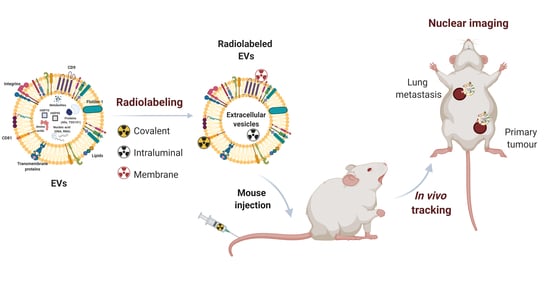
In Vivo Tracking of Extracellular Vesicles by Nuclear Imaging: Advances in Radiolabeling Strategies
Abstract
1. Introduction
2. Imaging Modalities for In Vivo Tracking of EVs
3. Radiolabeling Methods for Extracellular Vesicles
3.1. Covalent Binding
3.2. Encapsulation or Intraluminal Radiolabeling
3.3. Membrane Radiolabeling
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [PubMed]
- Jabalee, J.; Towle, R.; Garnis, C. The role of extracellular vesicles in cancer: Cargo, function, and therapeutic implications. Cells 2018, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef]
- Simeone, P.; Bologna, G.; Lanuti, P.; Pierdomenico, L.; Guagnano, M.T.; Pieragostino, D.; Del Boccio, P.; Vergara, D.; Marchisio, M.; Miscia, S. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020, 21, 2514. [Google Scholar] [CrossRef]
- Jurj, A.; Zanoaga, O.; Braicu, C.; Lazar, V.; Tomuleasa, C.; Irimie, A.; Berindan-Neagoe, I. A comprehensive picture of extracellular vesicles and their contents. Molecular Transfer to Cancer Cells. Cancers 2020, 12, 298. [Google Scholar] [CrossRef]
- Yang, B.; Chen, Y.; Shi, J. Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv. Mater. 2019, 31, 1802896. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.; Korbie, D.; Hill, M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.P.; Holme, M.N.; Stevens, M.M. Re-engineering extracellular vesicles as smart nanoscale therapeutics. Acs Nano 2017, 11, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.-M.; Quan, L.; Liu, J. Tracking exosomes in vitro and in vivo to elucidate their physiological functions: Implications for diagnostic and therapeutic nanocarriers. Acs Appl. Nano Mater. 2018, 1, 2438–2448. [Google Scholar] [CrossRef]
- Lorenc, T.; Chrzanowski, J.; Olejarz, W. Current Perspectives on Clinical Use of Exosomes as a Personalized Contrast Media and Theranostics. Cancers 2020, 12, 3386. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef]
- Zheng, Y.; Hasan, A.; Babadaei, M.M.N.; Behzadi, E.; Nouri, M.; Sharifi, M.; Falahati, M. Exosomes: Multiple-targeted multifunctional biological nanoparticles in the diagnosis, drug delivery, and imaging of cancer cells. Biomed. Pharmacother. 2020, 129, 110442. [Google Scholar] [CrossRef]
- Di Rocco, G.; Baldari, S.; Toietta, G. Towards Therapeutic Delivery of Extracellular Vesicles: Strategies for. Stem Cells Int. 2016. [Google Scholar] [CrossRef]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Pharmacokinetics of exosomes—an important factor for elucidating the biological roles of exosomes and for the development of exosome-based therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Adem, B.; Melo, S.A. Animal models in exosomes research: What the future holds. Nov. Implic. Exosomes Diagn. Treat. Cancer Infect. Dis. 2017, 53. [Google Scholar] [CrossRef]
- Wu, M.; Shu, J. Multimodal molecular imaging: Current status and future directions. Contrast Media Mol. Imaging 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kothandan, V.K.; Kim, H.W.; Kim, K.S.; Kim, J.Y.; Cho, H.J.; Lee, Y.-k.; Lee, D.-E.; Hwang, S.R. Noninvasive Assessment of Exosome Pharmacokinetics In Vivo: A Review. Pharmaceutics 2019, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Chuo, S.T.-Y.; Chien, J.C.-Y.; Lai, C.P.-K. Imaging extracellular vesicles: Current and emerging methods. J. Biomed. Sci. 2018, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Hong, C.M.; Ahn, B.-C. Current perspectives on in vivo noninvasive tracking of extracellular vesicles with molecular imaging. Biomed. Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Rhim, W.-K.; Kim, M.; Hartman, K.L.; Kang, K.W.; Nam, J.-M. Radionuclide-labeled nanostructures for in vivo imaging of cancer. Nano Converg. 2015, 2, 1–13. [Google Scholar] [CrossRef][Green Version]
