Highly Efficient, Tripodal Ion-Pair Receptors for Switching Selectivity between Acetates and Sulfates Using Solid–Liquid and Liquid–Liquid Extractions
Abstract
1. Introduction
2. Results and Discussion
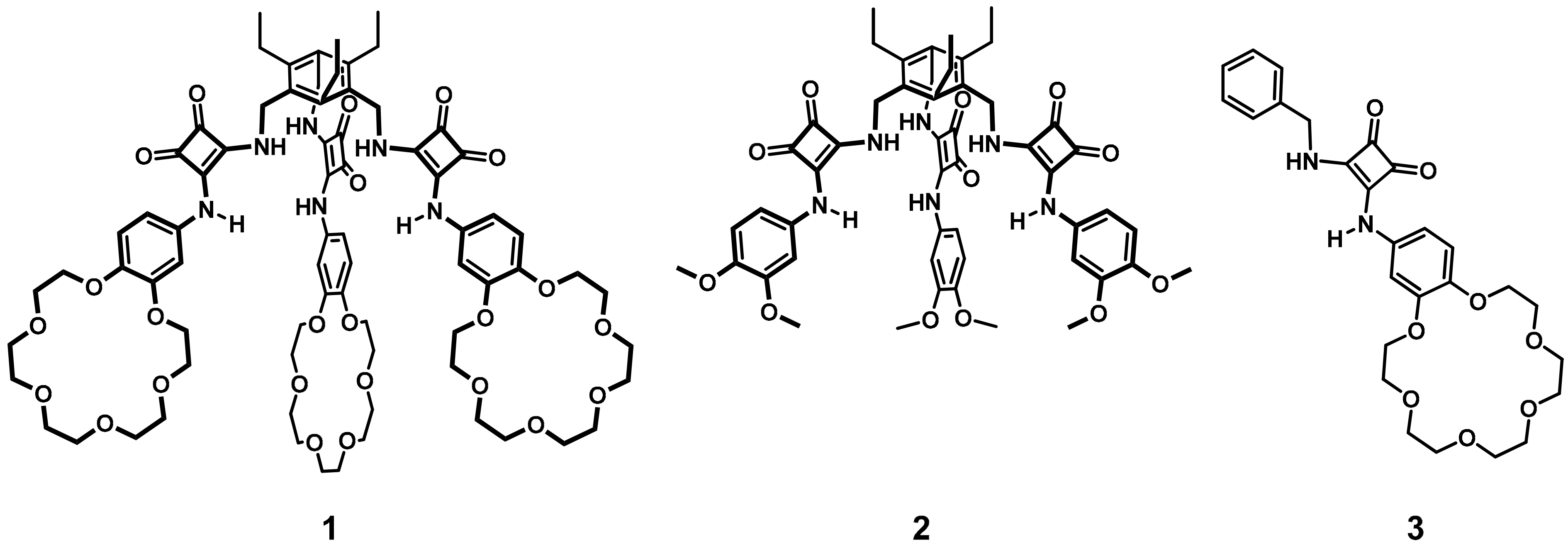
