High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing
Abstract
1. Introduction
2. Results
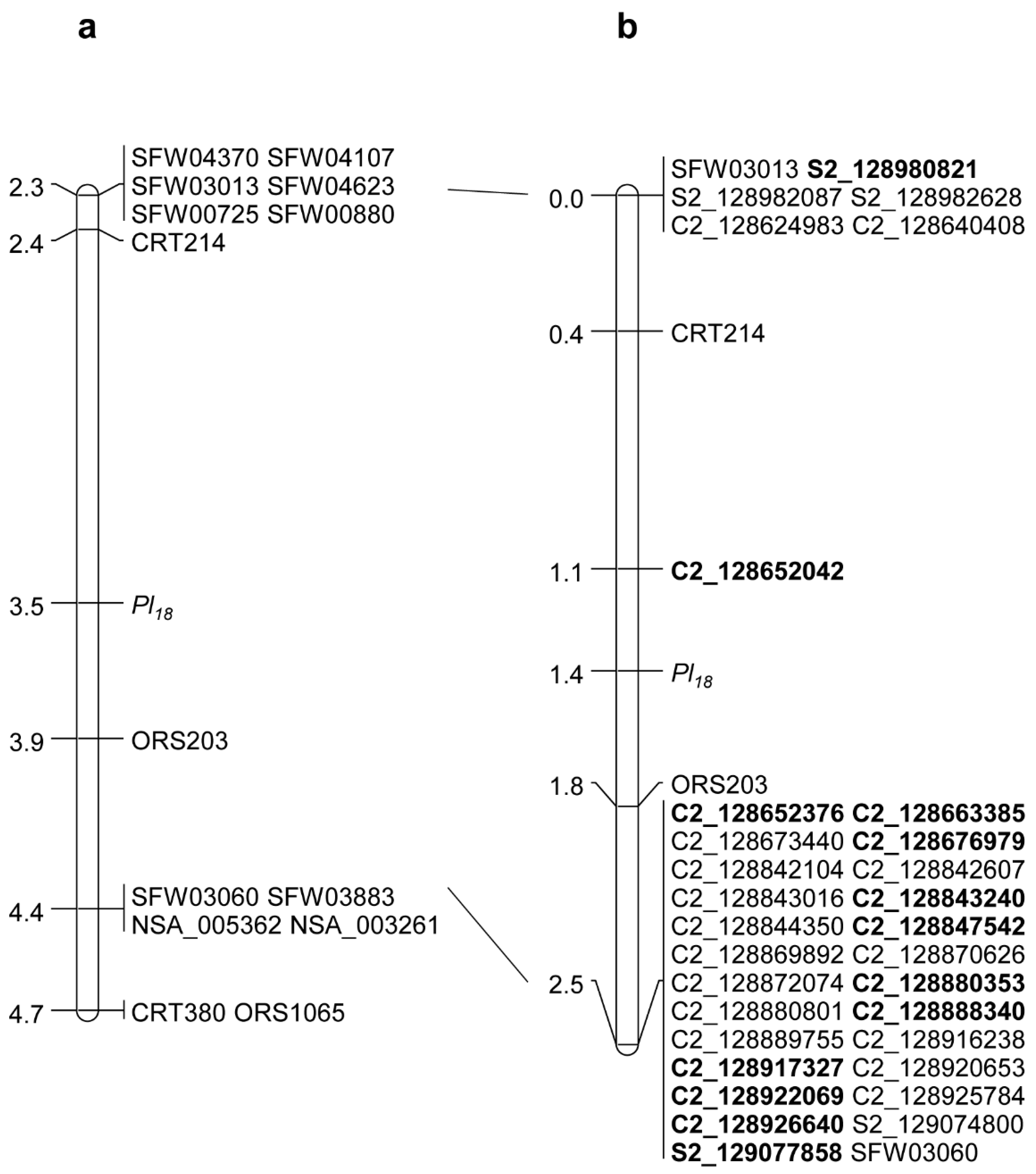
2.1. Saturation Mapping of Pl18
2.2. Saturation and Fine Mapping of Pl20
2.3. Identification of Candidate Genes for Pl18 and Pl20
2.4. Development of Diagnostic Markers for Pl18 and Pl20
3. Discussion
4. Materials and Methods
4.1. Mapping Populations and Evaluation Panel
4.2. SNP Marker Development from Whole-Genome Resequencing
4.3. Genotyping of PCR-Based SNP Markers and Linkage Analysis
4.4. Phenotypic Evaluation of Recombinants
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ma, G.J.; Markell, S.G.; Song, Q.J.; Qi, L.L. Genotyping-by-sequencing targeting of a novel downy mildew resistance gene Pl20 from wild Helianthus argophyllus for sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2017, 130, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Vear, F. Breeding disease-resistant sunflowers. CAB Rev. 2017, 12, 1–11. [Google Scholar] [CrossRef]
- Gulya, T.J.; Buetow, R.; Kandel, H. National sunflower association survey. In Proceedings of the 40th Sunflower Research Forum, Fargo, ND, USA, 10–11 January 2018. [Google Scholar]
- Gilley, M.; Gulya, T.; Seiler, G.J.; Underwood, W.; Hulke, B.S.; Misar, C.; Markell, S. Determination of virulence phenotypes of Plasmopara halstedii in the United States. Plant Dis. 2020, 104, 2823–2831. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Gulya, T.J. Evaluation of wild annual Helianthus species for resistance to downy mildew and Sclerotinia stalk rot. In Proceedings of the 27th Sunflower Research Forum, Fargo, ND, USA, 12–13 January 2005. [Google Scholar]
- Qi, L.L.; Foley, M.E.; Cai, X.W.; Gulya, T.J. Genetics and mapping of a novel downy mildew resistance gene, Pl18, introgressed from wild Helianthus argophyllus into cultivated sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2016, 129, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.L.; Seiler, G.J. Registration of an oilseed sunflower germplasm HA-DM1 resistant to sunflower downy mildew. J. Plant. Reg. 2016, 10, 195–199. [Google Scholar] [CrossRef]
- Slabaugh, M.B.; Yu, J.-K.; Tang, S.; Heesacker, A.; Hu, X.; Lu, G.; Bidney, D.; Han, F.; Knapp, S.J. Haplotyping and mapping a large cluster of downy mildew resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotechnol. J. 2003, 1, 167–185. [Google Scholar] [CrossRef]
- Yue, B.; Vick, B.A.; Cai, X.; Hu, J. Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus L.) by SSR and TRAP markers. Plant Breed. 2010, 129, 24–28. [Google Scholar] [CrossRef]
- Franchel, J.; Bouzidi, M.F.; Bronner, G.; Vear, F.; Nicolas, P.; Mouzeyar, S. Positional cloning of a candidate gene for resistance to the sunflower downy mildew, Plasmopara halstedii race 300. Theor. Appl. Genet. 2013, 126, 359–367. [Google Scholar] [CrossRef]
