Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]
Abstract
1. Introduction
2. Results
2.1. Overall Transcriptome Profiling and Mapping Statistics
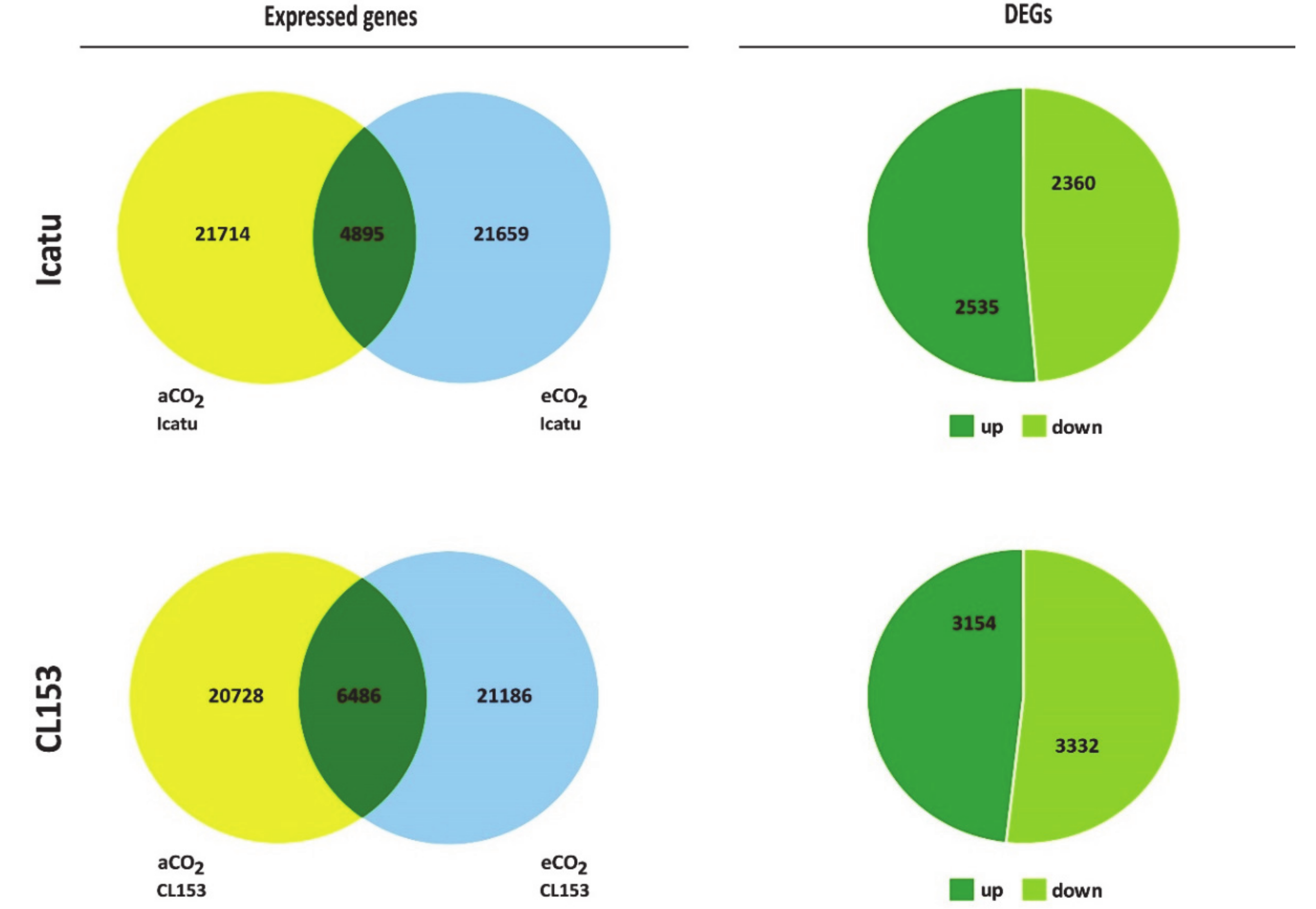
2.2. Differential Gene Expression in Response to eCO2
2.3. Major CO2-Responsive DEGs in the Two Genotypes
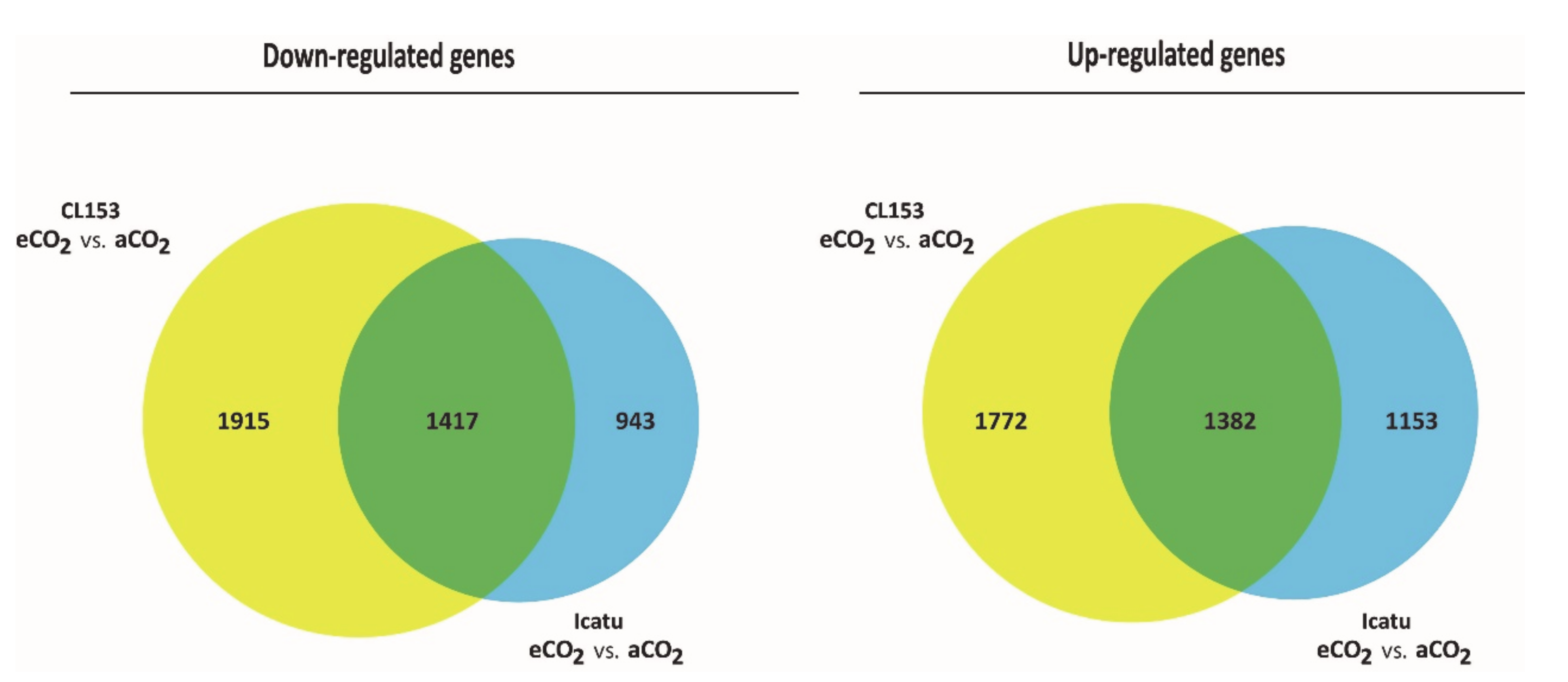
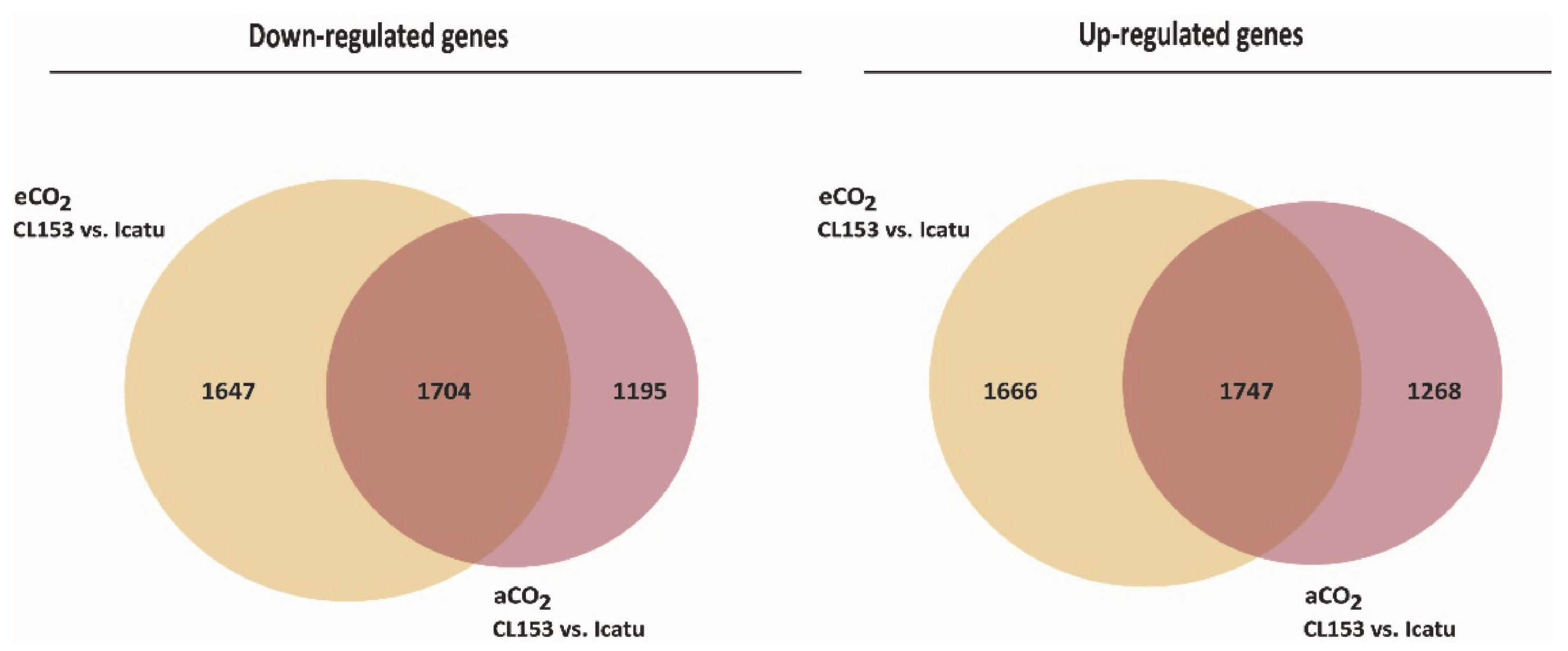
2.4. Differential Gene Expression Responses between Icatu and CL153
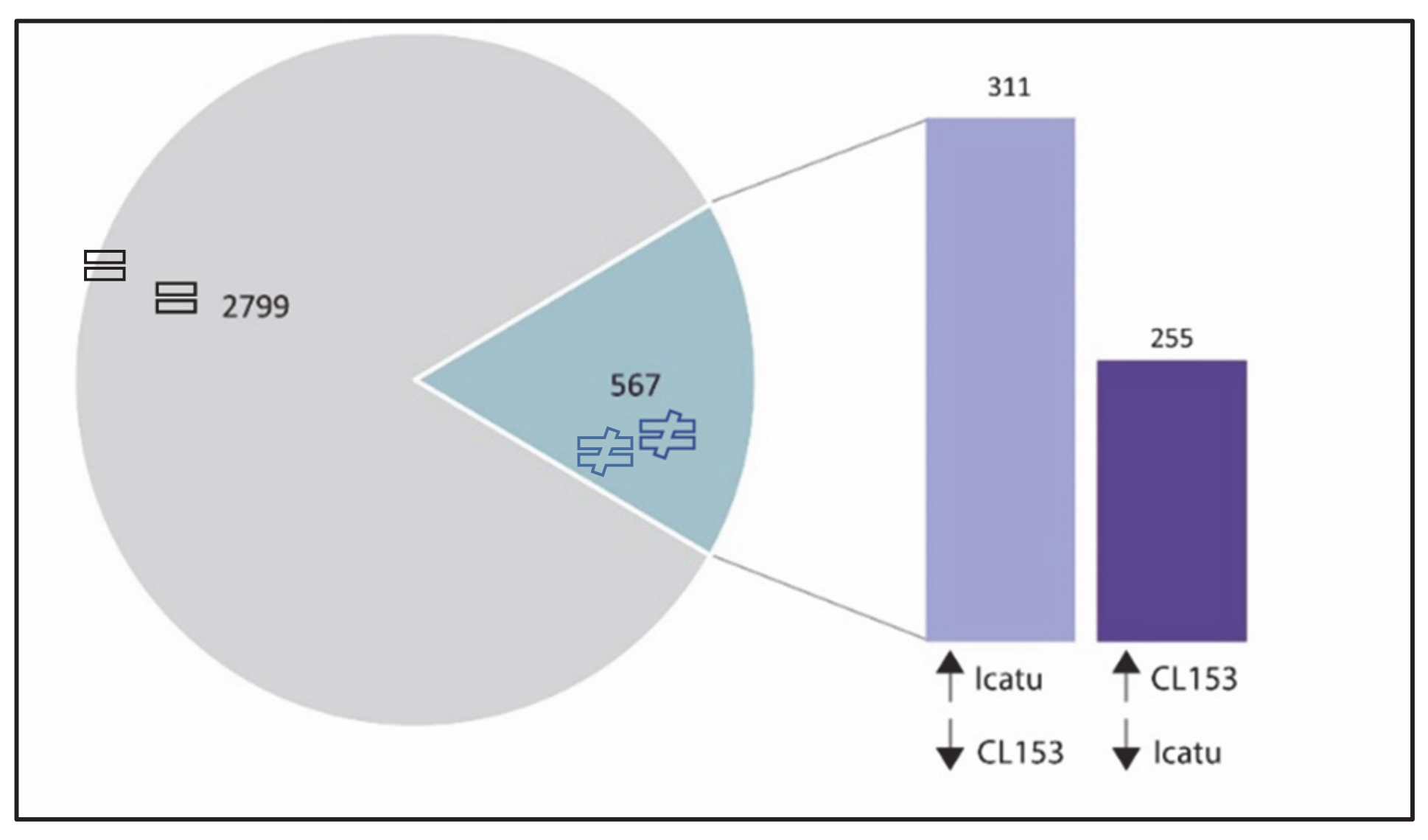
2.5. Common DEGs to Both Genotypes but with Opposite Patterns of Expression to eCO2
2.6. Significantly Enriched GO Categories of the Two Genotypes in Response to eCO2
2.7. Significant Metabolic Pathways
2.8. Identification of Photosynthetic and Other Biochemical-Related Responsive DEGs
3. Discussion
3.1. Transcriptomic Responses of Icatu to eCO2
3.2. Transcriptomic Responses of CL153 to eCO2
3.3. Main Transcriptomic Differences between the Two Genotypes
3.4. Transcriptomic Responses Associated to Physiological and Biochemical Behavior of Coffee
3.5. Adapting Coffee Crop for eCO2: Links to Future Research
3.6. Conclusions
4. Materials and Methods
4.1. Plant Material and Experimental Design
4.2. Total RNA, Poly(A) RNA Isolation, and Library Preparation
4.3. Processing and Mapping of Illumina Reads
4.4. Identification of Differentially Expressed Genes
4.5. Functional Annotation and Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DaMatta, F.M.; Avila, R.T.; Cardoso, A.A.; Martins, S.C.V.; Ramalho, J.C. Physiological and agronomic performance of the coffee crop in the context of climate change and global warming: A review. J. Agric. Food Chem. 2018, 66, 5264–5274. [Google Scholar] [CrossRef] [PubMed]
- Osorio, N. The Global Coffee Crisis: A Threat to Sustainable Development; International Coffee Organization: London, UK, 2002; p. 4. [Google Scholar]
- Semedo, J.N.; Rodrigues, W.P.; Dubberstein, D.; Martins, M.Q.; Martins, L.D.; Pais, I.P.; Rodrigues, A.P.; Leitão, A.E.; Partelli, F.L.; Campostrini, E.; et al. Coffee Responses to Drought, Warming and High [CO2] in a Context of Future Climate Change Scenarios. In Theory and Practice of Climate Adaptation; Climate Change Management Series; Alves, F., Leal, W., Azeiteiro, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 465–477. ISBN 978-3-319-72874-2. [Google Scholar] [CrossRef]
- Lashermes, P.; Combes, M.C.; Robert, J. Molecular characterisation and origin of the Coffea arabica L. Genome. Mol. Gen. Genet. 1999, 261, 259–266. [Google Scholar] [CrossRef]
- Tesfaye, K.; Borsch, T.; Govers, K.; Bekele, E. Characterization of Coffea chloroplast microsatellites and evidence for the recent divergence of C. arabica and C. eugenioides chloroplast genomes. Genome 2007, 50, 1112–1129. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Combes, M.C.; Lashermes, P. Genome evolution in diploid and tetraploid Coffea species as revealed by comparative analysis of orthologous genome segments. Plant Mol. Biol. 2012, 78, 135–145. [Google Scholar] [CrossRef]
- Combes, M.-C.; Dereeper, A.; Severac, D.; Bertrand, B.; Lashermes, P. Contribution of subgenomes to the transcriptome and their intertwined regulation in the allopolyploid Coffea arabica grown at contrasted temperatures. New Phytol. 2013, 200, 251–260. [Google Scholar] [CrossRef]
- Scalabrin, S.; Toniutti, L.; Di Gaspero, G.; Scaglione, D.; Magris, G.; Vidotto, M.; Pinosio, S.; Cattonaro, F.; Magni, F.; Jurman, I.; et al. A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci. Rep. 2020, 10, 4642. [Google Scholar] [CrossRef]
- Stocker, T.F.; Qin, D.; Plattner, G.-K.; Tignor, M.M.B.; Allen, S.K.; Boschung, J.; Nauels, A.; Xia, Y.; Bex, V.; Midgley, P.M. Climate Change 2013. Physical Science Basis; Summary for Policymakers, Technical Summary and Frequent Asked Questions. Part of the Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2013; p. 203. ISBN 978-92-9169-138-8. [Google Scholar]
- IPCC. Climate Change 2014. In Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Edenhofer, O., Pichs-Madruga, R., Sokona, Y., Farahani, E., Kadner, S., Seyboth, K., Adler, A., Baum, I., Brunner, S., Eickemeier, P., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014; p. 1435. ISBN 978-1-107-65481-5. [Google Scholar]
