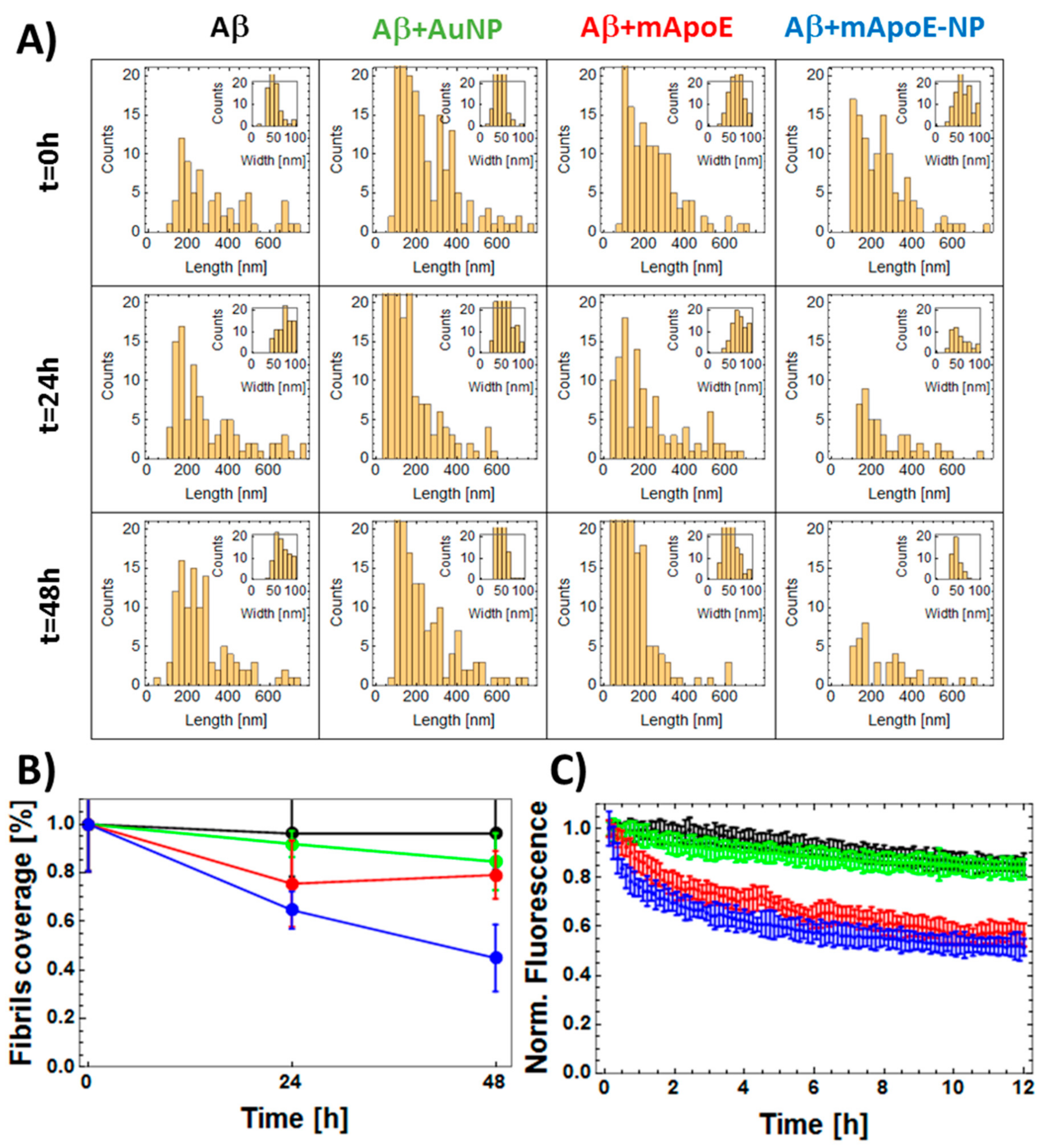
2.3. Disaggregation Effect of mApoE-AuNP on Aβ Preformed Fibrils
3. Discussion
4. Materials and Methods
4.1. AuNP Synthesis
4.2. Preparation of Aβ Samples
4.3. Aβ Aggregation and Disaggregation Process
4.4. Atomic Force Microscopy and Transmission Electron Microscopy Imaging
4.5. Morphological Analysis of AuNP and Fibril Recognition
4.6. Evaluation of Aβ Aggregation Kinetics by Fluorescence Experiments
4.7. Evaluation of Aβ Fibril Disaggregation by Thioflavin T Fluorescence Assay
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Aβ | Amyloid-β |
| AD | Alzheimer’s disease |
| BBB | Blood brain barrier |
| AFM | Atomic force microscopy |
| TEM | Transmission Electron Microscopy |
| ThT | Thioflavin T |
| mApoE | Peptide CWG-LRKLRKRLLR |
| ApoE | Apolipoprotein E |
| AuNPDLS | Gold nanoparticlesDynamic Light Scattering |
References
- Aleksis, R.; Oleskovs, F.; Jaudzems, K.; Pahnke, J.; Biverstål, H. Structural studies of amyloid-β peptides: Unlocking the mechanism of aggregation and the associated toxicity. Biochimie 2017, 140, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Scheper, W.; Hoozemans, J.J. The unfolded protein response in neurodegenerative diseases: A neuropathological perspective. Acta Neuropathol. 2015, 130, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Tosi, G.; Pederzoli, F.; Belletti, D.; Vandelli, M.A.; Forni, F.; Duskey, J.T.; Ruozi, B. Nanomedicine in Alzheimer’s disease: Amyloid beta targeting strategy. Prog. Brain Res. 2019, 245, 57–88. [Google Scholar] [CrossRef] [PubMed]
- Laurén, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid- oligomers. Nature 2009, 457, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Lomakin, A.; Chung, D.S.; Benedek, G.B.; Kirschner, D.A.; Teplow, D.B. On the nucleation and growth of amyloid -protein fibrils: Detection of nuclei and quantification of rate constants. Proc. Natl. Acad. Sci. USA 1996, 93, 1125–1129. [Google Scholar] [CrossRef]
- Shen, C.-L.; Scott, G.L.; Merchant, F.; Murphy, R.M. Light scattering analysis of fibril growth from the amino-terminal fragment (1-28) of -amyloid peptide. Biophys. J. 1993, 65, 2383–2395. [Google Scholar] [CrossRef]
- Shen, C.-L.; Murphy, R.M. Solvent effects on self-assembly of -amyloid peptide. Biophys. J. 1995, 69, 640–651. [Google Scholar] [CrossRef]
- Lue, L.-F.; Kuo, Y.-M.; Roher, A.E.; Brachova, L.; Shen, Y.; Sue, L.; Beach, T.; Kurth, J.H.; Rydel, R.E.; Rogers, J. Soluble amyloid peptide concentration as a predictor of synaptic change in Alzheimer disease. Am. J. Pathol. 1999, 155, 853–862. [Google Scholar] [CrossRef]
- Shankar, G.M.; Leissring, M.A.; Adame, A.; Sun, X.; Spooner, E.; Masliah, E.; Selkoe, D.J.; Lemere, C.A.; Walsh, D.M. Biochemical and immunohistochemical analysis of an Alzheimer’s disease mouse model reveals the presence of multiple cerebral A assembly forms throughout life. Neurobiol. Dis. 2009, 36, 293–302. [Google Scholar] [CrossRef]