- Betzer, O.; Barnoy, E.; Sadan, T.; Elbaz, I.; Braverman, C.; Liu, Z.; Popovtzer, R. Advances in imaging strategies for in vivo tracking of exosomes. Wiley Interdiscip. Rev. Nanomed. NanoBiotechnol. 2020, 12, e1594. [Google Scholar] [CrossRef]
- Kourembanas, S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015, 77, 13–27. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Serpooshan, V.; Laurent, S. Engineered nanoparticles for biomolecular imaging. Nanoscale 2011, 3, 3007–3026. [Google Scholar] [CrossRef]
- Youn, H.; Hong, K.-J. In vivo non invasive molecular imaging for immune cell tracking in small animals. Immune Netw. 2012, 12, 223–229. [Google Scholar] [CrossRef]
- Yi, Y.W.; Lee, J.H.; Kim, S.-Y.; Pack, C.-G.; Ha, D.H.; Park, S.R.; Youn, J.; Cho, B.S. Advances in analysis of biodistribution of exosomes by molecular imaging. Int. J. Mol. Sci. 2020, 21, 665. [Google Scholar] [CrossRef]
- Kim, J.E.; Kalimuthu, S.; Ahn, B.-C. In vivo cell tracking with bioluminescence imaging. Nucl. Med. Mol. Imaging 2015, 49, 3–10. [Google Scholar] [CrossRef]
- Li, Y.-J.; Wu, J.-Y.; Wang, J.-M.; Hu, X.-B.; Xiang, D.-X. Emerging strategies for labeling and tracking of extracellular vesicles. J. Control. Release 2020, 328, 141–159. [Google Scholar] [CrossRef]
- Gangadaran, P.; Hong, C.M.; Ahn, B.-C. An update on in vivo imaging of extracellular vesicles as drug delivery vehicles. Front. Pharmacol. 2018, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; C Yang, P. Iron Oxide Labeling and Tracking of Extracellular Vesicles. Magnetochemistry 2019, 5, 60. [Google Scholar] [CrossRef]
- Busato, A.; Bonafede, R.; Bontempi, P.; Scambi, I.; Schiaffino, L.; Benati, D.; Malatesta, M.; Sbarbati, A.; Marzola, P.; Mariotti, R. Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: A new method to obtain labeled exosomes. Int. J. Nanomed. 2016, 11, 2481. [Google Scholar]
- Cassidy, P.J.; Radda, G.K. Molecular imaging perspectives. J. R. Soc. Interface 2005, 2, 133–144. [Google Scholar] [CrossRef]
- Kim, J.; Lee, N.; Hyeon, T. Recent development of nanoparticles for molecular imaging. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2017, 375, 20170022. [Google Scholar] [CrossRef] [PubMed]
- Mirshojaei, S.F.; Ahmadi, A.; Morales-Avila, E.; Ortiz-Reynoso, M.; Reyes-Perez, H. Radiolabelled nanoparticles: Novel classification of radiopharmaceuticals for molecular imaging of cancer. J. Drug Target. 2016, 24, 91–101. [Google Scholar] [CrossRef]
- Gambhir, S.S. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer 2002, 2, 683–693. [Google Scholar] [CrossRef]
- Dobrucki, L.W.; Sinusas, A.J. PET and SPECT in cardiovascular molecular imaging. Nat. Rev. Cardiol. 2010, 7, 38. [Google Scholar] [CrossRef]
- Gomes, C.M.; Abrunhosa, A.J.; Ramos, P.; Pauwels, E.K. Molecular imaging with SPECT as a tool for drug development. Adv. Drug Deliv. Rev. 2011, 63, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Cherry, S.R. Fundamentals of positron emission tomography and applications in preclinical drug development. J. Clin. Pharmacol. 2001, 41, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Stéen, E.J.L.; Edem, P.E.; Nørregaard, K.; Jørgensen, J.T.; Shalgunov, V.; Kjaer, A.; Herth, M.M. Pretargeting in nuclear imaging and radionuclide therapy: Improving efficacy of theranostics and nanomedicines. Biomaterials 2018, 179, 209–245. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Sano, K.; Kato, K.; Yamashita, T.; Imai, T.; Saji, H.; Takakura, Y. Quantitative analysis of tissue distribution of the B16BL6-derived exosomes using a streptavidin-lactadherin fusion protein and iodine-125-labeled biotin derivative after intravenous injection in mice. J. Pharm. Sci. 2015, 104, 705–713. [Google Scholar] [CrossRef]