- Ma, G.J.; Song, Q.J.; Markell, S.G.; Qi, L.L. High-throughput genotyping-by-sequencing facilitates molecular tagging of a novel rust resistance gene, R15, in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2018, 131, 1423–1432. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Radwan, O. Identification of non-TIRNBS-LRR markers linked to the Pl5/Pl8 locus for resistance to downy mildew in sunflower. Theor. Appl. Genet. 2003, 106, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Liu, X.L.; Dai, L.Y.; Wang, G.L. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genom. 2007, 34, 765–776. [Google Scholar] [CrossRef]
- Bachlava, E.; Radwan, O.E.; Abratti, G.; Tang, S.; Gao, W.; Heesacker, A.F.; Bazzalo, M.E.; Zambelli, A.; Leon, A.J.; Knapp, S.J. Downy mildew (Pl8 and Pl14) and rust (RAdv) resistance genes reside in close proximity to tandemly duplicated clusters of non-TIR-like NBS-LRR-encoding genes on sunflower chromosomes 1 and 13. Theor. Appl. Genet. 2011, 122, 1211–1221. [Google Scholar] [CrossRef]
- Wersch, S.V.; Li, X. Stronger when together: Clustering of plant NLR disease resistance genes. Trends Plant Sci. 2019, 24, 8. [Google Scholar] [CrossRef]
- Sagi, M.S.; Deokar, A.A.; Tar’an, B. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to Ascochyta blight infection. Front. Plant Sci. 2017, 8, 838. [Google Scholar] [CrossRef]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019, 99, 95–111. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, F.; Zhu, S.; Li, X. The Maize NBS-LRR Gene ZmNBS25 Enhances Disease Resistance in Rice and Arabidopsis. Front. Plant Sci. 2018, 9, 1033. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, S.; Abate, Z.; Nirmala, J.; Rouse, M.N.; Dubcovsky, J. Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. USA 2017, 114, 45. [Google Scholar] [CrossRef]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef]
- Roulin, A. The fate of duplicated genes in a polyploidy plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Long, Y.; Song, Q.; Talukder, Z.I.; Shamimuzzaman, M.; Qi, L. Map and sequence-based chromosome walking toward cloning of the male fertility restoration gene Rf5 linked to the rust resistance gene R11 in sunflower. Sci. Rep. 2020. accepted. [Google Scholar]
- Pecrix, Y.; Penouilh-Suzette, C.; Muños, S.; Vear, F.; Godiard, L. Ten broad spectrum resistances to downy mildew physically mapped on the sunflower genome. Front. Plant Sci. 2018, 9, 1780. [Google Scholar] [CrossRef] [PubMed]
- Radwan, O.; Gandhi, S.; Heesacker, A.; Whitaker, B.; Taylor, C.; Plocik, A.; Kesseli, R.; Kozik, A.; Michelmore, R.W.; Knapp, S.J. Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol. Gen. Genom. 2008, 280, 111–125. [Google Scholar] [CrossRef]
- Zimmer, D.E.; Kinman, M.L. Downy mildew resistance in cultivated sunflower and its inheritance. Crop Sci. 1972, 12, 749–751. [Google Scholar] [CrossRef]
- Vear, F. Origins of major genes for downy mildew resistance in sunflower. In Proceedings of the 17th International Sunflower Conference, Córdoba, Spain, 8–12 June 2008; pp. 125–130. [Google Scholar]