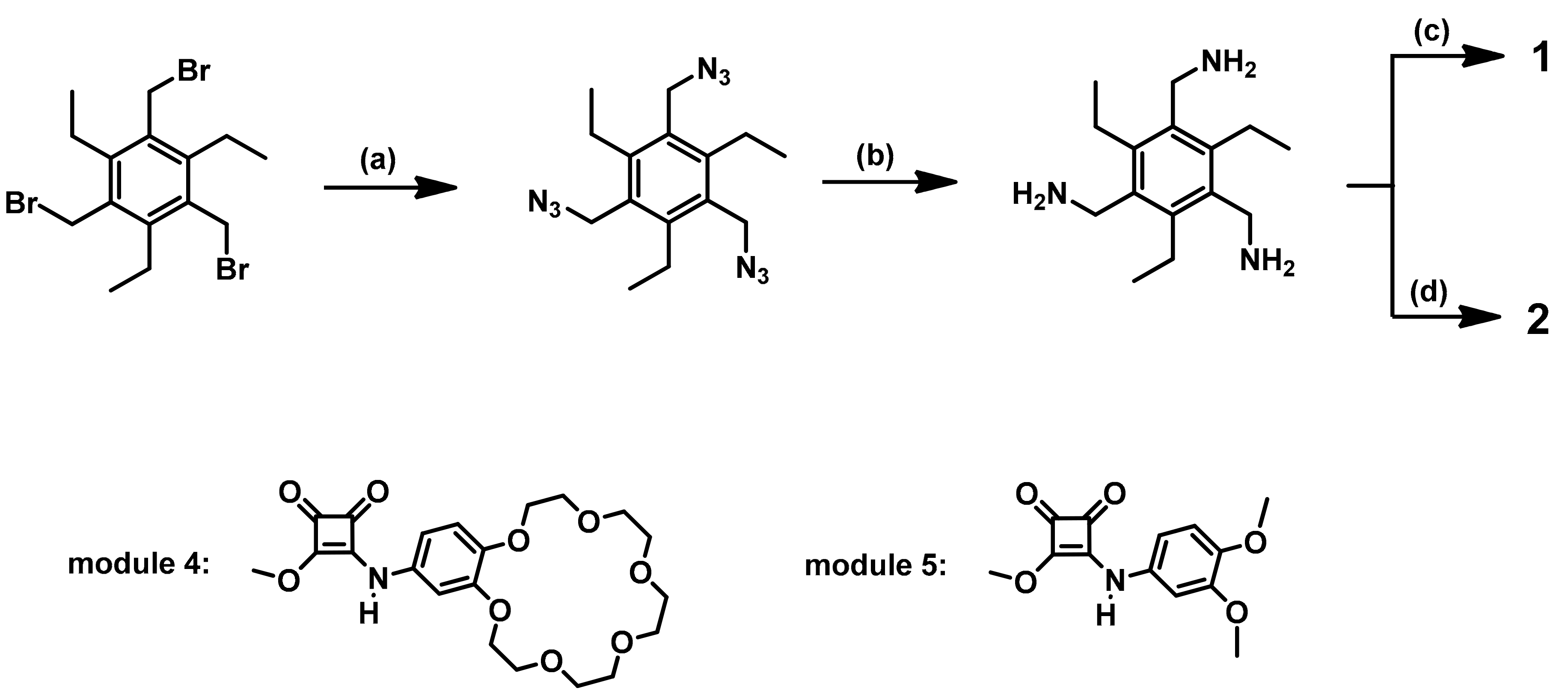
2.1. Receptor Design and Synthesis
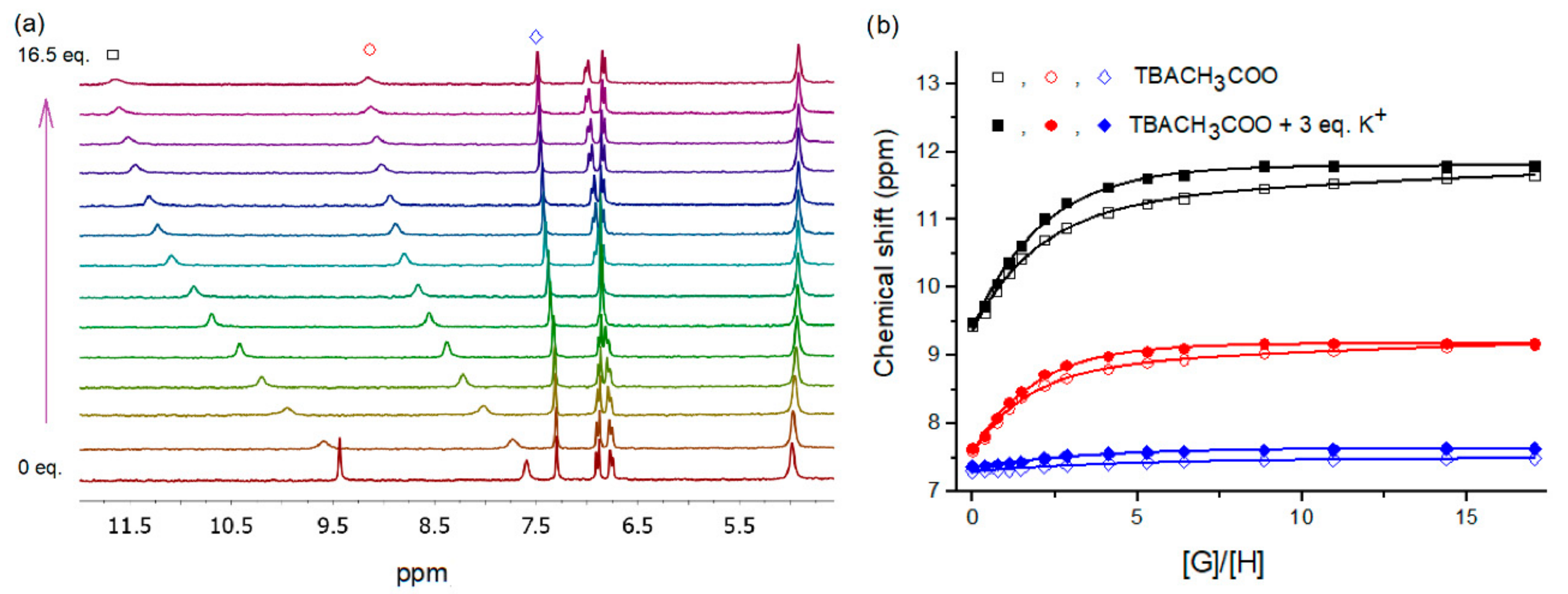
2.2. Binding Studies
2.3. Extraction Studies
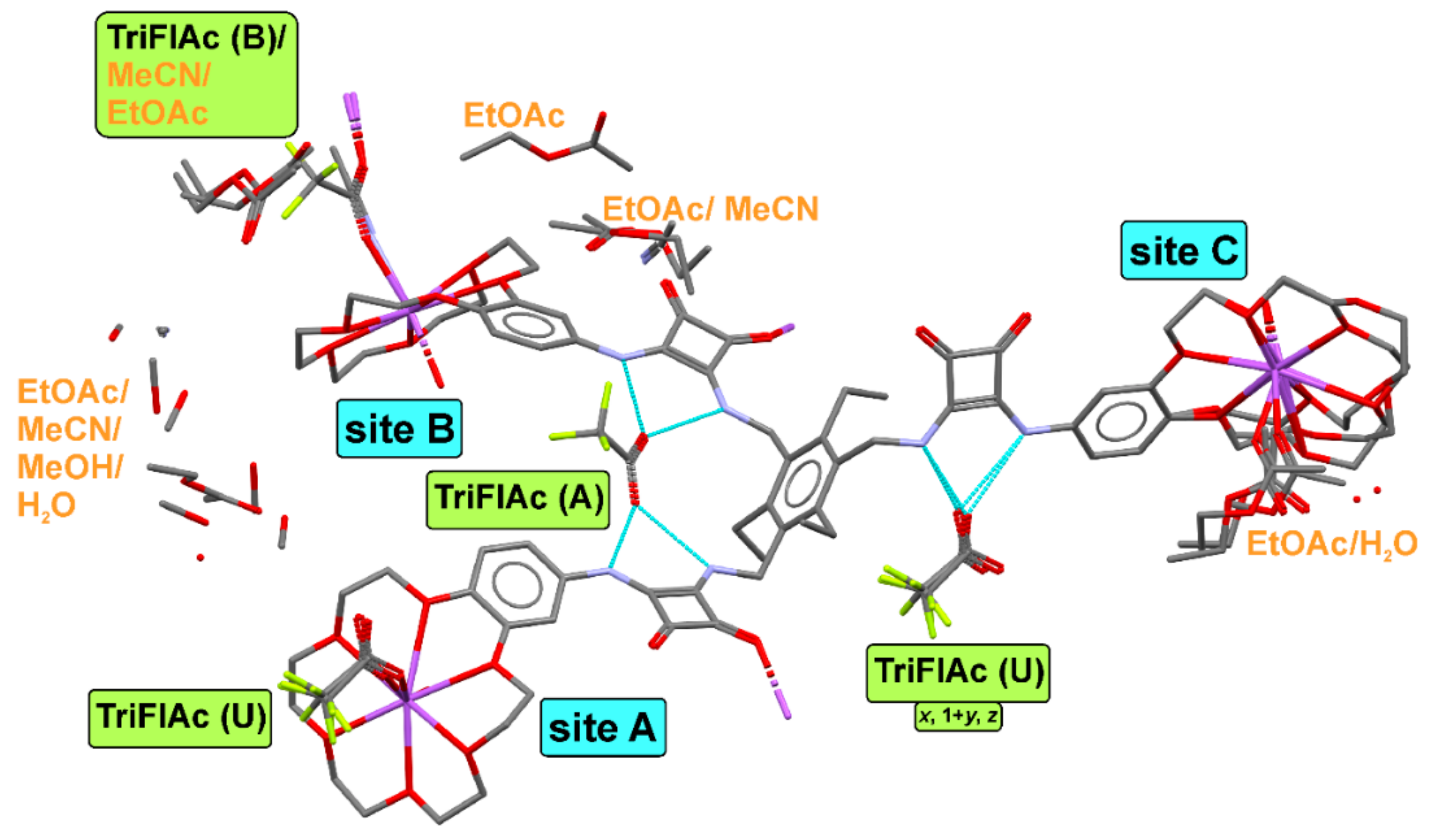
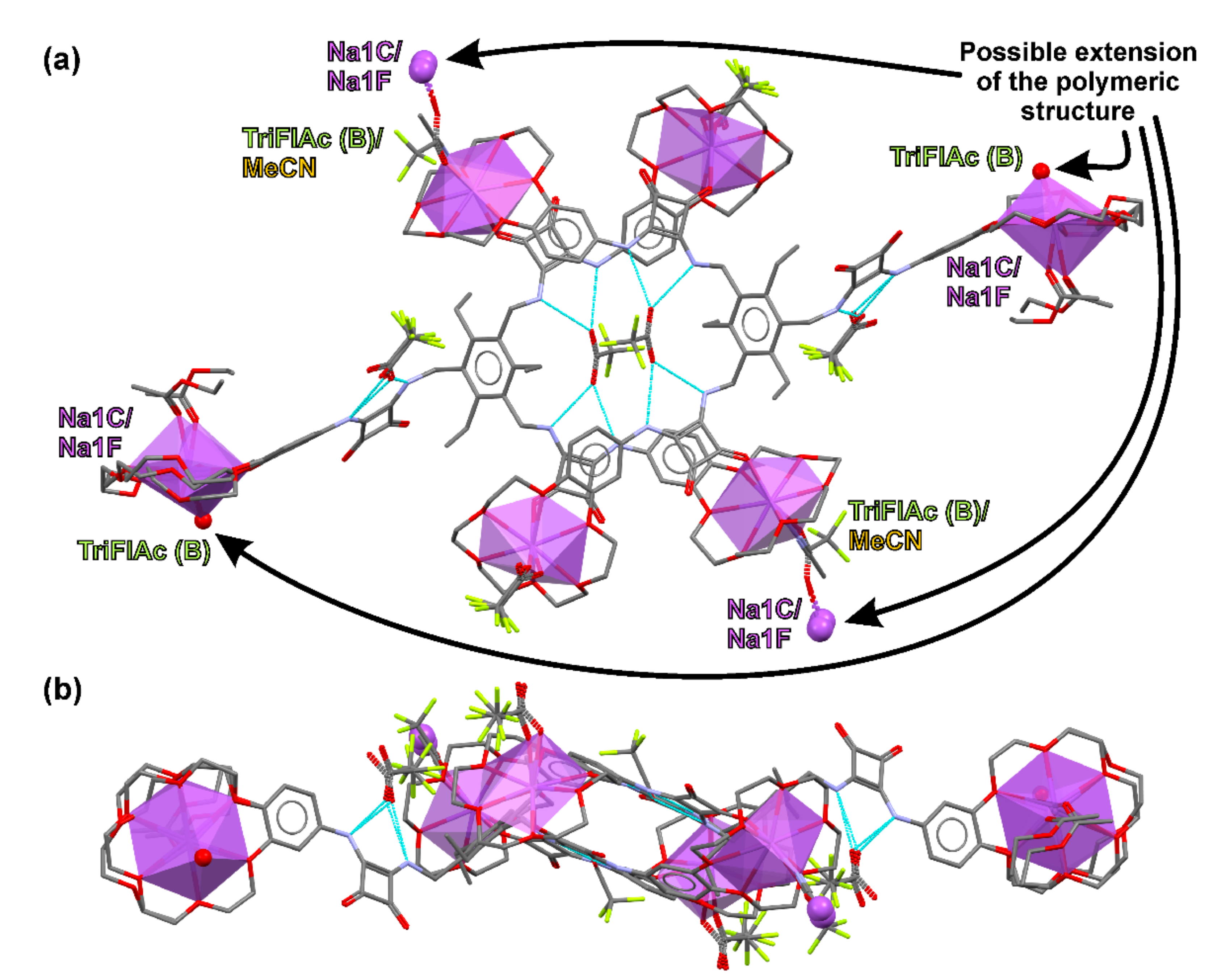