- Tubiello, F.N.; Soussana, J.-F.; Howden, M. Crop and pasture response to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19686–19690. [Google Scholar] [CrossRef]
- Wassmann, R.; Nelson, G.C.; Peng, S.B.; Sumfleth, K.; Jagadish, S.V.K.; Hosen, Y.; Rosegrant, M.W. Rice and global climate change. In Rice in the Global Economy: Strategic Research and Policy Issues for Food Security; Pandey, S., Byerlee, D., Dawe, D., Dobermann, A., Mohanty, S., Rozelle, S., Hardy, B., Eds.; International Rice Research Institute: Manila, Philippines, 2010; pp. 411–432. [Google Scholar]
- Ghini, R.; Torre-Neto, A.; Dentzien, A.F.M.; Guerreiro-Filho, O.; Iost, R.; Patrício, F.R.A.; Prado, J.S.M.; Thomaziello, R.A.; Bettiol, W.; DaMatta, F.M. Coffee growth, pest and yield responses to free air CO2 enrichment. Clim. Chang. 2015, 132, 307–320. [Google Scholar] [CrossRef]
- Magrach, A.; Ghazoul, J. Climate and pest-driven geographic shifts in global coffee production: Implications for forest cover, biodiversity and carbon storage. PLoS ONE 2015, 10, e0133071. [Google Scholar] [CrossRef]
- Polley, H.W. Implications of atmospheric and climate change for crop yield. Crop Sci. 2002, 42, 131–140. [Google Scholar] [CrossRef]
- Long, S.P.; Ainsworth, E.A.; Rogers, A.; Ort, D.R. Rising atmospheric carbon dioxide: Plants FACE the future. Annu. Rev. Plant Biol. 2004, 55, 591–628. [Google Scholar] [CrossRef]
- Kirschbaum, M.U.F. Does enhanced photosynthesis enhance growth? Lessons learned from CO2 enrichment studies. Plant Physiol. 2011, 155, 117–124. [Google Scholar] [CrossRef]
- Rodrigues, W.P.; Martins, M.Q.; Fortunato, A.S.; Rodrigues, A.P.; Semedo, J.N.; Simoes-Costa, M.C.; Pais, I.P.; Leitao, A.E.; Colwell, F.; Goulao, L.; et al. Long-term elevated air [CO2] strengthens photosynthetic functioning and mitigates the impact of supra-optimal temperatures in tropical Coffea arabica and C. canephora species. Glob. Chang. Biol. 2016, 22, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Lam, S.K.; Hao, X.; Li, F.Y.; Zong, Y.; Wang, H.; Li, P. Elevated CO2 reduces the adverse effects of drought stress on a high-yielding soybean (Glycine max (L.) Merr.) cultivar by increasing water use efficiency. Plant Physiol. Biochem. 2018, 132, 660–665. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Rodrigues, A.P.; Semedo, J.N.; Pais, I.P.; Martins, L.D.; Simões-Costa, M.C.; Leitã, A.E.; Fortunato, A.S.; Batista-Santos, P.; Palos, I.M.; et al. Sustained photosynthetic performance of Coffea spp. under long-term enhanced [CO2]. PLoS ONE 2013, 8, e82712. [Google Scholar] [CrossRef]
- Rakocevic, M.; Ribeiro, R.V.; Marchiori, P.E.R.; Filizola, H.F.; Batista, E.R. Structural and functional changes in coffee trees after 4 years under free air CO2 enrichment. Ann. Bot. 2018, 121, 1065–1078. [Google Scholar] [CrossRef]
- Avila, R.T.; Cardoso, A.; de Almeida, L.; Costa, L.C.; Machado, K.L.G.; Barbosa, M.L.; de Souza, R.P.B.; Oliveira, L.A.; Batista, D.S.; Martins, S.C.V.; et al. Coffee plants respond to drought and elevated [CO2] through changes in stomatal function, plant hydraulic conductance, and aquaporin expression. Environ. Exp. Bot. 2020, 177, 104148. [Google Scholar] [CrossRef]
- Martins, M.Q.; Rodrigues, W.P.; Fortunato, A.S.; Leitao, A.E.; Rodrigues, A.P.; Pais, I.P.; Martins, L.D.; Silva, M.J.; Reboredo, F.H.; Partelli, F.L.; et al. Protective response mechanisms to heat stress in interaction with high [CO2] conditions in Coffea spp. Front. Plant Sci. 2016, 7, 947. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Pais, I.P.; Leitão, A.E.; Guerra, M.; Reboredo, F.H.; Máguas, C.; Carvalho, M.L.; Scotti-Campos, P.; Ribeiro-Barros, A.I.; Lidon, F.C.; et al. Can elevated air [CO2] conditions mitigate the predicted warming impact on the quality of coffee bean? Front. Plant Sci. 2018, 9, 287. [Google Scholar] [CrossRef]
- Bunn, C.; Läderach, P.; Rivera, O.O.; Kirschke, D.A. Bitter cup: Climate change profile of global production of Arabica and Robusta coffee. Clim. Chang. 2015, 129, 89–101. [Google Scholar] [CrossRef]
- Tallis, M.J.; Lin, Y.; Rogers, A.; Zhang, J.; Street, N.R.; Miglietta, F.; Karnosky, D.F.; Angelis, P.D.; Calfapietra, C.; Taylor, G. The transcriptome of Populus in elevated CO2 reveals increased anthocyanin biosynthesis during delayed autumnal senescence. New Phytol. 2010, 186, 415–428. [Google Scholar] [CrossRef]