- Miller, J.F.; Gulya, T.J. Inheritance of resistance to race 3 downy mildew in sunflower. Crop Sci. 1987, 27, 210–212. [Google Scholar] [CrossRef]
- Miller, J.F.; Gulya, T.J. Inheritance of resistance to race 4 of downy mildew derived from interspecific crosses in sunflower. Crop Sci. 1991, 31, 40–43. [Google Scholar] [CrossRef]
- De Romano, A.B.; Romano, C.; Bulos, M.; Altieri, E.; Sala, C. A new gene for resistance to downy mildew in sunflower. In Proceedings of the International Symposium “Sunflower breeding on resistance to diseases”, Krasnodar, Russia, 23–24 June 2010. [Google Scholar]
- Qi, L.L.; Long, Y.M.; Jan, C.C.; Ma, G.J.; Gulya, T.J. Pl17 is a novel gene independent of known downy mildew resistance genes in the cultivated sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2015, 128, 757–767. [Google Scholar] [CrossRef]
- Long, Y.M.; Chao, W.S.; Ma, G.J.; Xu, S.S.; Qi, L.L. An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor. Appl. Genet. 2017, 130, 597–607. [Google Scholar] [CrossRef]
- Ma, G.; Song, Q.; Underwood, W.R.; Zhang, Z.; Fiedler, J.D.; Li, X.; Qi, L. Molecular dissection of resistance gene cluster and candidate gene identification of Pl17 and Pl19 in sunflower by whole-genome resequencing. Sci. Rep. 2019, 9, 14974. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Gulya, T.J.; Draper, M.; Harbour, J.; Holen, C.; Knodel, J.; Lamey, A.; Mason, P. Metalaxyl resistance in sunflower downy mildew in North America. In Proceedings of the 21st Sunflower Research Forum, Fargo, ND, USA, 14–15 January 1999; pp. 118–123. [Google Scholar]
- Vrânceanu, V.; Stoenescu, F. Immunity to sunflower downy mildew due to a single dominant gene. Probleme. Agric. 1970, 22, 34–40. [Google Scholar]
- Gedil, M.A. Candidate disease resistance genes in sunflower cloned using conserved nucleotide binding site motifs: Genetic mapping and linkage to downy mildew resistance gene Pl1. Genome 2001, 44, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Vear, F.; Gentzbittel, L.; Philippon, J.; Mouzeyar, S.; Mestries, E.; Roeckel-Drevet, P.; De Labrouhe, D.T.; Nicolas, P. The genetics of resistance to five races of downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 1997, 95, 584–589. [Google Scholar] [CrossRef]
- Vear, F.; Leclercq, P. Deux nouvea ux genes resistance au mildiou du tournesol. Ann. Amelior. Plant 1971, 21, 251–255. [Google Scholar]
- Vear, F. Studies on resistance to downy mildew in sunflower (Helianthus annuus L.). In Proceedings of the 6th International Sunflower Conference, Bucharest, Romania, 22–24 July 1974; pp. 297–302. [Google Scholar]
- Bert, P.F.; De Labrouhe, D.T.; Philippon, J.; Mouzeyar, S.; Jouan, I.; Nicolas, P.; Vear, F. Identification of a second linkage group carrying genes controlling resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2001, 103, 992–997. [Google Scholar] [CrossRef]
- Roeckel-Drevet, P. Colocation of downy mildew (Plasmopara halstedii) resistance genes in sunflower (Helianthus annuus L.). Euphytica 1996, 91, 225–228. [Google Scholar]