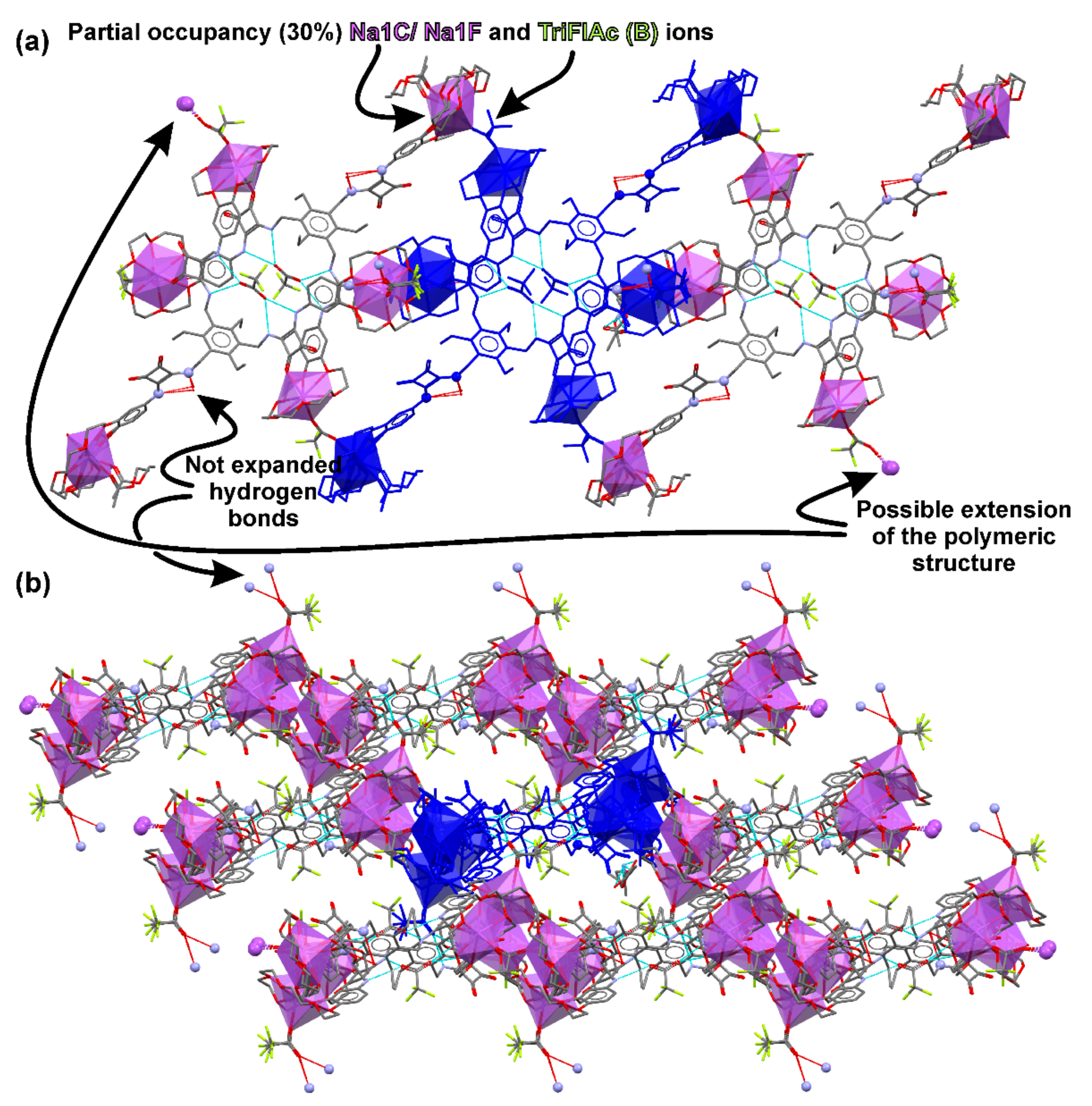
2.4. Crystallization and Single-Crystal X-ray Diffraction
3. Materials and Methods
3.1. General Methods
3.2. Synthetic Details
- HRMS (ESI): calculated for C15H21N9 (M + H) + 327.1909, found: 327.1915.