- Taylor, G.; Street, N.R.; Tricker, P.J.; Sjodin, A.; Graham, L.; Skogstrom, O.; Calfapietra, C.; Scarascia-Mugnozza, G.; Jansson, S. The transcriptome of Populus in elevated CO2. New Phytol. 2005, 167, 143–154. [Google Scholar] [CrossRef]
- De Souza, A.P.; Gasper, M.; Da Silva, E.A. Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ. 2008, 31, 1116–1127. [Google Scholar] [CrossRef]
- Leakey, A.D.B.; Ainsworth, E.A.; Bernacchi, C.J.; Rogers, A.; Long, S.P.; Ort, D.R. Elevated CO2 effects on plant carbon, nitrogen, and water relations: Six important lessons from FACE. J. Exp. Bot. 2009, 60, 2859–2876. [Google Scholar] [CrossRef] [PubMed]
- Grossman, J.D.; Rice, K.J. Contemporary evolution of an invasion grass in response to elevated atmospheric CO2 at a Mojave desert FACE site. Ecol. Lett. 2014, 17, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Horgan-Kobelski, T.; Matesanz, S.; Sultan, S.E. Limits to future adaptation in the invasive plant Polygonum cespitosum: Expression of functional and fitness traits at elevated CO2. J. Hered. 2016, 107, 1–9. [Google Scholar] [CrossRef][Green Version]
- Watson-Lazowski, A.; Lin, Y.; Miglietta, F.; Edwards, R.J.; Chapman, M.A.; Taylor, G. Plant adaptation or acclimation to rising CO2? Insight from first multigenerational RNA?Seq transcriptome. Glob. Chang. Biol. 2016, 22, 3760–3773. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181. [Google Scholar] [CrossRef]
- Kane, K.; Dahal, K.P.; Badawi, M.A.; Houde, M.; Hüner, N.P.A.; Sarhan, F. Long-term growth under elevated CO2 suppresses biotic stress genes in non-acclimated, but not cold-acclimated winter wheat. Plant Cell Physiol. 2013, 54, 1751–1768. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, Y.; Zhou, G. Response and adaptation of photosynthesis, respiration, and antioxidant systems to elevated CO2 with environmental stress in plants. Front. Plant Sci. 2015, 6, 701. [Google Scholar] [CrossRef]
- Guedes, F.A.; Nobres, P.; Rodrigues, D.C.F.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Ivamoto, S.T.; Reis, O.; Júnior Domingues, D.S.; dos Santos, T.B.; de Oliveira, F.F.; Pot, D.; Leroy, T.; Esteves Vieira, L.G.; Carazzolle, M.F.; Pereira, G.A.G.; et al. Transcriptome analysis of leaves, flowers and fruits perisperm of Coffea arabica L. reveals the differential expression of genes involved in raffinose Biosynthesis. PLoS ONE 2017, 12, e0169595. [Google Scholar] [CrossRef]
- Iannicelli, J.; Guariniello, J.; Tossi, V.E.; Regalado, J.J.; Di Ciaccio, L.; van Baren, C.M.; Álvarez, S.I.P.; Escandón, A.S. The “polyploid effect” in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- Adams, K.L.; Cronn, R.; Percifield, R.; Wendel, J.F. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 2003, 100, 4649–4654. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gómez-Gómez, L. The carotenoid cleavage dioxygenase CCD2 catalyzing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2015, 209, 650–663. [Google Scholar] [CrossRef]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in plants: Occurrence, regulation, and role. Front Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef]
- Scotti-Campos, P.; Pais, I.P.; Ribeiro-Barros, A.I.; Martins, L.D.; Tomaz, M.A.; Rodrigues, W.P.; Campostrini, E.; Semedo, J.N.; Fortunato, A.S.; Martins, M.Q.; et al. Lipid profile adjustments may contribute to warming acclimation and to heat impact mitigation by elevated [CO2] in Coffea spp. Environ. Exp. Bot. 2019, 167, 103856. [Google Scholar] [CrossRef]
- Liu, D.; Gong, Q.; Ma, Y.; Li, P.; Li, J.; Yang, S.; Yuan, L.; Yu, Y.; Pan, D.; Xu, F.; et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J. Exp. Bot. 2010, 61, 1655–1669. [Google Scholar] [CrossRef]
- Kato, Y.; Kouso, T.; Sakamoto, W. Variegated tobacco leaves generated by chloroplast FtsH suppression: Implication of FtsH function in the maintenance of thylakoid membranes. Plant Cell Physiol. 2012, 53, 391–404. [Google Scholar] [CrossRef]
- Kaldenhoff, R.; Kai, L.; Uehlein, N. Aquaporins and membrane diffusion of CO2 in living organisms. Biochim. Biophys. Acta 2014, 1840, 1592–1595. [Google Scholar] [CrossRef]
- Karve, T.M.; Cheema, A.K. Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J. Amino Acids 2011, 2011, 1–13. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef]
