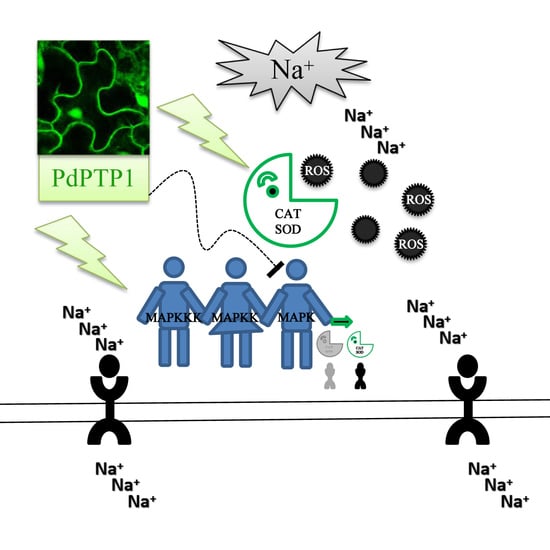
Poplar PdPTP1 Gene Negatively Regulates Salt Tolerance by Affecting Ion and ROS Homeostasis in Populus
Abstract
1. Introduction
2. Results
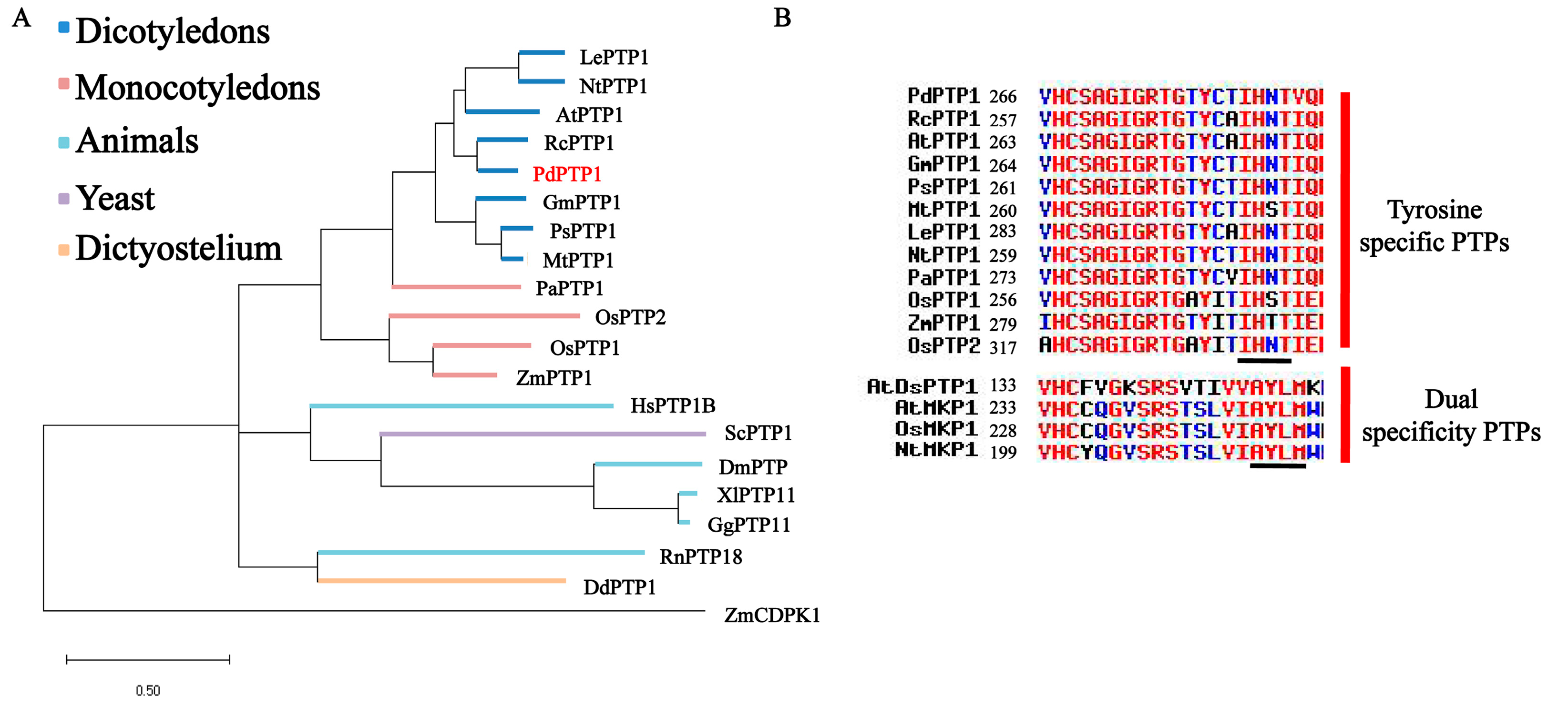
2.1. PdPTP1 Encodes a Tyr-Specific PTP in Populus
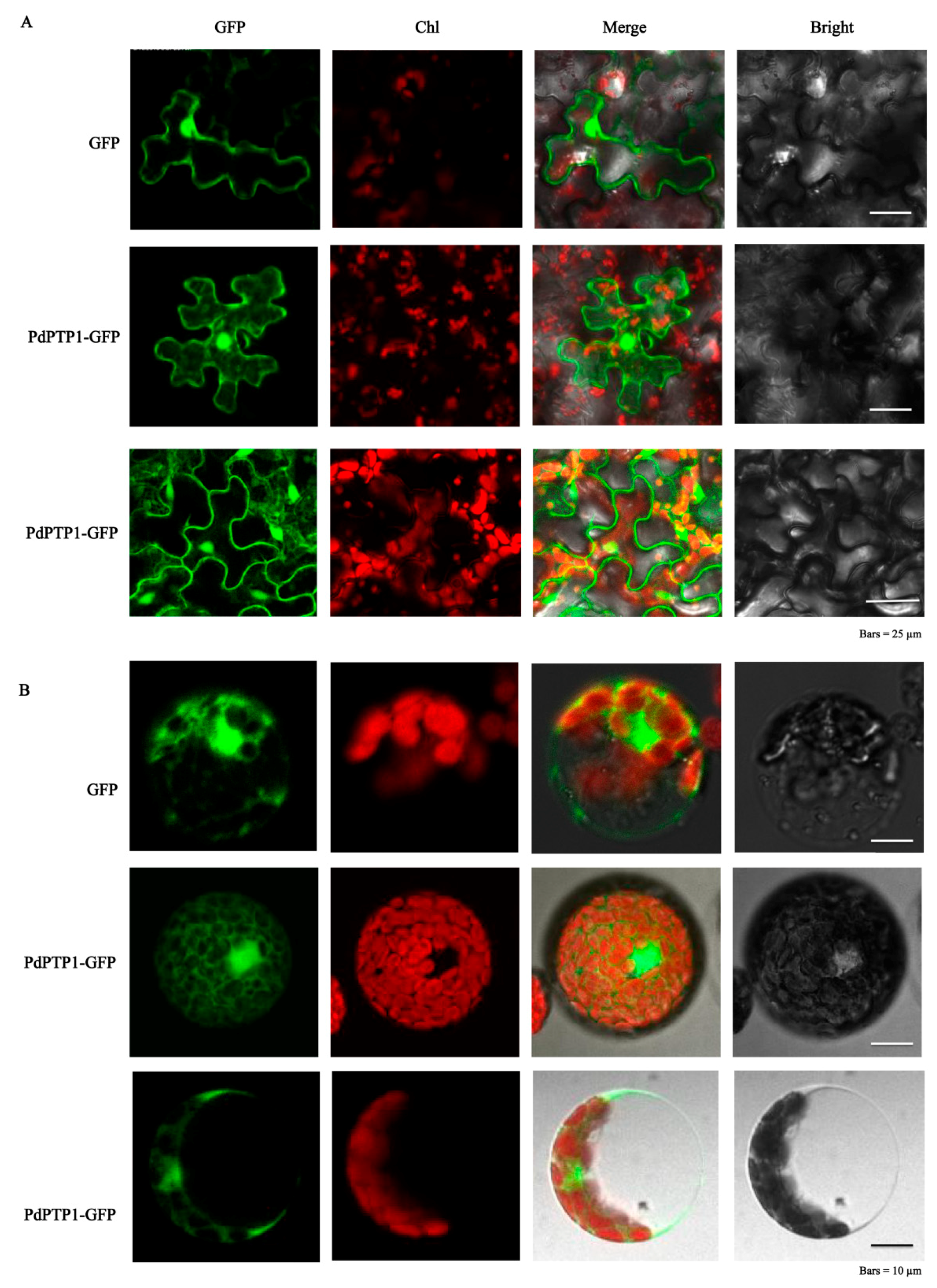
2.2. PdPTP1 Localized to both the Cytosol and Nucleus
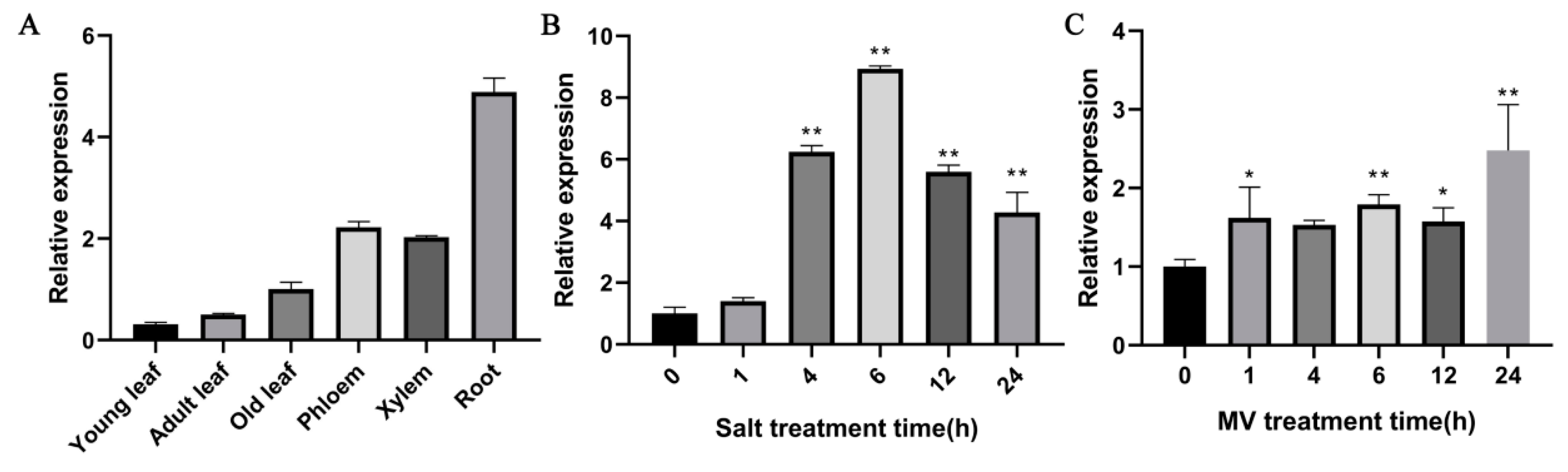
2.3. Expression Patterns of PdPTP1 in Response to Salt and Oxidative Stress
2.4. Identification of Transgenic Poplar
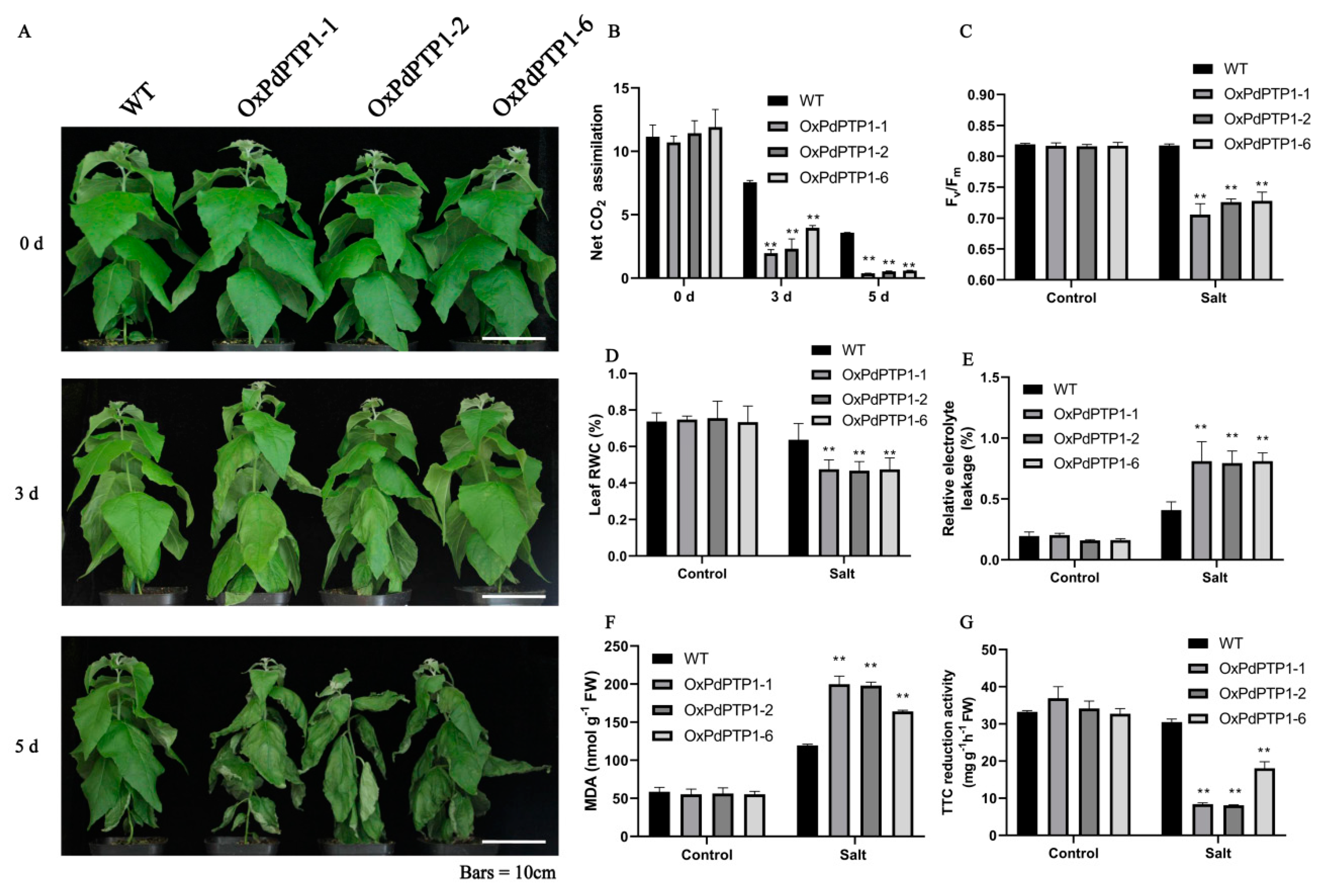
2.5. Overexpression of PdPTP1 Increased Sensitivity to NaCl in Transgenic Poplar
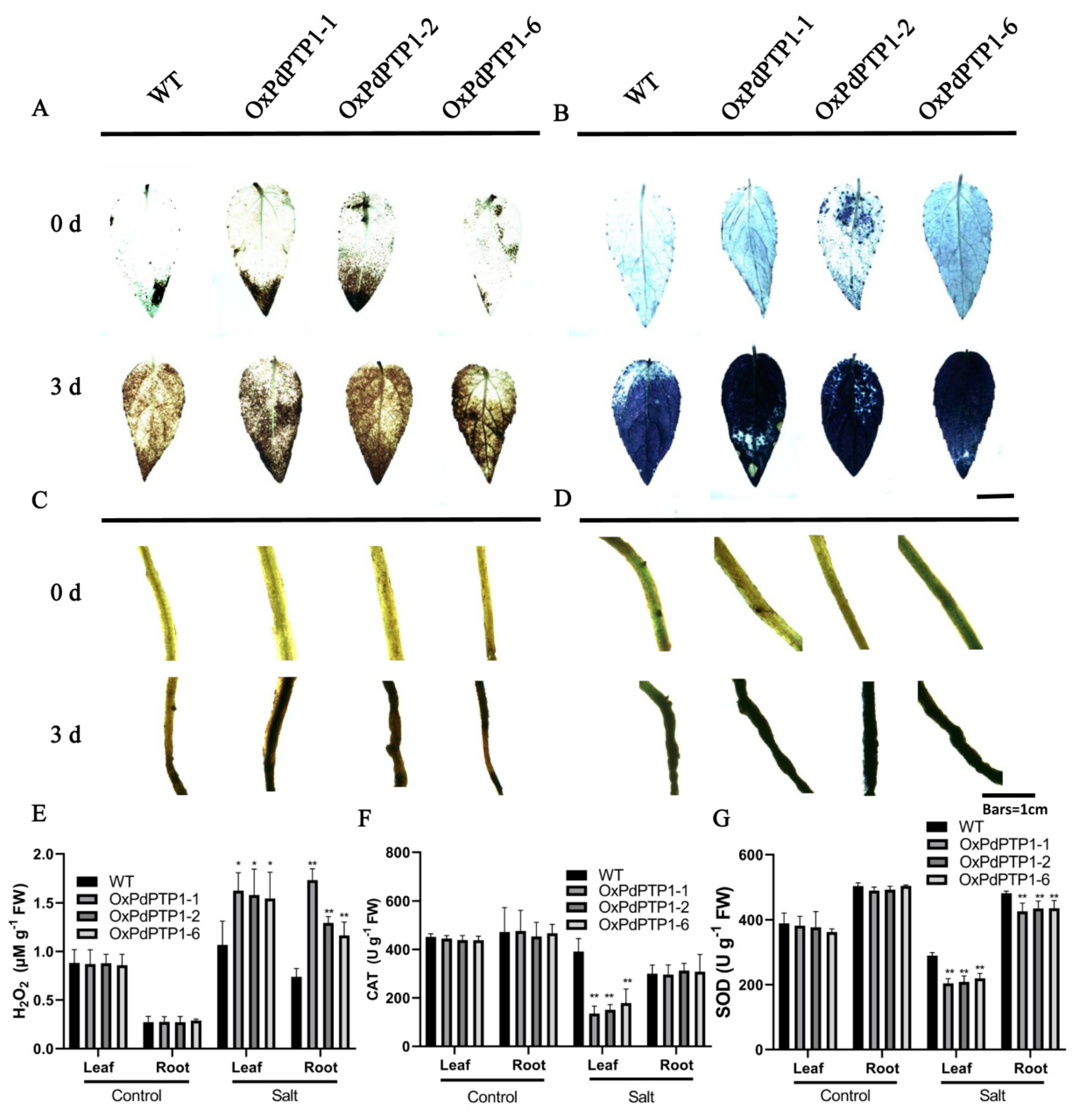
2.6. Overexpression of PdPTP1 Poplar Increased ROS Levels and Inhibited ROS Scavenger Activities in Response to Salt Stress
2.7. Overexpression of PdPTP1 Polar Compromised Ion Homeostasis in Response to Salt Stress
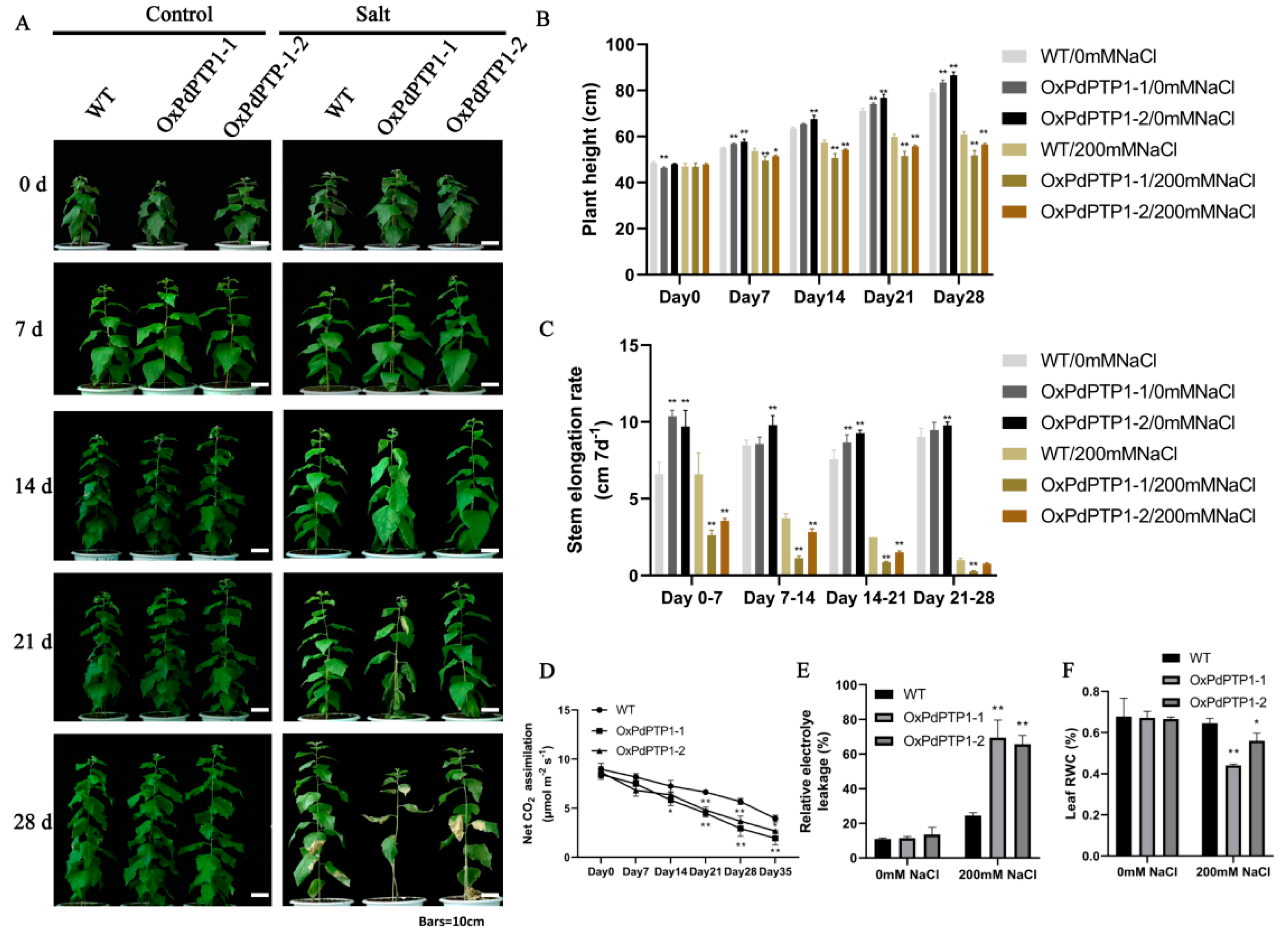
2.8. Overexpression of PdPTP1 Reduced Salt-Stress Tolerance in Transgenic Poplar under Long-Term Salt Stress
2.9. PdPTP1 Interacts with PdMPK3, PdMAPK6 In Vivo and In Vitro
3. Discussion
3.1. PdPTP1 is a Tyr-Specific PTP in Poplar
3.2. PdPTP1 Overexpression Reduced Tolerance to Salt Stress in Poplar due to the Impaired Ion and ROS Homeostasis
3.3. PdPTP1 Overexpression Reduced Salt Tolerance in Poplar due to the Impaired ROS Homeostasis
3.4. PdPTP1 Overexpression Reduced Salt Tolerance in Poplar due to the Impaired Ion Homeostasis
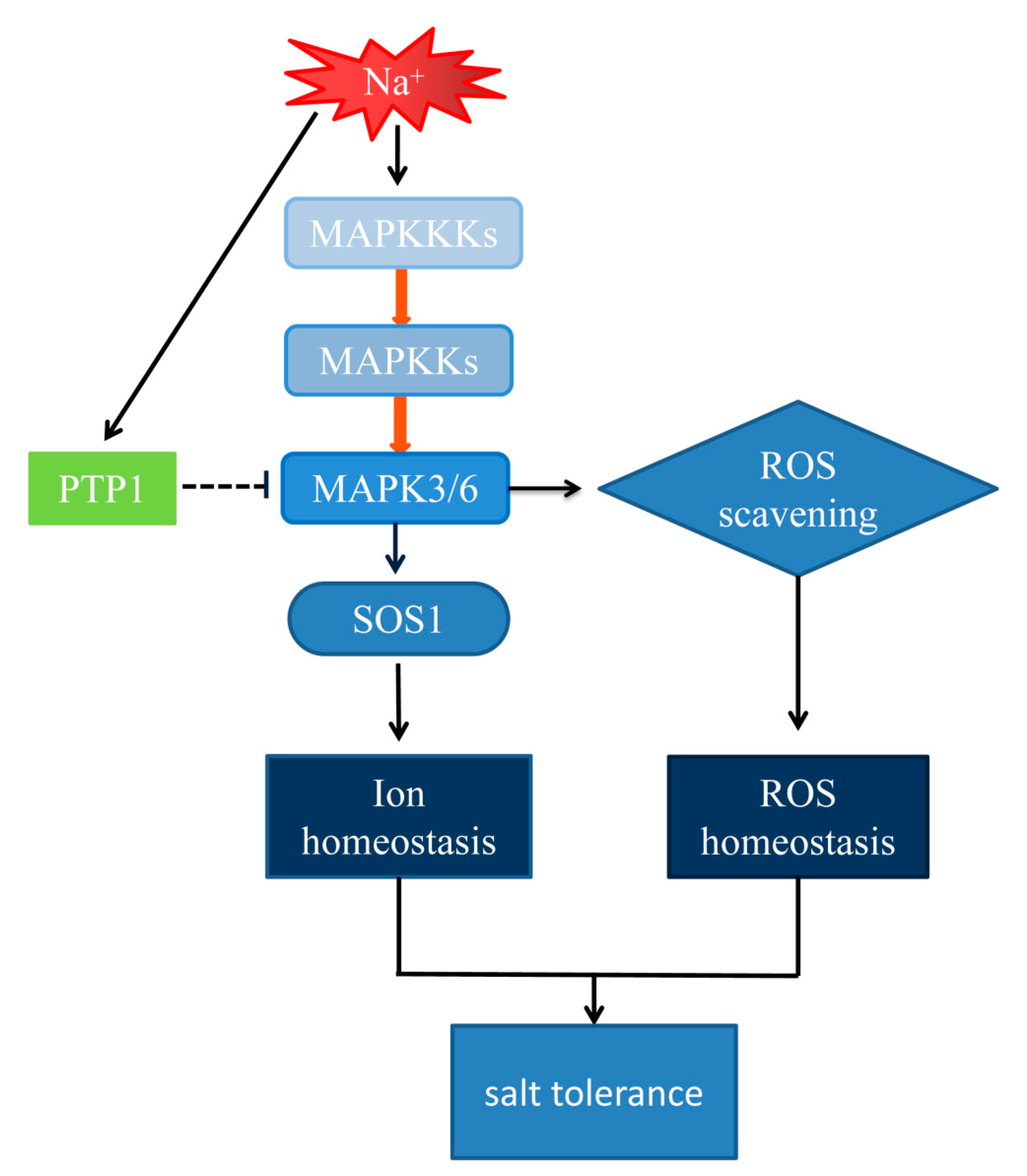