- Varga, Z.; Gyurkó, I.; Pálóczi, K.; Buzás, E.I.; Horváth, I.; Hegedűs, N.; Máthé, D.; Szigeti, K. Radiolabeling of extracellular vesicles with 99mTc for quantitative in vivo imaging studies. Cancer Biother. Radiopharm. 2016, 31, 168–173. [Google Scholar] [CrossRef]
- Royo, F.; Cossío, U.; de Angulo, A.R.; Llop, J.; Falcon-Perez, J.M. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale 2019, 11, 1531–1537. [Google Scholar] [CrossRef]
- Rashid, M.H.; Borin, T.F.; Ara, R.; Angara, K.; Cai, J.; Achyut, B.R.; Liu, Y.; Arbab, A.S. Differential in vivo biodistribution of 131I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102072. [Google Scholar] [CrossRef]
- González, M.I.; Martín-Duque, P.; Desco, M.; Salinas, B. Radioactive Labeling of Milk-Derived Exosomes with 99mTc and In Vivo Tracking by SPECT Imaging. Nanomaterials 2020, 10, 1062. [Google Scholar] [CrossRef]
- Hwang, D.W.; Choi, H.; Jang, S.C.; Yoo, M.Y.; Park, J.Y.; Choi, N.E.; Oh, H.J.; Ha, S.; Lee, Y.-S.; Jeong, J.M.; et al. Noninvasive imaging of radiolabeled exosome-mimetic nanovesicle using 99m Tc-HMPAO. Sci. Rep. 2015, 5, 15636. [Google Scholar] [CrossRef]
- Lynch, S.; Santos, S.G.; Campbell, E.C.; Nimmo, A.M.; Botting, C.; Prescott, A.; Antoniou, A.N.; Powis, S.J. Novel MHC class I structures on exosomes. J. Immunol. 2009, 183, 1884–1891. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Faruqu, F.N.; Wang, J.T.-W.; Xu, L.; McNickle, L.; Chong, E.M.-Y.; Walters, A.; Gurney, M.; Clayton, A.; Smyth, L.A.; Hider, R. Membrane Radiolabelling of Exosomes for Comparative Biodistribution Analysis in Immunocompetent and Immunodeficient Mice-A Novel and Universal Approach. Theranostics 2019, 9, 1666. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Hong, C.M.; Oh, J.M.; Rajendran, R.L.; Kalimuthu, S.; Son, S.H.; Gopal, A.; Zhu, L.; Baek, S.H.; Jeong, S.Y. In vivo non-invasive imaging of radio-labeled exosome-mimetics derived from red blood cells in mice. Front. Pharmacol. 2018, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Brechbiel, M.W. Bifunctional chelates for metal nuclides. Q. J. Nucl. Med. Mol. Imaging 2008, 52, 166–173. [Google Scholar]
- Choi, H. Endogenous Radionanomedicine: Radiolabeling. In Radionanomedicine; Springer: New York, NY, USA, 2018; pp. 141–152. [Google Scholar]
- Salunkhe, S.; Dheeraj; Basak, M.; Chitkara, D.; Mittal, A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J. Control. Release 2020, 326, 599–614. [Google Scholar]
- Banerjee, A.; Alves, V.; Rondão, T.; Sereno, J.; Neves, Â.; Lino, M.; Ribeiro, A.; Abrunhosa, A.J.; Ferreira, L.S. A positron-emission tomography (PET)/magnetic resonance imaging (MRI) platform to track in vivo small extracellular vesicles. Nanoscale 2019, 11, 13243–13248. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Li, T.; Wen, X.; Wu, S.Y.; Xiong, C.; Zhao, J.; Lincha, V.R.; Chow, D.S.; Liu, Y.; Sood, A.K. Copper-64 labeled PEGylated exosomes for in vivo positron emission tomography and enhanced tumor retention. Bioconjugate Chem. 2019, 30, 2675–2683. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.O.; Kim, Y.-H.; Chung, S.-J.; Lee, C.-H.; Rhee, S.; Pratx, G.; Chung, J.-K.; Youn, H. Identification of Lymphatic and Hematogenous Routes of Rapidly Labeled Radioactive and Fluorescent Exosomes through Highly Sensitive Multimodal Imaging. Int. J. Mol. Sci. 2020, 21, 7850. [Google Scholar] [CrossRef] [PubMed]