- Shankar, G.M.; Li, S.; Metha, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid- protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef]
- Puzzo, D.; Privitera, L.; Leznik, E.; Fa, M.; Staniszewski, A.; Palmeri, A.; Arancio, O. Picomolar amyloid- positively modulates synaptic plasticity and memory in hippocampus. J. Neurosci. 2008, 28, 14537–14545. [Google Scholar] [CrossRef] [PubMed]
- Townsend, M.; Shankar, G.M.; Metha, T.; Walsh, D.M.; Selkoe, D.J. Effects of secreted oligomers of amyloid beta-protein on hippocampal synaptic plasticity: A potent role for trimers. J. Physiol. 2006, 572, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Kowalewski, T.; Holtzman, D.M. In situ atomic force microscopy study of Alzheimer’s -amyloid peptide on different substrates: New insights into mechanism of -sheet formation. Proc. Natl. Acad. Sci. USA 1999, 96, 3688–3693. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Kistler, J.; Aebi, U.; Arvinte, T.; Cooper, G.J.S. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol. 1999, 253, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury, P.T., Jr. Observation of metastable A amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997, 4, 119–125. [Google Scholar] [CrossRef]
- Harper, J.D.; Lieber, C.M.; Lansbury, P.T., Jr. Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer’s disease amyloid-protein. Chem. Biol. 1997, 4, 951–959. [Google Scholar] [CrossRef]
- Stine, W.B., Jr.; Dahlgren, K.N.; Krafft, G.A.; LaDu, M.J. In vitro characterization of conditions for amyloid- peptide oligomerization and fibrillogenesis. J. Biol. Chem. 2003, 278, 11612–11622. [Google Scholar] [CrossRef]
- Muller, D.J.; Dufrene, Y.F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotech. 2008, 3, 261–269. [Google Scholar] [CrossRef]
- Bacskai, B.J.; Kajdasz, S.T.; Christie, R.H.; Carter, C.; Games, D.; Seubert, P.; Schenk, D.; Hyman, B.T. Imaging of amyloid-beta deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy. Nat. Med. 2001, 7, 369–372. [Google Scholar] [CrossRef]
- Meyer-Luehmann, M.; Spires-Jones, T.L.; Prada, C.; Garcia-Alloza, M.; de Calignon, A.; Rozkalne, A.; Koenigsknecht-Talboo, J.; Holtzman, D.M.; Bacskai, B.J.; Hyman, B.T. Rapid appearance and local toxicity of amyloid- plaques in a mouse model of Alzheimer’s disease. Nature 2008, 451, 720–725. [Google Scholar] [CrossRef]
- Bolder, S.G.; Sagis, L.M.C.; Venema, P.; van der Linden, E. Thioflavin T and birefringence assays to determine the conversion of proteins into fibrils. Langmuir 2007, 23, 4144–4147. [Google Scholar] [CrossRef] [PubMed]