3.5. The Hypersensitivity of OxPdPTP1 Poplar to Salt Is Associated with PdMPK3/6
4. Material and Methods
4.1. Plant Materials and Stress Treatments
4.2. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
4.3. Salt-Stress Tolerance Evaluation of Transgenic Poplars
4.4. Measurement of Na+ and K+ Contents
4.5. Subcellular Localization Analysis
4.6. Yeast Two-Hybrid
4.7. Bimolecular Fluorescence Complementation (BiFC) Assays
4.8. Phylogenetic and Domain Analysis of PdPTP1
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Kim, S.H.; Woo, D.H.; Kim, J.M.; Lee, S.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys. Res. Commun. 2011, 412, 150–154. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Hunter, T. Tyrosine-phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Yu, L.; Nie, J.; Cao, C.; Jin, Y.; Yan, M.; Wang, F.; Liu, J.; Xiao, Y.; Liang, Y.; Zhang, W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Perez-Salamo, I.; Papdi, C.; Rigo, G.; Zsigmond, L.; Vilela, B.; Lumbreras, V.; Nagy, I.; Horvath, B.; Domoki, M.; Darula, Z.; et al. The Heat Shock Factor A4A Confers Salt Tolerance and Is Regulated by Oxidative Stress and the Mitogen-Activated Protein Kinases MPK3 and MPK6. Plant Physiol. 2014, 165, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q. Molecular Characterization of a Tyrosine-Specific Protein Phosphatase Encoded by a Stress-Responsive Gene in Arabidopsis. Plant Cell 1998, 10, 849–858. [Google Scholar] [CrossRef]
- Kerk, D.; Templeton, G.; Moorhead, G.B.G. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef]