- Erice, G.; Aranjuelo, I.; Irigoyen, J.J.; Sánchez-Díaz, M. Effect of elevated CO2, temperature and limited water supply on antioxidant status during regrowth of nodulated alfalfa. Physiol. Plant. 2007, 130, 33–45. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Farfan-Vignolo, A.R.; Vos, D.; Asard, H. Elevated CO2 mitigates drought and temperature-induced oxidative stress differently in grasses and legumes. Plant Sci. 2015, 231, 1–10. [Google Scholar] [CrossRef]
- Watanabe, C.K.; Sato, S.; Yanagisawa, S.; Uesono, Y.; Terashima, I.; Noguchi, K. Effects of elevated CO2 on levels of primary metabolites and transcripts of genes encoding respiratory enzymes and their diurnal patterns in Arabidopsis thaliana: Possible relationships with respiratory rates. Plant Cell Physiol. 2014, 55, 341–357. [Google Scholar] [CrossRef]
- Vega, F.E.; Ziska, L.H.; Simpkins, A.; Infante, F.; Davis, A.P.; Rivera, J.A.; Barnaby, J.Y.; Wolf, J. Early growth phase and caffeine content response to recent and projected increases in atmospheric carbon dioxide in coffee (Coffea arabica and C. canephora). Sci Rep. 2020, 10, 5875. [Google Scholar] [CrossRef]
- Loreto, F.; Mannozzi, M.; Maris, C.; Nascetti, P.; Ferranti, F.; Pasqualini, S. Ozone quenching properties of isoprene and its antioxidant role in leaves. Plant Physiol. 2001, 126, 993–1000. [Google Scholar] [CrossRef]
- Sasaki, K.; Saito, T.; Lamsa, M.; Oksman-Caldentey, K.-M.; Suzuki, M.; Ohyama, K.; Muranaka, T.; Ohara, K.; Yazaki, K. Plants utilize isoprene emission as a thermo-tolerance mechanism. Plant Cell Physiol. 2007, 48, 1254–1262. [Google Scholar] [CrossRef]
- Belchínavarro, S.; Almagro, L.; Sabaterjara, A.B.; Fernández Pérez, F.; Bru, R.; Pedreño, M.A. Induction of trans-resveratrol and extracellular pathogenesis-related proteins in elicited suspension cultured cells of Vitis vinifera cv Monastrell. J. Plant Physiol. 2013, 170, 258–264. [Google Scholar] [CrossRef]
- Zhu, Q.G.; Gong, Z.Y.; Wang, M.M.; Li, X.; Grierson, D.; Yin, X.R.; Chen, K.S. A transcription factor network responsive to high CO2/hypoxia is in-volved in deastringency in persimmon fruit. J. Exp. Bot. 2018, 69, 2061–2070. [Google Scholar] [CrossRef]
- Wilkinson, M.J.; Monson, R.K.; Trahan, N.; Lee, S.; Brown, E.; Jackson, R.B.; Polley, H.W.; Fay, P.A.; Fall, R. Leaf isoprene emission rate as a function of atmospheric CO2 concentration. Glob. Chang. Biol. 2009, 15, 1189–1200. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef]
- Gray, J.E.; Holroyd, G.H.; van der Lee, F.M.; Bahrami, A.R.; Sijmons, P.C.; Woodward, F.I.; Schuch, W.; Hetherington, A.M. The HIC signalling pathway links CO2 perception to stomatal development. Nature 2000, 408, 713. [Google Scholar] [CrossRef]
- Marsh, E.N.; Waugh, M.W. Aldehyde Decarbonylases: Enigmatic enzymes of hydrocarbon biosynthesis. ACS Catal. 2013, 3, 2515–2521. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signalling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Uchida, A.; Jagendorf, A.T.; Hibino, T.; Takabe, T.; Takabe, T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002, 163, 515–523. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Yamaguchi, H.; Yuasa, T.; Inwaya-Inoue, M.; Arima, S.; Zheng, S. Hydrogen peroxide spraying alleviates drought stress in soybean plants. J. Plant Physiol. 2011, 168, 1562–1567. [Google Scholar] [CrossRef]
- Hossain, M.A.; Fujita, M. Hydrogen peroxide priming stimulates drought tolerance in mustard (Brassica juncea L.) seedlings. Plant Gene Trait 2013, 4, 109–123. [Google Scholar] [CrossRef]
- Verbancic, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef]
- Ezquer, I.; Salameh, I.; Colombo, L.; Kalaitzis, P. Plant cell walls tackling climate change: Biotechnological strategies to improve crop adaptations and photosynthesis in response to global warming. Plants 2020, 9, 212. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Long, S.P. What have we learned from 15 years of free air-CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 2005, 165, 351–372. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Mondego, J.M.C.; Vidal, R.O.; Carazzolle, M.F. An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Biol. 2011, 11, 30. [Google Scholar] [CrossRef]