- 1H NMR (300 MHz, CDCl3) δ 4.50 (s, 6H), 2.84 (q, 6H), 1.25 (t, 9H).
- 13C NMR (75 MHz, CDCl3) δ 145.0, 129.8, 47.9, 23.2 15.8.
- HRMS (ESI): calculated for C15H27N3 (M + H) + 250.2283, found: 250.2290.
- 1H NMR (300 MHz, CDCl3) δ 3.85 (s, 6H), 2.80 (q, 6H), 1.40 (s, 6H), 1.25 (t, 9H).
- 13C NMR (75 MHz, CDCl3) δ 140.4, 137.5, 39.7, 22.5, 16.8.
- HRMS (ESI): calculated for C21H27NO9Na (M + Na)+: 460.1584, found: 460.1588.
- 1H NMR (300 MHz, DMSO-d6) δ 10.64 (s, 1H), 7.18–6.96 (m, 1H), 6.96–6.84 (m, 1H), 6.83–6.78 (m, 1H), 4.34 (s, 3H), 4.10–3.95 (m, 4H), 3.80–3.65 (m, 4H), 3.62–3.44 (m, 12H).
- 13C NMR (75 MHz, DMSO-d6) δ 189.3, 184.1, 178.9, 169.1, 148.9, 145.6, 132.0, 114.0, 112.1, 106.7, 70.5, 70.3, 70.2, 69.6, 69.4, 69.1, 68.7, 61.0.
- HRMS (ESI): calculated for C13H13NO5Na (M + Na)+: 286.0692, found: 286.0702.
- u1H NMR (300 MHz, DMSO-d6) δ 10.63 (s, 1H), 7.20–6.65 (m, 3H), 4.37 (s, 3H), 3.85–3.65 (m, 6H).
- 13C NMR (75 MHz, DMSO-d6) δ 189.1, 183.8, 178.6, 169.2, 149.4, 146.2, 131.8, 112.6, 111.9, 105.3, 60.9, 56.2, 55.9.
- HRMS (ESI): calculated for C75H96N6O24Na (M + Na)+:1487.6373, found: 1487.6333.
- 1H NMR (300 MHz, DMSO-d6) δ 9.43 (s, 1H), 7.59 (s, 1H), 7.30 (s, 1H), 6.92–6.73 (m, 2H), 4.98 (s, 2H), 4.12–4.00 (m, 4H), 3.80–3.72 (m, 4H), 3.62–3.50 (m, 12H), 2.85 (s, 2H), 1.19 (m, 3H).
- 13C NMR (75 MHz, DMSO-d6) δ 183.8, 180.7, 168.5, 163.9, 148.6, 144.4, 144.0, 133.1, 132.5, 113.3, 110.0, 104.3, 70.0, 69.9, 69.0, 68.8, 67.9, 67.7, 65.4, 42.0, 23.1, 16.8.
- HRMS (ESI): calculated for C51H54N6O12Na (M + Na)+: 965.3698, found: 965.3712.
- 1H NMR (300 MHz, DMSO-d6) δ 9.44 (s, 1H), 7.59 (s, 1H), 7.31 (s, 1H), 6.94-6.76 (m, 2H), 4.99 (s, 2H), 3.79–3.68 (m, 6H), 2.88–2.84 (m, 2H), 1.19 (m, 3H).
- 13C NMR (75 MHz, DMSO-d6) δ 183.7, 180.7, 168.4, 163.9, 149.8, 145.1, 144.3, 133.1, 132.6, 113.1, 109.9, 103.9, 56.3, 55.9, 42.0, 23.1, 16.8.
- HRMS (ESI): calculated for C27H32N2O8Na (M + Na)+: 535.2056, found: 535.2043.
- 1H NMR (300 MHz, DMSO-d6) δ 9.56 (s, 1H), 7.90 (s, 1H), 7.45–7.30 (m, 5H), 7.23 (s, 1H), 7.00–6.80 (m, 1H), 6.80–6.71 (m, 1H), 4.80 (s, 2H), 4.12–3.95 (m, 4H), 3.79–3.68 (m, 4H), 3.66–3.49 (m, 12 H).
- 13C NMR (75 MHz, DMSO-d6) δ 183.9, 181.2, 168.9, 164.2, 149.2, 144.6, 139.0, 133.1, 129.2, 128.1, 128.1, 114.5, 110.4, 105.1, 70.3, 69.2, 69.1, 69.0, 68.4, 47.6.