- Nardo, L.; Re, F.; Brioschi, S.; Cazzaniga, E.; Orlando, A.; Minniti, S.; Lamperti, M.; Gregori, M.; Cassina, V.; Brogioli, D.; et al. Fluorimetric detection of the earliest events in amyloid beta oligomerization and its inhibition by pharmacologically active liposomes. Biochim. Biophys. Acta 2016, 1860, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Drummond, E.; Goñi, F.; Liu, S.; Prelli, F.; Scholtzova, H.; Wisniewski, T. Potential Novel Approaches to Understand the Pathogenesis and Treat Alzheimer’s Disease. J. Alzheimers Dis. 2018, 64, S299–S312. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Saraf, S.; Saraf, S.; Antimisiaris, S.G.; Hamano, N.; Li, S.D.; Chougule, M.; Shoyele, S.A.; Gupta, U.; Ajazuddin Alexander, A. Recent advancements in the field of nanotechnology for the delivery of anti-Alzheimer drug in the brain region. Expert Opin. Drug Deliv. 2018, 15, 589–617. [Google Scholar] [CrossRef] [PubMed]
- Austen, B.M.; Paleologou, K.E.; Ali, S.A.; Qureshi, M.M.; Allsop, D.; El-Agnaf, O.M. Designing peptide inhibitors for oligomerization and toxicity of Alzheimer’s beta-amyloid peptide. Biochemistry 2008, 47, 1984–1992. [Google Scholar] [CrossRef]
- Wisniewski, T.; Golabek, A.; Matsubara, E.; Ghiso, J.; Frangione, B. Apolipoprotein E: Binding to soluble Alzheimer’s beta-amyloid. Biochem. Biophys. Res. Commun. 1993, 192, 359–365. [Google Scholar] [CrossRef]
- Dal Magro, R.; Ornaghi, F.; Cambianica, I.; Beretta, S.; Re, F.; Musicanti, C.; Rigolio, R.; Donzelli, E.; Canta, A.; Ballarini, E.; et al. ApoE-modified solid lipid nanoparticles: A feasible strategy to cross the blood-brain barrier. J. Control. Release 2017, 249, 103–110. [Google Scholar] [CrossRef]
- Mancini, S.; Minniti, S.; Gregori, M.; Sancini, G.; Cagnotto, A.; Couraud, P.O.; Ordóñez-Gutiérrez, L.; Wandosell, F.; Salmona, M.; Re, F. The hunt for brain Aβ oligomers by peripherally circulating multi-functional nanoparticles: Potential therapeutic approach for Alzheimer disease. Nanomedicine 2016, 12, 43–52. [Google Scholar] [CrossRef]
- Cox, A.; Andreozzi, P.; Dal Magro, R.; Fiordaliso, F.; Corbelli, A.; Talamini, L.; Chinello, C.; Raimondo, F.; Magni, F.; Tringali, M.; et al. Evolution of Nanoparticle Protein Corona across the Blood-Brain Barrier. ACS Nano 2018, 12, 7292–7300. [Google Scholar] [CrossRef]
- Sot, J.; Mendanha-Neto, S.A.; Busto, J.V.; García-Arribas, A.B.; Li, S.; Burgess, S.W.; Shaw, W.A.; Gil-Carton, D.; Goñi, F.M.; Alonso, A. The interaction of lipid-liganded gold clusters (Aurora ™) with lipid bilayers. Chem. Phys. Lipids 2019, 218, 40–46. [Google Scholar] [CrossRef]
- Gregori, M.; Cassina, V.; Brogioli, D.; Salerno, D.; De Kimpe, L.; Scheper, W.; Masserini, M.; Mantegazza, F. Stability of Aβ(1-42) peptide fibrils as consequence of environmental modifications. Eur. Biophys. J. 2010, 39, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Dal Magro, R.; Simonelli, S.; Cox, A.; Formicola, B.; Corti, R.; Cassina, V.; Nardo, L.; Mantegazza, F.; Salerno, D.; Grasso, G.; et al. The Extent of Human Apolipoprotein A-I Lipidation Strongly Affects the β-Amyloid Efflux Across the Blood-Brain Barrier in vitro. Front. Neurosci. 2019, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Persson, N.E.; McBride, M.A.; Grover, M.A.; Reichmanis, E. Automated Analysis of Orientational Order in Images of Fibrillar Materials. Chem. Mater. 2017, 29, 3–14. [Google Scholar] [CrossRef]