- Yuyama, P.M.; Júnior, O.R.; Ivamoto, S.T.; Silva Domingues, D.; Carazzolle, M.F.; Guimarães Pereira, G.A.; Charmetant, P.; Leroy, T.; Protasio Pereira, L.F. Transcriptome analysis in Coffea eugenioides, an Arabica coffee ancestor, reveals differentially expressed genes in leaves and fruits. Mol. Genet. Genomics 2016, 291, 323–336. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Ramalho, J.D.C. Impacts of drought and temperature stress on coffee physiology and production: A review. Braz. J. Plant Physiol. 2006, 18, 55–81. [Google Scholar] [CrossRef]
- Ramalho, J.C.; DaMatta, F.M.; Rodrigues, A.P.; Scotti-Campos, P.; Pais, I.; Batista-Santos, P.; Partelli, F.L.; Ribeiro, A.; Lidon, F.C.; Leitão, A.E. Cold impact and acclimation response of Coffea spp. plants. Theor. Exp. Plant Physiol. 2014, 26, 5–18. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Alves, G.; Torres, L.F.; Déchamp, E.; Breitler, J.C.; Joët, T.; Gatineau, F.; Andrade, A.C.; Bertrand, B.; Marraccini, P.; Etienne, H. Differential fine-tuning of gene expression regulation in coffee leaves by CcDREB1D promoter haplotypes under water deficit. J. Exp. Bot. 2017, 68, 3017–3031. [Google Scholar] [CrossRef]
- Torres, L.F.; Reichel, T.; Déchamp, E.; de Aquino, S.O.; Duarte, K.E.; Alves, G.S.C.; Silva, A.T.; Cotta, M.G.; Costa, T.S.; Diniz, L.E.C. Expression of DREB-Like genes in Coffea canephora and C. arabica subjected to various types of abiotic stress. Trop. Plant Biol. 2019, 12, 98–116. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dédaldéchamp, F.; Perez-Garcia, M.D.; Ogé, L.; Hamama, L.; Atanassova, R. The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef]
- Fukayama, H.; Sugino, M.; Fukuda, T.; Masumoto, C.; Taniguchi, Y.; Okada, M.; Sameshima, R.; Hatanaka, T.; Misoo, S.; Hasegawa, T.; et al. Gene expression profiling of rice grown in free air CO2 enrichment (FACE) and elevated soil temperature. Field Crop. Res. 2011, 121, 195–199. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Jaafar, H.Z.E.; Rahmat, A. Elevated carbon dioxide increases contents of flavonoids and phenolic compounds, and antioxidant activities in Malaysian young ginger (Zingiber officinale Roscoe.) varieties. Molecules 2010, 15, 7907–7922. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Shimizu, H.; Ito, S.; Yagasaki, Y.; Zou, C.J.; Zhou, G.S.; Zheng, Y. Effects of elevated CO2, warming and precipitation change on plant growth, photosynthesis and peroxidation in dominant species from North China grassland. Planta 2014, 239, 421–435. [Google Scholar] [CrossRef]
- Zinta, G.; AbdElgawad, H.; Domagalska, M.A.; Vergauwen, L.; Knapen, D.; Nijs, I.; Janssens, I.A.; Beemster, G.T.S.; Asard, H. Physiological, biochemical, and genome-wide transcriptional analysis reveals that elevated CO2 mitigates the impact of combined heat wave and drought stress in Arabidopsis thaliana at multiple organizational levels. Glob. Chang. Biol. 2014, 20, 3670–3685. [Google Scholar] [CrossRef]
- Pritchard, S.G.; Ju, Z.; Santen, E.; Qiu, J.; Weaver, D.B.; Prior, S.A.; Rogers, H.H. The influence of elevated CO2 on the activities of antioxidative enzymes in two soybean genotypes. Aust. J. Plant Physiol. 2000, 27, 1061–1068. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Campos, P.S.; Teixeira, M.; Nunes, M.A. Nitrogen dependent changes in antioxidant systems and in fatty acid composition of chloroplast membranes from Coffea arabica L. plants submitted to high irradiance. Plant Sci. 1998, 135, 115–124. [Google Scholar] [CrossRef]
- Semedo, J.N.; Rodrigues, A.P.; Lidon, F.C.; Pais, I.P.; Marques, I.; Gouveia, D.; Armengaud, J.; Martins, S.; Semedo, M.C.; Silva, M.J.; et al. Intrinsic non-stomatal resilience to drought of the photosynthetic apparatus in Coffea spp. can be strengthened by elevated Air CO2. Tree Physiol. 2020, tpaa158. [Google Scholar] [CrossRef]
- Ijaq, J.; Chandrasekharan, M.; Poddar, R.; Bethi, N.; Sundararajan, V.S. Annotation and curation of uncharacterized proteins-challenges. Front. Genet. 2015, 6, 119. [Google Scholar] [CrossRef]
- Naveed, M.; Chaudhry, Z.; Ali, Z.; Amjad, M.; Zulfiqar, F.; Numan, A. Annotation and curation of hypothetical proteins: Prioritizing targets for experimental study. Adv. Life Sci. 2018, 5, 73–87. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 12 November 2019).