3.3. NMR Titration Procedure
3.4. UV-Vis Titration Procedure
3.5. Crystallographic Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Evans, N.H.; Beer, P.D. Advances in Anion Supramolecular Chemistry: From Recognition to Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 11716–11754. [Google Scholar] [CrossRef] [PubMed]
- Busschaert, N.; Caltagirone, C.; Van Rossom, W.; Gale, P.A. Applications of supramolecular anion recognition. Chem. Rev. 2015, 115, 8038–8155. [Google Scholar] [CrossRef] [PubMed]
- Carter, K.P.; Young, A.M.; Palmer, A.E. Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 2014, 114, 4564–4601. [Google Scholar] [CrossRef] [PubMed]
- Gale, P.A. From anion receptors to transporters. Acc. Chem. Res. 2011, 44, 216–226. [Google Scholar] [CrossRef]
- He, Q.; Seung, G.I.V.; Kim, H.; Kim, S.K.; Sessler, J.L. Macrocycles as Ion Pair Receptors. Chem. Rev. 2019, 119, 9753–9835. [Google Scholar] [CrossRef]
- Kneeland, D.M.; Ariga, K.; Lynch, V.M.; Huang, C.Y.; Anslyn, E.V. Bis(Alkylguanidinium) Receptors for Phosphodiesters—Effect of Counterions, Solvent Mixtures, and Cavity Flexibility on Complexation. J. Am. Chem. Soc. 1993, 115, 10042–10055. [Google Scholar] [CrossRef]
- Shukla, R.; Kida, T.; Smith, B.D. Effect of competing alkali metal cations on neutral host’s anion binding ability. Org. Lett. 2000, 2, 3099–3102. [Google Scholar] [CrossRef]
- Arduini, A.; Brindani, E.; Giorgi, G.; Pochini, A.; Secchi, A. Anion effects on the recognition of ion pairs by calix[4]arene-based heteroditopic receptors. J. Org. Chem. 2002, 67, 6188–6194. [Google Scholar] [CrossRef]
- Bohmer, V.; Cort, A.D.; Mandolini, L. Counteranion effect on complexation of quats by a neutral calix[5]arene receptor. J. Org. Chem. 2001, 66, 1900–1902. [Google Scholar] [CrossRef]
- Berthault, P.; Desvaux, H.; Wendlinger, T.; Gyejacquot, M.; Stopin, A.; Brotin, T.; Dutasta, J.P.; Boulard, Y. Effect of pH and Counterions on the Encapsulation Properties of Xenon in Water Soluble Cryptophanes. Chem. Eur. J. 2010, 16, 12941–12946. [Google Scholar] [CrossRef]
- Kim, S.K.; Sessler, J.L. Ion pair receptors. Chem. Soc. Rev. 2010, 39, 3784–3809. [Google Scholar] [CrossRef] [PubMed]
- McConnell, A.J.; Beer, P.D. Heteroditopic receptors for ion-pair recognition. Angew. Chem. Int. Ed. 2012, 51, 5052–5061. [Google Scholar] [CrossRef] [PubMed]
- Zdanowski, S.; Romański, J. Ion pair binding by an L-tyrosine-based polymerizable molecular receptor. N. J. Chem. 2015, 39, 6216–6222. [Google Scholar] [CrossRef]
- Gale, P.A.; Busschaert, N.; Haynes, C.J.E.; Karagiannidis, L.E.; Kirby, I.L. Anion receptor chemistry: Highlights from 2011 and 2012. Chem. Soc. Rev. 2014, 43, 205–241. [Google Scholar] [CrossRef]
- Gale, P.A.; Caltagirone, G. Anion sensing by small molecules and molecular ensembles. Chem. Soc. Rev. 2015, 44, 4212–4227. [Google Scholar] [CrossRef]
- Martínez-Máñez, R.; Sancenón, F. Fluorogenic and Chromogenic Chemosensors and Reagents for Anions. Chem. Rev. 2003, 103, 4419–4476. [Google Scholar] [CrossRef]
- Pletnev, I. Macrocyclic Chemistry: Current Trends and Future Perspectives; Gloe, K., Ed.; Springer: Dordrecht, The Netherlands, 2005; p. 450. ISBN 101402033648. [Google Scholar]
- Kuswandi, B.; Nuriman, N.; Verboom, W.; Reinhoudt, D.N. Tripodal Receptors for Cation and Anion Sensors. Sensors 2006, 6, 978–1017. [Google Scholar] [CrossRef]
- Berocal, M.J.; Cruz, A.; Badr, I.H.A.; Bachas, L.G. Tripodal ionophore with sulphate recognition properties for anion-selective electrode. Anal. Chem. 2000, 72, 5295–5299. [Google Scholar] [CrossRef]
- Ravikumar, I.; Ghosh, P. Recognition and separation of sulfate anions. Chem. Soc. Rev. 2012, 41, 3077–3098. [Google Scholar] [CrossRef]
- Dey, S.K.; Basu, A.; Chutia, R.; Das, G. Anion coordinated capsules and pseudocapsules of tripodal amide, urea and thiurea scaffolds. RSC Adv. 2016, 6, 26568–26589. [Google Scholar] [CrossRef]
- Sato, K.; Arail, S.; Yamagishi, T.A. New tripodal anion receptor with C-H—X hydrogen bonding. Tetrahedron Lett. 1999, 40, 5219–5222. [Google Scholar] [CrossRef]
- Ballester, P.; Costa, A.; Deya, P.M.; Vega, M.; Morey, J. Influence of remote intramolecular hydrogen bonds on the thermodynamics of molecular recognition of cis-1,3,5-cyclohexanetricarboxylic acid. Tetrahedron Lett. 1999, 40, 171–174. [Google Scholar] [CrossRef]
- Fan, A.L.; Hong, H.K.; Valiyaveettil, S.; Vittal, J.J. A urea-incorporated receptor for aromatic carboxylate anion recognition. J. Supramol. Chem. 2002, 2, 247–254. [Google Scholar] [CrossRef]
- Schmuck, C.; Schwegmann, M. A Molecular Flytrap for the Selective Binding of Citrate and Other Tricarboxylates in Water. J. Am. Chem. Soc. 2005, 127, 3373–3379. [Google Scholar] [CrossRef]
- Li, Z.; Yu, X.-H.; Chen, Y.; Yuan, D.-Q.; Chen, W.-H. Synthesis, Anion Recognition, and Transmembrane Anionophoric Activity of Tripodal Diaminocholoyl Conjugates. J. Org. Chem. 2017, 82, 13368–13375. [Google Scholar] [CrossRef]
- Hennrich, G.; Anslyn, E.V. Novel C3-Symmetric Molecular Scaffolds with Potential Facial Differentiation. Chem. Eur. J. 2002, 8, 2218–2224. [Google Scholar] [CrossRef]
- Jonah, T.M.; Mathivathanan, L.; Morozov, A.N.; Mebel, A.M.; Raptis, R.G.; Kavallieratos, K. Remarkably selective NH4+ binding and fluorescence sensing by tripodal tris(pyrazolyl) receptors derived from 1,3,5-triethylbenzene: Structural and theoretical insights on the role of ion pairing. N. J. Chem. 2017, 41, 14835–14838. [Google Scholar] [CrossRef]
- Schulze, M.M.; Koch, N.; Seicheter, W.; Mazik, M. Crystalline ammonium complexes of Trimethyl- and triethylbenzene-based tripodal compounds bearing pyrazole and indazole groups. Eur. J. Org. Chem. 2018, 2018, 4317–4330. [Google Scholar] [CrossRef]