- Tonks Nicholas, K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Barsam, B.; Veretnik, S.; Gribskov, M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002, 129, 908–925. [Google Scholar] [CrossRef]
- Yu, L.; Feng, D.; Li, W.; Wang, H.; Bing, L. Comprehensive and evolutionary analysis of protein tyrosine phosphatases (PTP) in the green plants. Plant. OMICS 2013, 6, 215–223. [Google Scholar]
- Gupta, R.; Luan, S. Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 2003, 132, 1149–1152. [Google Scholar] [CrossRef]
- Ulm, R.; Revenkova, E.; di Sansebastiano, G.P.; Bechtolad, N.; Paszkowski, J. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Gene Dev. 2001, 15, 699–709. [Google Scholar] [CrossRef]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Quettier, A.L.; Bertrand, C.; Habricot, Y.; Miginiac, E.; Maldiney, R. The phs1-3 mutation in a putative dual-specificity protein tyrosine phosphatase gene provokes hypersensitive responses to abscisic acid in Arabidopsis thaliana. Plant J. 2006, 47, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Camps, M.; Nichols, A.; Arkinstall, S. Dual specificity phosphatases: A gene family for control of MAP kinase function. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000, 14, 6–16. [Google Scholar] [CrossRef]
- Luan, S. Tyrosine phosphorylation in plant cell signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 11567–11569. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, H.; Gupta, R.; Morris, P.C.; Luan, S.; Kieber, J.J. ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 2000, 122, 1301–1310. [Google Scholar] [CrossRef]