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000 Res. 2018, 7, 1338. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 genome project data processing subgroup. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, G. Gffread: GFF/GTF Utility Providing Format Conversions, Region Filtering, FASTA Sequence Extraction and More. 2015. Available online: https://github.com/gpertea/gffread (accessed on 24 November 2019).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 14 December 2019).
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Sci. 2000, 25, 60–83. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Götz, S. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 21 June 2020).
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef]
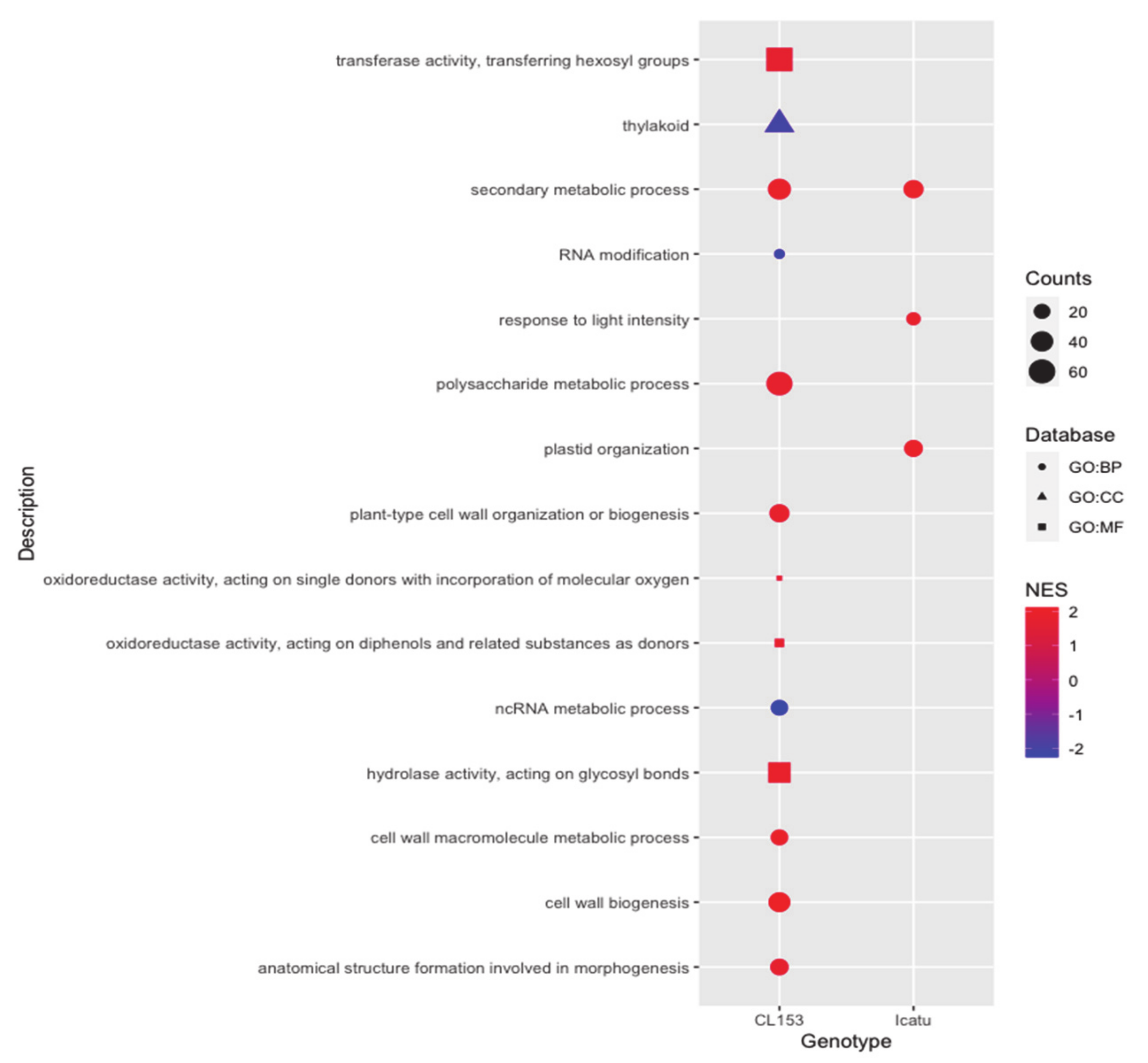
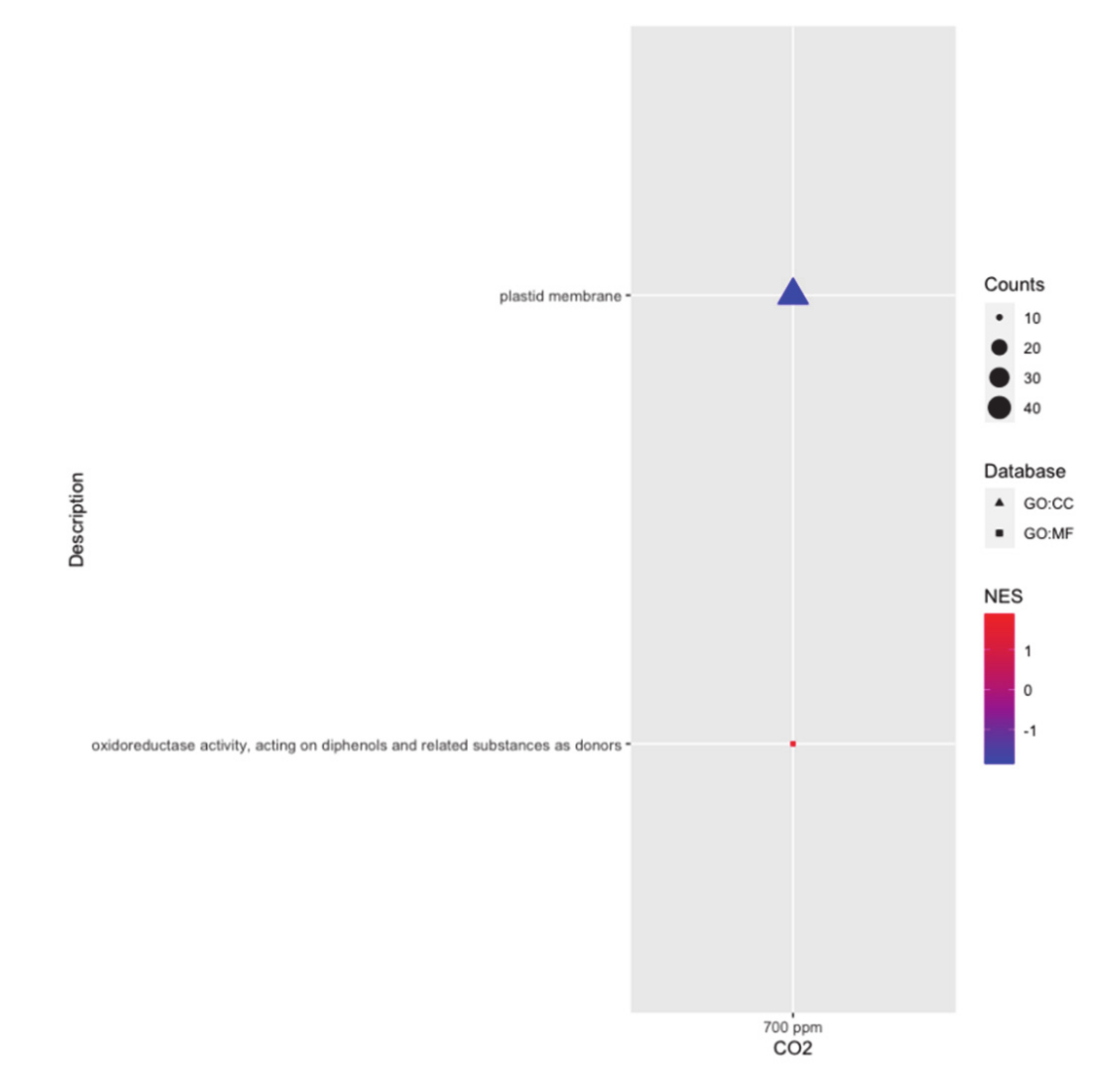
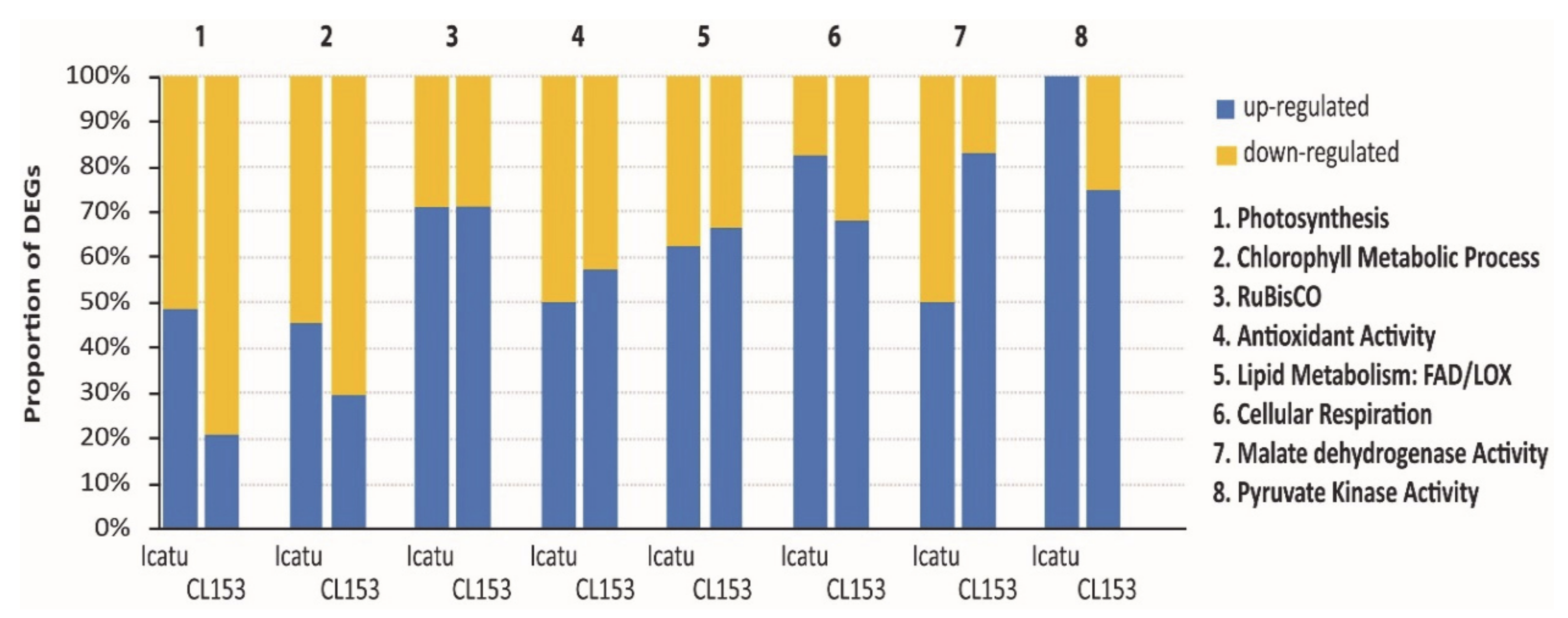
| Gene | Protein Name | FC | Biological Processes |
|---|---|---|---|
| Top up-regulated | |||
| Cc01_g09180 | Flavonol 7-O-beta-glucosyltransferase | 6.11 | acid metabolic process |
| Cc10_g08360 | Uncharacterized protein | 5.60 | |
| Cc01_g05170 | Omega-6 fatty acid endoplasmic reticulum isozyme 2 | 5.60 | unsaturated fatty acid biosynthetic process |
| Cc07_g12510 | ATP-dependent zinc metalloprotease | 5.19 | photosystem II repair, proteolysis, thylakoid membrane organization |
| Cc02_g07390 | Uncharacterized protein | 5.18 | |
| Cc05_g12830 | Zinc finger Constans-Like 10 | 4.86 | metal binding |
| Cc08_g16300 | Uncharacterized protein At5g39865 | 4.83 | cell redox homeostasis |
| Cc07_g04750 | Zinc finger Constans-Like 2 | 4.63 | chloroplast organization |
| Cc07_g12820 | LHY | 4.42 | circadian rhythm, regulation of transcription, response to stress |
| Cc02_g05510 | Aquaporin PIP subfamily | 4.36 | water transport |
| Cc04_g17080 | Aquaporin, MIP family, PIP subfamily | 4.07 | water transport |
| Cc09_g02890 | Uncharacterized protein | 4.05 | |
| Top down-regulated | |||
| Cc02_g03420 | Ethylene-responsive transcription factor ERF025 | −5.11 | DNA-binding transcription factor activity |
| Cc02_g03180 | Uncharacterized protein | −5.21 | |
| Cc06_g10420 | E3 ubiquitin- ligase CIP8 | −5.46 | protein ubiquitination |
| Cc11_g07700 | Uncharacterized protein | −5.62 | |
| Cc02_g27160 | Vicianin hydrolase (Fragment) | −5.69 | carbohydrate catabolic process |
| Cc11_g07710 | Two-component response regulator | −5.77 | carbohydrate catabolic process |
| Cc06_g08260 | Umecyanin | −5.83 | electron transfer activity, metal ion binding |
| Cc09_g06040 | Pentatricopeptide repeat-containing At2g35130 | −6.30 | electron transfer activity, metal ion binding |
| Cc11_g12580 | Uncharacterized protein | −7.69 | |
| Cc01_g14750 | Chloroplast import apparatus 2 | −8.08 | protein ubiquitation |
| Gene | Protein Name | FC | Biological Process |
|---|---|---|---|
| Top up-regulated | |||
| Cc05_g05400 | Isoprene chloroplastic | 9.88 | isoprene synthase activity |
| Cc05_g04940 | Isocitrate lyase | 8.60 | glyoxylate cycle, tricarboxylic acid cycle |