- Tomas, S.; Prohens, R.; Vega, M.; Rotger, M.C.; Deya, P.M.; Ballester, P.; Costa, A. Squaramido-based receptors: Design, synthesis, and application to the recognition of tetraalkylammonium compounds. J. Org. Chem. 1996, 61, 9394–9401. [Google Scholar] [CrossRef]
- Hao, J.; Hiratani, K.; Kameta, N.; Oba, T. Synthesis of a novel tripodal having 3-hydroxy-2-naphthoeic amide groups and its anion recognition ability. J. Incl. Phenom. Macrocycl. Chem. 2009, 65, 257–262. [Google Scholar] [CrossRef]
- Turner, D.R.; Paterson, M.J.; Steed, J.W. A conformationally flexible, urea-based tripodal anion receptor: Solid-state, solution, and theoretical studies. J. Org. Chem. 2006, 71, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, K.; Kitchen, J.A.; Blasco, S.; Boyle, E.M.; Fitzpatrick, B.; Feeney, M.; Kruger, P.E.; Gunnlaugsson, T. Unexpected self-sorting self-assembly formation of a [4:4] sulfate: Ligand cage from a preorganized tripodal urea ligand. Angew. Chem. Int. Ed. 2015, 54, 4566–4570. [Google Scholar] [CrossRef] [PubMed]
- Valkenier, H.; Dias, C.M.; Porter Goff, K.L.; Jureček, O.; Puttreddy, R.; Rissanen, K.; Davis, A.P. Sterically geared tris-thiureas; transmembrane chloride transporters with unusual activity and accessibility. Chem. Commun. 2015, 51, 14235–14238. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; Kim, J.H.; Kim, D.S.; Han, H.J.; Lynch, V.M.; Sessler, J.L.; Kim, S.K. Synthesis and Anion Recognition Features of a Molecular Cage Containing Both Hydrogen Bond Donors and Acceptors. Org. Lett. 2019, 21, 4336–4339. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-J.; Yoo, J.; Jeong, S.-D.; Lee, C.-H. Convenient synthesis of tripodal-pyrrole receptor and anion binding properties. J. Incl. Phenom. Macrocycl. Chem. 2010, 66, 209–212. [Google Scholar] [CrossRef]
- Bill, N.L.; Kim, D.S.; Kim, S.K.; Park, J.S.; Lynch, V.M.; Young, N.J.; Hay, B.P.; Yang, Y.; Anslyn, E.V.; Sessler, J.L. Oxoanion recognition by benzene-based tripodal pyrrolic receptors. Supramol. Chem. 2012, 24, 72–76. [Google Scholar] [CrossRef]
- Tomàs, S.; Prohens, R.; Deslongchamps, G.; Ballester, P.; Costa, A. An effective fluorescent sensor for choline-containing phospholipids. Angew. Chem. Int. Ed. 1999, 38, 2208–2211. [Google Scholar] [CrossRef]
- Soberats, B.; Martínez, L.; Sanna, E.; Sampedro, A.; Rotger, C.; Costa, A. Janus-like Squaramide-based hosts: Dual mode of binding and conformational transitions driven by ion-pair recognition hosts dual mode of binding and conformational transitions driven by ion-pair recognition. Chem. Eur. J. 2012, 18, 7533–7542. [Google Scholar] [CrossRef]
- Jain, A.; Jain, Y.; Gupta, R.; Agarwal, M. Trifluoromethyl group containing C3 symmetric coumarin-triazole based fluorometric tripodal receptors for selective fluoride ion recognition: A theoretical and experimental approach. J. Fluor. Chem. 2018, 212, 153–160. [Google Scholar] [CrossRef]
- Brynatsev, V.S.; Hay, B.P. Influence of substituents on the strength of aryl C-H···Anion hydrogen bonds. Org. Lett. 2005, 7, 5031–5034. [Google Scholar] [CrossRef]
- Esipenko, N.A.; Koutnik, P.; Minami, T.; Mosca, L.; Lynch, V.M.; Zyryanov, G.V.; Anzenbacher, P. First supramolecular sensors for phosphonate anions. Chem. Sci. 2013, 4, 3617–3623. [Google Scholar] [CrossRef]
- Minami, T.; Liu, Y.; Akdeniz, A.; Koutnik, P.; Esipenko, N.A.; Nishiyabu, R.; Kubo, Y.; Anzenbacher, P. Interamolecular indicator displacement assay for anions: Supramolecular sensor for glyphosate. J. Am. Chem. Soc. 2014, 136, 11396–11401. [Google Scholar] [CrossRef] [PubMed]
- Metzger, A.; Anslyn, E.V. A Chemosensor for Citrate in Beverages. Angew. Chem. Int. Ed. 1998, 37, 649–652. [Google Scholar] [CrossRef]
- Lee, M.; Zali-Boeini, H.; Li, F.; Lindoy, L.F.; Jolliffe, K.A. Synthesis of tris-(azacrown) ethers for carboxylic acid recognition. Tetrahedron 2013, 69, 38–42. [Google Scholar] [CrossRef]
- Turner, D.R.; Paterson, M.J.; Steed, J.W. Conformational control by ‘zipping-up’ an anion-binding unimolecular capsule. Chem. Commun. 2008, 1395–1397. [Google Scholar] [CrossRef]
- Arunachalam, M.; Ghosh, P. Recognition and complexation of hydrated fluoride anion: F2(H2O)62- templated formation of a dimeric capsule of a tripodal amide. Chem. Commun. 2009, 5389–5391. [Google Scholar] [CrossRef]
- Arunachalam, M.; Ghosh, P. Formation of a nitrate zipped dimeric capsule and un-zipping by chloride doping. Chem. Commun. 2009, 3184–3186. [Google Scholar] [CrossRef]
- Beer, P.D.; Hopkins, P.K.; McKinney, J.D. Cooperative halide, perrhenate anion-sodium cation binding and pertechnetate extraction and transport by a novel tripodal tris(amido benzo-15-crown-5) ligand. Chem. Commun. 1999, 1253–1254. [Google Scholar] [CrossRef]
- Marchetti, L.A.; Kumawat, L.K.; Mao, N.; Stephens, J.C.; Elmes, R.B.P. Versatility of Squaramides: From Supramolecular Chemistry to Chemical Biology. Chem 2019, 5, 1–88. [Google Scholar] [CrossRef]
- Wurm, F.R.; Klok, H.-A. Be squared: Expanding the Horizon of Squaric Acid-Mediated Conjugations. Chem. Soc. Rev. 2013, 42, 8220–8236. [Google Scholar] [CrossRef]
- Storer, R.I.; Aciro, C.; Jones, L.H. Squaramides: Physical Properties, Synthesis and Applications. Chem. Soc. Rev. 2011, 40, 2330–2346. [Google Scholar] [CrossRef] [PubMed]
- Jagleniec, D.; Siennicka, S.; Dobrzycki, Ł.; Karbarz, M.; Romański, J. Recognition and extraction of sodium chloride by a squaramide-based ion pair receptor. Inorg. Chem. 2018, 57, 12941–12952. [Google Scholar] [CrossRef] [PubMed]