- Bartels, S.; Anderson, J.C.; González Besteiro, M.A.; Carreri, A.; Hirt, H.; Buchala, A.; Métraux, J.; Peck, S.C.; Ulm, R. Map Kinase Phosphatase1 and Protein Tyrosine Phosphatase1 Are Repressors of Salicylic Acid Synthesis and SNC1-Mediated Responses inArabidopsis. Plant. Cell 2009, 21, 2884–2897. [Google Scholar] [CrossRef]
- Kitajima, M.; Butler, W.L. Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim Biophys Acta 1975, 376, 105–115. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Bu, J.J.; Wang, S.F.; Hu, G.H.; Wang, Q.L. Effect of coronatine on cotton root activity determined by TTC assay at different levels of potassium. Plant Physiol. J. 2015, 51, 695–701. [Google Scholar]
- Brunner, A.M.; Busov, V.B.; Strauss, S.H. Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci. 2004, 9, 49–56. [Google Scholar] [CrossRef]
- Ma, J.; Wan, D.; Duan, B.; Bai, X.; Bai, Q.; Chen, N.; Ma, T. Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 2019, 17, 451–460. [Google Scholar] [CrossRef]
- Mansfield, S.D.; Kang, K.Y.; Chapple, C. Designed for deconstruction poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 2012, 194, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hawighorst, P.; Sun, J.; Polle, A. Salt tolerance in Populus: Significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environ. Exp. Bot. 2014, 107, 113–124. [Google Scholar] [CrossRef]