| Cc01_g05170 | Uncharacterized protein | 8.46 | |
| Cc05_g00220 | Uncharacterized protein | 8.26 | |
| Cc02_g25790 | Very-long-chain aldehyde decarbonylase CER3 | 8.00 | alkane biosynthetic process |
| Cc02_g20300 | Uncharacterized protein | 7.83 | |
| Cc02_g03420 | Ethylene-responsive transcription factor ERF025 | 7.79 | defense response to fungus, ethylene-activated signaling pathway |
| Cc03_g12230 | Desiccation-related PCC13-62 | 7.59 | cellular response to desiccation, cellular response to salt |
| Cc06_g13950 | EG45-like domain containing 1 | 7.56 | cellular response to hypoxia |
| Cc03_g15270 | Desiccation-related PCC13-62 | 7.55 | cellular response to desiccation, cellular response to salt |
| Cc01_g08230 | Glucomannan 4-beta-mannosyltransferase 9 | 6.96 | cell wall organization |
| Cc01_g09500 | Cytochrome P450 94A1 | 6.89 | oxidoreductase activity |
| Cc09_g03820 | 12-oxophytodienoate reductase 1 | 6.88 | oxylipin biosynthetic process |
| Cc02_g08150 | Uncharacterized protein | 6.54 | |
| Cc00_g12860 | Uncharacterized protein | 6.40 | |
| Cc10_g12850 | Stearoyl | 6.13 | cellular response to hypoxia |
| Cc00_g11660 | Uncharacterized protein | 6.12 | |
| Cc05_g13060 | (-)-germacrene D synthase | 6.11 | terpenoid biosynthetic process |
| Cc07_g14440 | Uncharacterized protein | 6.08 | |
| Top down-regulated | |||
| Cc11_g04610 | Momilactone A synthase | -5.01 | phytoalexins biosynthesis |
| Cc06_g10420 | Uncharacterized protein | -5.09 | |
| Cc11_g07700 | Uncharacterized protein | -5.22 | |
| Cc07_g08450 | Detoxification 6 | -5.31 | transmembrane transporter activity |
| Gene | Protein Name | FC | Biological Processes |
|---|---|---|---|
| Top up-regulated | |||
| Cc11_g10870 | Uncharacterized protein | 14.23 | |
| Cc00_g10190 | Acyl- -binding domain-containing 4 | 12.51 | lipid transport, response to ethylene, response to jasmonic acid, response to light stimulus |
| Cc10_g14870 | Uncharacterized protein | 12.03 | |
| Cc00_g08140 | Homeobox-leucine zipper protein | 11.89 | root, shoot development, response to light, red, far-red light phototransduction |
| Cc05_g00740 | Acidic endochitinase | 11.87 | chitin catabolic process, polysaccharide catabolic process |
| Cc00_g25490 | 8-hydroxyquercetin 8-O-methyltransferase | 11.57 | flavonoid metabolic process, methylation |
| Cc03_g11280 | Cell division control 48 homolog B | 11.29 | cell cycle, cell division, protein transport |
| Cc07_g01500 | Uncharacterized protein | 11.05 | |
| Cc01_g01970 | F-box LRR-repeat At4g14096 | 11.00 | |
| Cc00_g17790 | Uncharacterized protein | 10.92 | |
| Cc00_g26610 | Uncharacterized protein | 10.87 | |
| Cc10_g14880 | Uncharacterized protein | 10.82 | |
| Cc09_g08250 | Uncharacterized protein | 10.71 | |
| Cc03_g09260 | Uncharacterized protein | 10.59 | |
| Cc00_g21940 | UTP--glucose-1-phosphate uridylyltransferase | 10.54 | UDP-glucose metabolic process |
| Cc01_g01360 | Uncharacterized protein | 10.54 | |
| Cc06_g16070 | Uncharacterized protein | 10.36 | |