- Gulya, T.J.; Sackston, W.E.; Viranyi, F.; Masirevic, S.; Rashid, K.Y. New races of the sunflower downy mildew pathogen (Plasmopara halstedii) in Europe and North and South America. J. Phytopathol. 1991, 132, 303–311. [Google Scholar] [CrossRef]
- Seiler, G.J. Registration of 13 downy mildew tolerant interspecific sunflower germplasm lines derived from wild annual species. Crop Sci. 1991, 31, 1714–1716. [Google Scholar]
- Dußle, C.M.; Hahn, V.; Knapp, S.J.; Bauer, E. PlArg from Helianthus argophyllus is unlinked to other known downy mildew resistance genes in sunflower. Theor. Appl. Genet. 2004, 109, 1083–1086. [Google Scholar] [CrossRef]
- Rahim, M.; Jan, C.C.; Gulya, T.J. Inheritance of resistance to sunflower downy mildew races 1, 2 and 3 in cultivated sunflower. Plant Breed. 2002, 121, 57–60. [Google Scholar] [CrossRef]
- Gulya, T.J. Registration of five disease-resistant sunflower germplasms. Crop Sci. 1985, 25, 719–720. [Google Scholar] [CrossRef]
- Mulpuri, S.; Liu, Z.; Feng, J.; Gulya, T.J.; Jan, C.C. Inheritance and molecular mapping of a downy mildew resistance gene, Pl13 in cultivated sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2009, 119, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gulya, T.J.; Seiler, G.J.; Vick, B.A.; Jan, C.C. Molecular mapping of the Pl16 downy mildew resistance gene from HA-R4 to facilitate marker-assisted selection in sunflower. Theor. Appl. Genet. 2012, 125, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Hulke, B.S.; Miller, J.F.; Gulya, T.J.; Vick, B.A. Registration of the oilseed sunflower genetic stocks HA 458, HA 459, and HA 460 possessing genes for resistance to downy mildew. J. Plant Reg. 2010, 4, 93–97. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Ma, G.J.; Zhao, J.; Markell, S.G.; Qi, L.L. Discovery and introgression of the wild sunflower-derived novel downy mildew resistance gene Pl19 in confection sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2017, 130, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Vincourt, P.; As-Sadi, F.; Bordat, A.; Langlade, N.B.; Gouzy, J.; Pouilly, N.; Lippi, Y.; Serre, F.; Godiard, L.; De Labrouhe, D.T.; et al. Consensus mapping of major resistance genes and independent QTL for quantitative resistance to sunflower downy mildew. Theor. Appl. Genet. 2012, 125, 909–920. [Google Scholar] [CrossRef]
- Pecrix, Y.; Buendia, L.; Penouilh-Suzette, C.; Maréchaux, M.; Legrand, L.; Bouchez, O.; Rengel, D.; Gouzy, J.; Cottret, L.; Vear, F.; et al. Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii. Plant J. 2018, 97, 730–748. [Google Scholar] [CrossRef]
- Jan, C.C.; Gulya, T.J. Registration of a sunflower germplasm resistant to rust, downy mildew and virus. Crop Sci. 2006, 46, 1829. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, L.; Ma, G.J.; Seiler, G.J.; Jan, C.C.; Qi, L.L. Molecular mapping of the downy mildew and rust resistance genes in a sunflower germplasm line TX16R. Mol. Breed. 2019, 39, 19. [Google Scholar] [CrossRef]