- Polle, A.; Chen, S. On the salty side of life: Molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant. Cell Environ. 2014, 38, 1794–1816. [Google Scholar] [CrossRef]
- Luan, S. Protein Phosphatases in Plants. Annu. Rev. Plant Biol. 2003, 54, 63. [Google Scholar] [CrossRef] [PubMed]
- Fordham-Skelton, A.P.; Chilley, P.; Lumbreras, V.; Reignoux, S.; Fenton, T.R.; Dahm, C.C.; Pages, M.; Gatehouse, J.A. A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant. J. Cell Mol. Biol. 2002, 29, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lin, C.; Kao, T.; Huang, D.; Huang, H. PaPTP1, a Gene Encoding Protein Tyrosine Phosphatase from Orchid, Phalaenopsis amabilis, is Regulated During Floral Development and Induced by Wounding. Plant Mol. Biol. Rep. 2011, 29, 106–116. [Google Scholar] [CrossRef]
- Shankar, A.; Agrawal, N.; Sharma, M.; Pandey, A.; Pandey, G.K. Role of Protein Tyrosine Phosphatases in Plants. Curr. Genom. 2015, 16, 224–236. [Google Scholar] [CrossRef]
- Desikan, R.; A-H-Mackerness, S.; Hancock, J.T.; Neill, S.J. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001, 127, 159–172. [Google Scholar] [CrossRef]
- Mu, C.; Zhou, L.; Shan, L.; Li, F.; Li, Z. Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). New Phytol. 2019, 223, 1856–1872. [Google Scholar] [CrossRef]
- Hernández, J.A.; Jiménez, A.; Mullineaux, P.; Sevilia, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Allakhverdiev, S.I. Salt Stress Inhibits the Repair of Photodamaged Photosystem II by Suppressing the Transcription and Translation of psbA Genes in Synechocystis. Plant Physiol. 