- Rotger, M.C.; Piña, M.N.; Frontera, A.; Martorell, G.; Ballester, P.; Deya, P.M.; Costa, A. Conformational preferences and self-template macrocyclization of squaramide-based foldable modules. J. Org. Chem. 2004, 69, 2302–2308. [Google Scholar] [CrossRef] [PubMed]
- Rotger, C.; Soberats, B.; Quiñonero, D.; Frontera, A.; Ballester, P.; Benet-Buchholz, J.; Deya, P.M.; Costa, A. Crystallographic and theoretical evidence of anion–p and hydrogen-bonding interactions in a squaramide–nitrate salt. Eur. J. Org. Chem. 2008, 2008, 1864–1868. [Google Scholar] [CrossRef]
- López, C.; Ximenis, M.; Orvay, F.; Rotger, C.; Costa, A. Supramolecular hydrogels based on minimalist amphiphilic squaramide–squaramates for controlled release of zwitterionic biomolecules. Chem. Eur. J. 2017, 23, 7590–7594. [Google Scholar] [CrossRef]
- Martínez-Crespo, L.; Escudero-Adán, E.C.; Costa, A.; Rotger, C. The role of N-methyl squaramides in a hydrogenbonding strategy to fold peptidomimetic compounds. Chem. Eur. J. 2018, 24, 17802–17813. [Google Scholar] [CrossRef]
- Martínez, L.; Martorell, G.; Sampedro, A.; Ballester, P.; Costa, C.; Rotger, C. Hydrogen bonded squaramide-based foldable module induces both b- and a-turns in hairpin structures of a-peptides in water. Org. Lett. 2015, 17, 2980–2983. [Google Scholar] [CrossRef]
- Olmo, F.; Rotger, C.; Ramírez-Macías, I.; Martínez, L.; Marín, C.; Carreras, L.; Urbanová, K.; Vega, M.; Chaves-Lemaur, G.; Sampedro, A.; et al. Synthesis and biological evaluation of N,N’-squaramides with high in vivo efficacy and low toxicity: Toward a lowcost drug against Chagas disease. J. Med. Chem. 2014, 57, 987–999. [Google Scholar] [CrossRef]
- Amendola, V.; Bergamaschi, G.; Boiocchi, M.; Fabbrizzi, L.; Milani, M. The Squaramide versus Urea Contest for Anion Recognition. Chem. Eur. J. 2010, 16, 4368–4380. [Google Scholar] [CrossRef]
- Cai, X.-J.; Li, Z.; Chen, W.-H. Synthesis, Anion Recognition and Transmembrane Anion-Transport Properties of Squaramides and Their Derivatives. Mini-Rev. Org. Chem. 2018, 15, 148–156. [Google Scholar] [CrossRef]
- Gale, P.A.; Davis, J.T.; Quesada, R. Anion transport and supramolecular medicinal chemistry. Chem. Soc. Rev. 2017, 46, 2497–2519. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, X.; Busschaert, N.; Furuta, H.; Gale, P.A. Dissecting the Chloride-Nitrate Anion Transport Assay. Chem. Commun. 2017, 53, 9230–9233. [Google Scholar] [CrossRef] [PubMed]
- Busschaert, N.; Kirby, I.L.; Young, S.; Coles, S.J.; Horton, P.N.; Light, M.E.; Gale, P.A. Squaramides as Potent Transmembrane Anion Transporters. Angew. Chem. Int. Ed. 2012, 51, 4426–4430. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.-J.; Li, Z.; Chen, W.-H. Tripodal Squaramide Conjugates as Highly Effective Transmembrane Anion Transporters. Bioorganic Med. Chem. Lett. 2017, 27, 1999–2002. [Google Scholar] [CrossRef]
- Wu, X.; Judd, L.W.; Howe, E.N.W.; Withecombe, A.M.; Soto-Cerrato, V.; Li, H.; Busschaert, N.; Valkenier, H.; Pérez-Tomás, R.; Sheppard, D.N.; et al. Nonprotonophoric Electrogenic Cl− Transport Mediated by Valinomycin-like Carries. Chem 2016, 1, 127–146. [Google Scholar] [CrossRef]
- Busschaert, N.; Park, S.-H.; Baek, K.-H.; Choi, Y.P.; Park, J.; Howe, E.N.W.; Hiscock, J.R.; Karagiannidis, L.E.; Marques, I.; Félix, V.; et al. A Synthetic Ion Transporter that Disrupts Autophagy and Induces Apoptosis by Perturbing Cellular Chloride Concentrations. Nat. Chem. 2017, 9, 667–675. [Google Scholar] [CrossRef]
- Elmes, R.B.P.; Yuen, K.; Jolliffe, K.A. Sulfate-Selective Recognition by Using Neutral Dipeptide Anion Receptors in Aqueous Solution. Chem. Eur. J. 2014, 20, 7373–7380. [Google Scholar] [CrossRef]
- Qin, L.; Hartley, A.; Turner, P.; Elmes, R.B.P.; Jolliffe, K.A. Macrocyclic squaramides: Anion receptors with high sulfate binding affinity and selectivity in aqueous media. Chem. Sci. 2016, 7, 4563–4572. [Google Scholar] [CrossRef]
- Frontera, A.; Orell, M.; Garau, C.; Quiñonero, D.; Molins, E.; Mata, I.; Morey, J. Preparation, Solid-State characterization, and Computational Study of a Crown Ether Attached to a Squaramide. Org. Lett. 2005, 7, 1437–1440. [Google Scholar] [CrossRef]
- Załubiniak, D.; Zakrzewski, M.; Piątek, P. Highly Effective Ion-Pair Receptors Based on 2,2-Bis(aminomethyl)-propionic Acid. Dalton Trans. 2016, 45, 15557–15564. [Google Scholar] [CrossRef]
- Zdanowski, S.; Piątek, P.; Romański, J. An Ion Pair Receptor Facilitating the Extraction of Chloride Salt from the Aqueous to the Organic Phase. N. J. Chem. 2016, 40, 7190–7196. [Google Scholar] [CrossRef]
- Zaleskaya, M.; Jagleniec, D.; Karbarz, M.; Dobrzycki, Ł.; Romański, J. Squaramide based ion pair receptors possessing ferrocene as a signalling unit. Inorg. Chem. Front. 2020, 7, 972–983. [Google Scholar] [CrossRef]
- Zaleskaya, M.; Karbarz, M.; Wilczek, M.; Dobrzycki, Ł.; Romański, J. Cooperative Transport and Selective Extraction of Sulfates by a Squaramide-Based Ion Pair Receptor: A Case of Adaptable Selectivity. Inorg. Chem. 2020, 59, 13749–13759. [Google Scholar] [CrossRef] [PubMed]
- Jagleniec, D.; Dobrzycki, Ł.; Karbarz, M.; Romański, J. Ion-pair induced supramolecular assembly formation for elective extraction and sensing of potassium sulfate. Chem. Sci. 2019, 10, 9542–9547. [Google Scholar] [CrossRef]
- Cabell, L.A.; Best, M.D.; Lavigne, J.J.; Schneider, S.E.; Perreault, D.M.; Monahan, M.-K.; Anslyn, E.V. Metal triggered fluorescence sensing of citrate using a synthetic receptor. J. Chem. Soc. Perkin Trans. 2001, 3, 315–323. [Google Scholar] [CrossRef]
- Capitán-Vallvey, L.F.; Arroyo-Guerrero, E.; Fernández-Ramos, M.D.; Santoyo-Gonzalez, F. Disposable Receptor-Based Optical Sensor for Nitrate. Anal. Chem. 2005, 77, 4459–4466. [Google Scholar] [CrossRef]
- APEX3 V2019; Bruker AXS, Inc.: Madison, WI, USA, 2019.