- Miller, J.F.; Gulya, T.J.; Seiler, G.J. Registration of five fertility restorer sunflower germplasms. Crop Sci. 2002, 42, 989–991. [Google Scholar] [CrossRef]
- Talukder, Z.I.; Ma, G.; Hulke, B.S.; Jan, C.C.; Qi, L.L. Linkage mapping and genome-wide association studies of the Rf gene cluster in sunflower (Helianthus annuus L.) and their distribution in world sunflower collections. Front. Genet. 2019, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.L.; Ma, G.J.; Li, X.H.; Seiler, G.J. Diversification of the downy mildew resistance gene pool by introgression of a new gene, Pl35, from wild Helianthus argophyllus into oilseed and confection sunflowers (Helianthus annuus L.). Theor. Appl. Genet. 2019, 132, 2553–2565. [Google Scholar] [CrossRef] [PubMed]


| Marker | No. Recombination | Genetic Distance (cM) | Physical Position on XRQ Assembly | Physical Position on HA412-HO Assembly | ||
|---|---|---|---|---|---|---|
| Start | End | Start | End | |||
| S2_128980821 | 0.0 | 128,510,328 | 128,510,728 | 128,980,621 | 128,981,021 | |
| S2_128982087 | 0 | 0.0 | 128,511,594 | 128,511,994 | 128,981,887 | 128,982,287 |
| SFW03013 | 0 | 0.0 | 128,511,854 | 128,511,770 | 128,982,147 | 128,982,063 |
| S2_128982628 | 0 | 0.0 | 128,512,135 | 128,512,535 | 128,982,428 | 128,982,828 |
| C2_128624983 | 0 | 0.0 | 128,624,783 | 128,625,183 | 132,735,799 | 132,736,199 |
| C2_128640408 | 0 | 0.0 | 128,640,208 | 128,640,608 | 132,741,177 | 132,741,577 |
| CRT214 | 1 | 0.4 | NA | NA | NA | NA |
| C2_128652042 | 2 | 0.7 | 128,651,842 | 128,652,242 | NA | NA |
| Pl18 | 1 | 0.3 | - | - | - | - |
| ORS203 | 1 | 0.4 | NA | NA | NA | NA |
| C2_128652376 | 2 | 0.7 | 128,652,176 | 128,652,576 | 132,753,709 | 132,753,885 |
| C2_128663385 | 0 | 0.7 | 128,663,185 | 128,663,585 | 97,034,302 | 97,033,902 |
| C2_128673440 | 0 | 0.7 | 128,673,240 | 128,673,640 | 97,024,972 | 97,024,572 |
| C2_128676979 | 0 | 0.7 | 128,676,779 | 128,677,179 | 97,021,432 | 97,021,031 |
| C2_128842104 | 0 | 0.7 | 128,841,904 | 128,842,304 | 129,433,726 | 129,434,126 |
| C2_128842607 | 0 | 0.7 | 128,842,407 | 128,842,807 | 129,434,224 | 129,434,624 |
| C2_128843016 | 0 | 0.7 | 128,842,816 | 128,843,216 | 129,434,633 | 129,435,033 |
| C2_128843240 | 0 | 0.7 | 128,843,040 | 128,843,440 | 129,434,857 | 129,435,257 |
| C2_128844350 | 0 | 0.7 | 128,844,150 | 128,844,550 | 129,435,967 | 129,436,367 |
| C2_128847542 | 0 | 0.7 | 128,847,342 | 128,847,742 | 129,646,291 | 129,646,691 |
| C2_128869892 | 0 | 0.7 | 128,869,692 | 128,870,092 | 129,451,579 | 129,451,979 |
| C2_128870626 | 0 | 0.7 | 128,870,426 | 128,870,826 | 129,452,313 | 129,452,713 |
| C2_128872074 | 0 | 0.7 | 128,871,874 | 128,872,274 | 129,670,656 | 129,670,912 |
| C2_128880353 | 0 | 0.7 | 128,880,153 | 128,880,553 | 129,545,140 | 129,545,540 |
| C2_128880801 | 0 | 0.7 | 128,881,001 | 128,881,401 | 129,461,631 | 129,462,020 |
| C2_128888340 | 0 | 0.7 | 128,888,140 | 128,888,540 | 129,468,821 | 129,469,221 |