- Mehta, D.; Jackson, R.; Paul, G.; Shi, J.; Sabbagh, M. Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010-2015. Expert Opin. Investig. Drugs 2017, 26, 735–739. [Google Scholar] [CrossRef] [PubMed]
- John, T.; Gladytz, A.; Kubeil, C.; Martin, L.L.; Risselada, H.J.; Abel, B. Impact of nanoparticles on amyloid peptide and protein aggregation: A review with a focus on gold nanoparticles. Nanoscale 2018, 10, 20894–20913. [Google Scholar] [CrossRef] [PubMed]
- Re, F.; Airoldi, C.; Zona, C.; Masserini, M.; La Ferla, B.; Quattrocchi, N.; Nicotra, F. Beta amyloid aggregation inhibitors: Small molecules as candidate drugs for therapy of Alzheimer’s disease. Curr. Med. Chem. 2010, 17, 2990–3006. [Google Scholar] [CrossRef]
- Gupta, J.; Fatima, M.T.; Islam, Z.; Khan, R.H.; Uversky, V.N.; Salahuddin, P. Nanoparticle formulations in the diagnosis and therapy of Alzheimer’s disease. Int. J. Biol. Macromol. 2019, 130, 515–526. [Google Scholar] [CrossRef]
- Bana, L.; Minniti, S.; Salvati, E.; Sesana, S.; Zambelli, V.; Cagnotto, A.; Orlando, A.; Cazzaniga, E.; Zwart, R.; Scheper, W.; et al. Liposomes bi-functionalized with phosphatidic acid and an ApoE-derived, peptide affect A beta aggregation features and cross the, blood-brain-barrier: Implications for therapy of Alzheimer disease. Nanomedicine 2014, 10, 1583–1590. [Google Scholar] [CrossRef]
- Re, F.; Cambianica, I.; Zona, C.; Sesana, S.; Gregori, M.; Rigolio, R.; La Ferla, B.; Nicotra, F.; Forloni, G.; Cagnotto, A.; et al. Functionalization of liposomes with ApoE-derived peptides at different density affects cellular uptake and drug transport across a blood-brain barrier model. Nanomedicine 2011, 7, 551–559. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.H.; Ryu, J.; Kim, D.J. Multivalent & multifunctional ligands to beta-amyloid. Curr. Pharm. Des. 2009, 15, 637–658. [Google Scholar] [CrossRef]
- Bartus, É.; Olajos, G.; Schuster, I.; Bozsó, Z.; Deli, M.A.; Veszelka, S.; Walter, F.R.; Datki, Z.; Szakonyi, Z.; Martinek, T.A.; et al. Structural Optimization of Foldamer-Dendrimer Conjugates as Multivalent Agents against the Toxic Effects of Amyloid Beta Oligomers. Molecules 2018, 23, 2523. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Uzun, O.; Hu, Y.; Han, H.S.; Watson, N.; Chen, S.; Irvine, D.J.; Stellacci, F. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat. Mater. 2008, 7, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Raghnaill, M.N.; Bramini, M.; Mahon, E.; Åberg, C.; Salvati, A.; Dawson, K.A. Nanoparticle accumulation and transcytosis in brain endothelial cell layers. Nanoscale 2013, 5, 11153–11165. [Google Scholar] [CrossRef] [PubMed]