| Imaging Modality | Detection | Spatial Resolution | Depth Penetration | Temporal Resolution | Sensitivity (M) | Examples |
|---|---|---|---|---|---|---|
| FLI and BLI | visible light | 2–5 µm | 1–2 cm | s to min | 10−9–10−12 | GFP, Luciferase |
| MRI | radiofrequency | 25–100 µm | no limit | min to h | 10−3–10−5 | SPION |
| CT | X-rays | 50–200 µm | no limit | min | - | Gold nanoparticle |
| PET | pairs of γ-rays (511 keV) | 2–4 mm | no limit | 10 s to min | 10−11–10−12 | 64Cu-DOTA |
| SPECT | single γ-ray (140 keV) | 4–6 mm | no limit | min | 10−10–10−11 | 99mTc-HMPAO |
| Labeling Method | Nuclear Imaging | Radionuclide | EVs (Markers) | Origin | Mouse Strain | Dose | Admin. Route | In Vitro Stability | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Covalent binding | - | 125I-IBB | Exosomes | B16BL6 | BALB/c | 37 KBq (4 µg) | I.V. | PBS 20% FBS | [44] |
| SPECT/CT | 99mTc-tricarbonyl complex | EVs | RBCs | BALB/c | 15 ± 2 MBq | I.V. | - | [45] | |
| PET | [124I]NaI | EVs | MLP29 | BALB/cJRj | 1.8 ± 0.5 MBq (120 ng) 0.6 ± 0.2 MBq (40 ng) | I.V. Hock | NaCl | [46] | |
| SPECT/CT | 131I | Exosomes (CD9, CD63) | 4T1; AT3 | BALB/c, C57BL/J6 | 350 ± 50 µCi | I.V. | 20% FBS | [47] | |
| SPECT/CT | 99mTc | Exosomes | Goat milk | BALB/c | 140–170 µCi (12 µg) 190–340 µCi (19 µg) 310–350 µCi (18 µg) | I.N. I.P. I.V. | PBS | [48] | |
| Encapsulation | SPECT/CT | 99mTc-HMPAO | ENVs (CD63) | Raw 264.7 | BALB/c | 7.4–14.8 MBq (29–64 µg) | I.V. | Serum or PBS | [49] |
| - | 111In-oxine | Exosomes (HSP 70, 90, 27; CD9) | MCF-7 PC3 | Nude Nu/J | 7.2 µCi (32 µg) 7.6 µCi (30 µg) | I.V. | - | [51] | |
| SPECT/CT | 111In via tropolone | Exosomes (CD81, CD9) | B16F10 | C57BL/6; NSG | 5–10 MBq (1 × 1011 part.) | I.V. | 50% FBS or PBS | [52] | |
| Gamma camera | 99mTc | Exosome mimetics | RBCs | C57BL/6 | 37 MBq | I.V. | PBS 20% FBS | [53] | |
| Membrane radiolabeling | SPECT/CT | 111In-DTPA | Exosomes (CD81, CD9) | B16F10 | C57BL/6; NSG | 5–10 MBq (1 × 1011 part.) | I.V. | 50% FBS or PBS | [52] |
| PET/MRI | 64Cu-DOTA | SEVs (CD9, CD63, CD45) | hUCB-MNCs | C57BL/6J | 100–150 µCi (2.5–3.5 × 1010 part.) | I.V. | PBS, serum | [57] | |
| PET | 64Cu-NOTA-PEG | Exosomes | 4T1 | BALB/c | 50 µCi | I.V. | PBS or 25% mouse serum | [58] | |
| PET | 64Cu (or 68Ga)-NOTA | Exosomes | 4T1 | BALB/c nu/nu | 0.74 MBq | I.V. S.C. | 10% exo-free FBS | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, S.; Santos, L.; Falcão, A.; Gomes, C.; Abrunhosa, A. In Vivo Tracking of Extracellular Vesicles by Nuclear Imaging: Advances in Radiolabeling Strategies. Int. J. Mol. Sci. 2020, 21, 9443. https://doi.org/10.3390/ijms21249443
Almeida S, Santos L, Falcão A, Gomes C, Abrunhosa A. In Vivo Tracking of Extracellular Vesicles by Nuclear Imaging: Advances in Radiolabeling Strategies. International Journal of Molecular Sciences. 2020; 21(24):9443. https://doi.org/10.3390/ijms21249443
Chicago/Turabian StyleAlmeida, Sara, Liliana Santos, Amílcar Falcão, Célia Gomes, and Antero Abrunhosa. 2020. "In Vivo Tracking of Extracellular Vesicles by Nuclear Imaging: Advances in Radiolabeling Strategies" International Journal of Molecular Sciences 21, no. 24: 9443. https://doi.org/10.3390/ijms21249443
APA StyleAlmeida, S., Santos, L., Falcão, A., Gomes, C., & Abrunhosa, A. (2020). In Vivo Tracking of Extracellular Vesicles by Nuclear Imaging: Advances in Radiolabeling Strategies. International Journal of Molecular Sciences, 21(24), 9443. https://doi.org/10.3390/ijms21249443