| C2_128889755 | 0 | 0.7 | 128,889,555 | 128,889,955 | 129,470,227 | 129,470,627 |
| C2_128916238 | 0 | 0.7 | 128,916,038 | 128,916,438 | 129,497,380 | 129,497,780 |
| C2_128917327 | 0 | 0.7 | 128,917,127 | 128,917,527 | 129,498,469 | 129,498,869 |
| C2_128920653 | 0 | 0.7 | 128,920,453 | 128,920,853 | 129,501,790 | 129,502,189 |
| C2_128922069 | 0 | 0.7 | 128,921,869 | 128,922,269 | 129,503,206 | 129,503,605 |
| C2_128925784 | 0 | 0.7 | 128,925,584 | 128,925,784 | 129,507,004 | 129,507,404 |
| C2_128926640 | 0 | 0.7 | 128,926,440 | 128,926,840 | 129,507,860 | 129,508,260 |
| S2_129074800 | 0 | 0.7 | 129,300,154 | 129,299,754 | 129,074,600 | 129,074,800 |
| S2_129077858 | 0 | 0.7 | 129,297,096 | 129,296,696 | 129,077,658 | 129,078,058 |
| SFW03060 | 0 | 0.7 | 129,292,083 | 129,292,202 | 129,082,571 | 129,082,452 |
| Marker | No. Recombination | Genetic Distance (cM) | Physical Position on XRQ Assembly (bp) | Physical Position on HA412-HO Assembly (bp) |
|---|---|---|---|---|
| SFW01920 † | 0.00 | 855,100–855,219 | 8,626,529–8,626,648 | |
| SFW09076 † | 73 | 1.47 | 6,259,434–6,259,553 | 9,317,681–9,317,800 |
| C8_7890010 | 73 | 1.47 | 7,889,810–7,890,210 | 11,328,554–11,328,954 |
| S8_11272025 † | 3 | 0.06 | 7,890,621–7,891,021 | 11,271,825–11,272,225 |
| S8_11272046 † | 0 | 0.00 | 7,890,600–7,891,000 | 11,271,846–11,272,246 |
| C8_7894021 | 8 | 0.16 | 7,893,821–7,894,221 | 11,268,522–11,268,922 |
| C8_7895128 | 0 | 0.00 | 7,894,928–7,895,328 | 11,323,742–11,324,142 |
| C8_7902182 | 0 | 0.00 | 7,901,982–7,902,382 | 11,262,375–11,262,775 |
| C8_7914784 | 4 | 0.08 | 7,914,584–7,914,984 | 11,251,806–11,252,206 |
| C8_7917284 | 2 | 0.04 | 7,917,084–7,917,484 | 11,249,306–11,249,706 |
| SFW04358 † | 1 | 0.02 | 6,438,630–6,438,749 | 10,072,538–100,72,657 |
| C8_7919216 | 0 | 0.00 | 7,919,016–7,919,416 | 11,247,370–11,247,773 |
| C8_7920504 | 0 | 0.00 | 7,920,304–7,920,704 | 11,246,082–11,246,482 |
| C8_7921819 | 0 | 0.00 | 7,921,619–7,922,019 | 11,244,767–11,245,167 |
| Pl20 | 3 | 0.06 | – | – |
| C8_8012577 | 7 | 0.14 | 8,012,377–8,012,777 | 9,689,417–9,689,678 |
| C8_8003402 | 1 | 0.02 | 8,003,202–8,003,602 | 9,680,535–9,680,935 |
| C8_8009207 | 0 | 0.00 | 8,009,007–8,009,407 | 10,853,046–10,853,441 |
| C8_8004678 | 1 | 0.02 | 8,004,478–8,004,878 | 9,681,811–9,682,211 |
| C8_8504355 | 0 | 0.00 | 8,504,155–8,504,555 | 10,905,758–10,906,158 |
| C8_8459708 | 1 | 0.02 | 8,459,508–8,459,908 | 12,109,436–12,109,838 |
| C8_8460626 | 1 | 0.02 | 8,460,426–8,460,826 | 12,226,272–12,226,515 |
| C8_8458330 | 1 | 0.02 | 8,458,130–8,458,530 | 12,108,058–12,108,458 |
| SFW02745 † | 1 | 0.02 | 8,456,520–8,456,639 | 11,614,201–11,614,082 |
| C8_8452446 | 1 | 0.02 | 8,452,246–8,452,646 | 11,620,100–11,620,414 |
| C8_8639656 | 26 | 0.52 | 8,639,456–8,639,856 | 12,827,866–12,828,204 |
| C8_8666606 | 1 | 0.02 | 8,666,406–8,666,806 | 12,796,693–12,797,102 |
| C8_8800366 | 9 | 0.18 | 8,800,166–8,800,566 | 13,920,131–13,920,531 |
| C8_8835415 | 0 | 0.00 | 8,835,215–8,835,615 | 13,960,633–13,961,033 |