2002, 130, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.; Bohnert, H.J. Plant Cellular and Molecular Responses To High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Yasar, F.; Kusvuran, S.; Ellialtioglu, S. Determination of anti-oxidant activities in some melon (Cucumis melo L.) varieties and cultivars under salt stress. J. Hortic. Sci. Biotech. 2006, 81, 627–630. [Google Scholar] [CrossRef]
- Hideg, E.; Schreiber, U. Parallel assessment of ROS formation and photosynthesis in leaves by fluorescence imaging. Photosynth. Res. 2007, 92, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Mapk, G. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Tena, G.; Asai, T.; Chiu, W.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Zeng, Q.; Chen, J.G.; Ellis, B.E. AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 2011, 67, 895–906. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Ahlfors, R.; Macioszek, V.; Rudd, J.; Brosché, M.; Schlichting, R.; Scheel, D.; Kangasjärvi, J. Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 2004, 40, 512–522. [Google Scholar] [CrossRef]
- Millar, J.B.; Buck, V.; Wilkinson, M.G. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995, 9, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Leple, J.C.; Brasileiro, A.C.M.; Michel, M.F.; Delmotte, F.; Jouanin, L. Transgenic poplars: expression of chimeric genes using four different constructs. Plant Cell Rep. 1992, 11, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M. Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB. Cold Spring Harb. Protoc. 2010, 2010, 1228–1229. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, H.; Li, H.; Su, Y.; Li, S.; Yang, Y.; Feng, C.; Yin, W.; Xia, X. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 2018, 16, 1514–1528. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.; Xia, X.; Yin, W. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 2016, 14, 849–860. [Google Scholar] [CrossRef]
- Yamauchi, T.; Watanabe, K.; Fukazawa, A.; Mori, H.; Abe, F.; Kawaguchi, K.; Oyanagi, A.; Nakazono, M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 2014, 65, 261–273. [Google Scholar] [CrossRef]
- Chen, S.L.; Li, J.K.; Wang, S.S.; Fritz, E.; Huttermann, A.; Altman, A. Effects of NaCl on shoot growth, transpiration, ion compartmentation, and transport in regenerated plants of Populus euphratica and Populus tomentosa. Can. J. For. Res.-Rev. Can. Rech. For. 2003, 33, 967–975. [Google Scholar] [CrossRef]
- Wang, F.; Deng, S.; Ding, M.; Sun, J.; Wang, M.; Zhu, H.; Han, Y.; Shen, Z.; Jing, X.; Zhang, F. Overexpression of a poplar two-pore K+ channel enhances salinity tolerance in tobacco cells. Plant Cell Tissue Organ Cult. 2013, 112, 19–31. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis Ubiquitin Conjugase UBC32 Is an ERAD Component That Functions in Brassinosteroid-Mediated Salt Stress Tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Shen, Z.; Pardington-Purtymun, P.E.; Comeaux, J.C.; Moyzis, R.K.; Chen, D.J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics 1996, 37, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Stolpe, T.; Süsslin, C.; Marrocco, K.; Nick, P.; Kretsch, T.; Kircher, S. In planta analysis of protein–protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 2005, 226, 137–146. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Su, W.; Bao, Y.; Wang, S.; He, F.; Wang, D.; Yu, X.; Yin, W.; Liu, C.; Xia, X. Poplar PdPTP1 Gene Negatively Regulates Salt Tolerance by Affecting Ion and ROS Homeostasis in Populus. Int. J. Mol. Sci. 2020, 21, 1065. https://doi.org/10.3390/ijms21031065
Lu Y, Su W, Bao Y, Wang S, He F, Wang D, Yu X, Yin W, Liu C, Xia X. Poplar PdPTP1 Gene Negatively Regulates Salt Tolerance by Affecting Ion and ROS Homeostasis in Populus. International Journal of Molecular Sciences. 2020; 21(3):1065. https://doi.org/10.3390/ijms21031065
Chicago/Turabian StyleLu, Yingying, Wanlong Su, Yu Bao, Shu Wang, Fang He, Dongli Wang, Xiaoqian Yu, Weilun Yin, Chao Liu, and Xinli Xia. 2020. "Poplar PdPTP1 Gene Negatively Regulates Salt Tolerance by Affecting Ion and ROS Homeostasis in Populus" International Journal of Molecular Sciences 21, no. 3: 1065. https://doi.org/10.3390/ijms21031065
APA StyleLu, Y., Su, W., Bao, Y., Wang, S., He, F., Wang, D., Yu, X., Yin, W., Liu, C., & Xia, X. (2020). Poplar PdPTP1 Gene Negatively Regulates Salt Tolerance by Affecting Ion and ROS Homeostasis in Populus. International Journal of Molecular Sciences, 21(3), 1065. https://doi.org/10.3390/ijms21031065