- SAINT V8.40A; Bruker AXS, Inc.: Madison, WI, USA, 2019.
- TWINABS V2012/1; Bruker AXS, Inc.: Madison, WI, USA, 2019.
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Cowley, J.M. International Tables for Crystallography. Volume C: Mathematical, Physical and Chemical Tables; Wilson, A.J.C., Ed.; Kluwer: Dordrecht, The Netherlands, 1992; pp. 223–245. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, P.A. Wood Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef]

| 1 | 1 + 3 Equivalent K+ | KK+/KTBA- | |
|---|---|---|---|
| Cl− | 283 | 373 | 1.32 |
| Br− | 40 | 100 | 2.50 |
| NO2− | 59 | 101 | 1.71 |
| NO3− (b) | − | − | - |
| PhCOO− | 415 | 500 | 1.20 |
| CH3COO− | 400 | 453 | 1.13 |
| SO42− (c) | − | - | - |
| H2PO4− (c) | - | - | - |
| 1 | 1 + 3 equiv. K+ | KK+/KTBA− | |
|---|---|---|---|
| Cl− | 3.68 × 104 | 4.60 × 104 | 1.25 |
| Br− | 2.27 × 104 | 3.61 × 104 | 1.60 |
| NO2− | 2.16 × 104 | 3.55 × 104 | 1.65 |
| NO3− | 5.26 × 103 | 7.84 × 103 | 1.49 |
| PhCOO− | 8.40 × 104 | 9.02 × 104 | 1.07 |
| CH3COO− | 6.14 × 104 | 7.59 × 104 | 1.24 |
| SO42− (b) | - | - | - |
| H2PO4− (b) | - | - | - |
| Formula | C102.01H140.02F6.91N7.59Na2.30O39.28, corresponding to approx.: 1 + 2.30 × (sodium trifluoroacetate) + 4.61 × (ethyl acetate) + 1.58 × acetonitrile + 0.80 × methanol * + 0.65 × H2O* |
|---|---|
| Mx/g∙mol−1 | 2288.94 |
| T/K | 130.0(5) |
| λ/Å | 0.71073 |
| Crystal size | 0.086 × 0.307 × 0.569 |
| Space group | P1 |
| Unit cell dimensions | a = 13.9030(10) Å α = 106.491(2)° b = 19.1752(13) Å β = 98.390(2)° c = 24.2017(16) Å γ = 105.570(2)° |
| V/Å3, Z | 5784.7(7), 2 |
| Dx/g∙cm−3 | 1.312 |
| μ/mm−1 | 0.114 |
| F(000) | 2414 |
| θmin, θmax | 2.86°, 25.05° |
| Index ranges (merged data) | −16 ≤ h ≤ 16, −23 ≤ k ≤ 22, 0 ≤ l ≤ 29 |
| Reflections collected/independent | 208133/ 21096 ** Rint = 0.0674 ** |
| Completeness | 99.1% |
| Absorption correction | Multi-Scan |
| Tmax, Tmin | 0.990, 0.938 |
| Structure solution technique | direct methods |
| Refinement method | Full-matrix LSQ on F2 |
| Data/restraints/parameters | 20322/603/1807 |
| GOF on F2 | 1.051 |
| Final R indices | 13630 data; I > 2σ(I) R1 = 0.0699, wR2 = 0.1894 all data R1 = 0.1085, wR2 = 0.2084 |
| Extinction coefficient | 0.0013(4) |
| Δρmax, Δρmin | 0.397, −0.396 e∙Å−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaleskaya, M.; Dobrzycki, Ł.; Romański, J. Highly Efficient, Tripodal Ion-Pair Receptors for Switching Selectivity between Acetates and Sulfates Using Solid–Liquid and Liquid–Liquid Extractions. Int. J. Mol. Sci. 2020, 21, 9465. https://doi.org/10.3390/ijms21249465
Zaleskaya M, Dobrzycki Ł, Romański J. Highly Efficient, Tripodal Ion-Pair Receptors for Switching Selectivity between Acetates and Sulfates Using Solid–Liquid and Liquid–Liquid Extractions. International Journal of Molecular Sciences. 2020; 21(24):9465. https://doi.org/10.3390/ijms21249465
Chicago/Turabian StyleZaleskaya, Marta, Łukasz Dobrzycki, and Jan Romański. 2020. "Highly Efficient, Tripodal Ion-Pair Receptors for Switching Selectivity between Acetates and Sulfates Using Solid–Liquid and Liquid–Liquid Extractions" International Journal of Molecular Sciences 21, no. 24: 9465. https://doi.org/10.3390/ijms21249465
APA StyleZaleskaya, M., Dobrzycki, Ł., & Romański, J. (2020). Highly Efficient, Tripodal Ion-Pair Receptors for Switching Selectivity between Acetates and Sulfates Using Solid–Liquid and Liquid–Liquid Extractions. International Journal of Molecular Sciences, 21(24), 9465. https://doi.org/10.3390/ijms21249465