| S8_100385559 † | 0 | 0.00 | 8,907,619–8,908,019 | 100,385,359–100,385,759 |
| Genes | Definition | Physical Position | Length (bp) |
|---|---|---|---|
| For Pl18 | |||
| HanXRQChr02g0048101 | Probable mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein | 128,508,926...128,514,561 | 5636 |
| HanXRQChr02g0048111 | Putative glycoside hydrolase family 17; Glycoside hydrolase superfamily | 128,551,551...128,553,503 | 1953 |
| HanXRQChr02g0048131 | Putative alcohol dehydrogenase superfamily, zinc-type; L-threonine 3-dehydrogenase; NAD(P)-binding domain | 128,639,137...128,639,993 | 857 |
| HanXRQChr02g0048141 | Putative alcohol dehydrogenase superfamily, zinc-type; GroES-like | 128,642,764...128,642,967 | 204 |
| HanXRQChr02g0048151 | Putative alcohol dehydrogenase superfamily, zinc-type; GroES-like | 128,651,738...128,652,649 | 912 |
| HanXRQChr02g0048171 | Putative NAC domain | 128,841,438...128,843,598 | 2161 |
| HanXRQChr02g0048181 | Putative NB-ARC; Powdery mildew resistance protein, RPW8 domain; P-loop containing nucleoside triphosphate hydrolase; Leucine-rich repeat domain, L domain-like | 128,920,787...128,926,787 | 6001 |
| For Pl20 | |||
| HanXRQChr08g0210011 | Probable GYF domain-containing protein | 7,910,378...7,917,771 | 7394 |
| HanXRQChr08g0210051 | Putative NB-ARC; Toll-like receptor; P-loop containing nucleoside triphosphate hydrolase; Leucine-rich repeat domain, L domain-like | 8,010,685...8,035,718 | 25,034 |
| HanXRQChr08g0210081 | Putative tify domain; CO/COL/TOC1, conserved site | 8,273,535...8,277,186 | 3652 |
| HanXRQChr08g0210111 | Putative bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain | 8,306,497...8,309,225 | 2729 |
| HanXRQChr08g0210131 | Probable CONTAINS InterPro DOMAIN/s: WW/Rsp5/WWP (InterPro:IPR001202) | 8,325,909...8,327,735 | 1827 |
| HanXRQChr08g0210141 | Probable B-box type zinc finger protein with CCT domain | 8,398,470...8,403,825 | 5356 |
| HanXRQChr08g0210151 | Putative leucine-rich repeat-containing N-terminal, plant-type; Leucine-rich repeat domain, L domain-like | 8,408,566...8,411,305 | 2740 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, G.; Song, Q.; Li, X.; Qi, L. High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing. Int. J. Mol. Sci. 2020, 21, 9571. https://doi.org/10.3390/ijms21249571
Ma G, Song Q, Li X, Qi L. High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing. International Journal of Molecular Sciences. 2020; 21(24):9571. https://doi.org/10.3390/ijms21249571
Chicago/Turabian StyleMa, Guojia, Qijian Song, Xuehui Li, and Lili Qi. 2020. "High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing" International Journal of Molecular Sciences 21, no. 24: 9571. https://doi.org/10.3390/ijms21249571
APA StyleMa, G., Song, Q., Li, X., & Qi, L. (2020). High-Density Mapping and Candidate Gene Analysis of Pl18 and Pl20 in Sunflower by Whole-Genome Resequencing. International Journal of Molecular Sciences, 21(24), 9571. https://doi.org/10.3390/ijms21249571