- Gobbi, M.; Re, F.; Canovi, M.; Beeg, M.; Gregori, M.; Sesana, S.; Sonnino, S.; Brogioli, D.; Musicanti, C.; Gasco, P.; et al. Lipid-based nanoparticles with high binding affinity for amyloid-β1-42 peptide. Biomaterials 2010, 31, 6519–6529. [Google Scholar] [CrossRef] [PubMed]
- Cassina, V.; Manghi, M.; Salerno, D.; Tempestini, A.; Iadarola, V.; Nardo, L.; Brioschi, S.; Mantegazza, F. Effects of cytosine methylation on DNA morphology: An atomic force microscopy study. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1–7. [Google Scholar] [CrossRef]
- Cassina, V.; Seruggia, D.; Beretta, G.L.; Salerno, D.; Brogioli, D.; Manzini, S.; Zunino, F.; Mantegazza, F. Atomic force microscopy study of DNA conformation in the presence of drugs. Euro. Biophys. J. 2011, 40, 59–68. [Google Scholar] [CrossRef]
- Ruggeri, F.S.; Charmet, J.; Kartanas, T.; Peter, Q.; Chia, S.; Habchi, J.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J. Microfluidic deposition for resolving single-molecule protein architecture and heterogeneity. Nat. Commun. 2018, 9, 3890. [Google Scholar] [CrossRef]
- Groenning, M. Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J. Chem. Biol. 2010, 3, 1–18. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corti, R.; Cox, A.; Cassina, V.; Nardo, L.; Salerno, D.; Marrano, C.A.; Missana, N.; Andreozzi, P.; Silva, P.J.; Stellacci, F.; et al. The Clustering of mApoE Anti-Amyloidogenic Peptide on Nanoparticle Surface Does Not Alter Its Performance in Controlling Beta-Amyloid Aggregation. Int. J. Mol. Sci. 2020, 21, 1066. https://doi.org/10.3390/ijms21031066
Corti R, Cox A, Cassina V, Nardo L, Salerno D, Marrano CA, Missana N, Andreozzi P, Silva PJ, Stellacci F, et al. The Clustering of mApoE Anti-Amyloidogenic Peptide on Nanoparticle Surface Does Not Alter Its Performance in Controlling Beta-Amyloid Aggregation. International Journal of Molecular Sciences. 2020; 21(3):1066. https://doi.org/10.3390/ijms21031066
Chicago/Turabian StyleCorti, Roberta, Alysia Cox, Valeria Cassina, Luca Nardo, Domenico Salerno, Claudia Adriana Marrano, Natalia Missana, Patrizia Andreozzi, Paulo Jacob Silva, Francesco Stellacci, and et al. 2020. "The Clustering of mApoE Anti-Amyloidogenic Peptide on Nanoparticle Surface Does Not Alter Its Performance in Controlling Beta-Amyloid Aggregation" International Journal of Molecular Sciences 21, no. 3: 1066. https://doi.org/10.3390/ijms21031066
APA StyleCorti, R., Cox, A., Cassina, V., Nardo, L., Salerno, D., Marrano, C. A., Missana, N., Andreozzi, P., Silva, P. J., Stellacci, F., Dal Magro, R., Re, F., & Mantegazza, F. (2020). The Clustering of mApoE Anti-Amyloidogenic Peptide on Nanoparticle Surface Does Not Alter Its Performance in Controlling Beta-Amyloid Aggregation. International Journal of Molecular Sciences, 21(3), 1066. https://doi.org/10.3390/ijms21031066