| Cc11_g00660 | Uncharacterized protein | 10.24 | |
| Cc09_g08930 | Uncharacterized protein | 10.22 | |
| Cc00_g07170 | UDP-glucose flavonoid 3-O-glucosyltransferase 3 | 10.00 | transferase activity |
| Top down-regulated | |||
| Cc07_g14720 | 1-aminocyclopropane-1-carboxylate oxidase 2 | −10.06 | stress response, ethylene biosynthetic process |
| Cc09_g05150 | Cytochrome P450 89A2 | −10.09 | oxidoreductase activity |
| Cc00_g29880 | 8-hydroxyquercetin 8-O-methyltransferase | −10.14 | flavonoid metabolic process, methylation |
| Cc08_g08870 | BURP domain RD22 | −10.25 | response to salt stress |
| Cc11_g04040 | Uncharacterized protein | −10.28 | |
| Cc10_g04410 | Uncharacterized protein | −10.40 | |
| Cc00_g23750 | Uncharacterized protein | −10.48 | |
| Cc10_g14350 | Uncharacterized protein | −10.66 | |
| Cc00_g20380 | FK506-binding 2 | −10.70 | stress response |
| Cc05_g05110 | Cytochrome P450 71D10 | −10.70 | oxidoreductase activity |
| Cc00_g17430 | Isoflavone reductase homolog P3 | −10.76 | response to cadmium ion, response to oxidative stress |
| Cc06_g14770 | Uncharacterized protein | −10.97 | |
| Cc11_g00400 | Uncharacterized protein | −11.08 | |
| Cc00_g29670 | Uncharacterized protein | −11.54 | |
| Cc04_g13840 | Uncharacterized protein | −11.60 | |
| Cc00_g30240 | Cysteinease inhibitor 5 | −13.36 | cellular response to heat, defense response |
| Gene | Protein Name | FC | Biological Processes |
|---|---|---|---|
| Top up-regulated | |||
| Cc00_g10190 | Acyl- -binding domain-containing 4 | 12.17 | lipid transport, response to ethylene, response to light stimulus |
| Cc10_g14870 | Uncharacterized protein | 11.60 | |
| Cc04_g16250 | Uncharacterized protein | 11.30 | |
| Cc08_g09540 | Carotenoid cleavage dioxygenase chloroplastic | 11.21 | leaf morphogenesis, secondary shoot formation |
| Cc05_g00740 | Uncharacterized protein | 10.67 | |
| Cc11_g08120 | Plant cadmium resistance 2 | 10.62 | response to oxidative stress |
| Cc01_g04210 | Homeobox-leucine zipper ATHB-8 | 10.57 | cell differentiation |
| Top down-regulated | |||
| Cc00_g06270 | Annexin D8 | −10.07 | response to cold, response to heat, response to salt stress, response to water deprivation |
| Cc00_g30240 | Cysteinease inhibitor 5 | −10.09 | cellular response to heat, defense response |
| Cc00_g17430 | Isoflavone reductase homolog P3 | −10.23 | response to oxidative stress |
| Cc02_g38970 | Uncharacterized protein | −10.28 | |
| Cc00_g19610 | Uncharacterized protein | −10.39 | |
| Cc00_g11770 | Tabersonine 16-O-methyltransferase | −10.44 | alkaloid biosynthetic process |
| Cc00_g01740 | Uncharacterized protein | −10.77 | |
| Cc05_g05110 | Cytochrome P450 71D10 | −10.99 | oxidoreductase activity |
| Database | ID | Description | Counts | NES | p-value | FDR |
| eCO2 vs. aCO2 | CL153 | ||||||
| KEGG | map00906 | carotenoid biosynthesis | 12 | 1.99 | <0.001 | 8.12 × 10−4 |
| map00073 | cutin, suberine and wax biosynthesis | 5 | 1.76 | 1.69 × 10−3 | 4.91 × 10−2 | |
| CL153 vs. Icatu | eCO2 | ||||||
| WikiPathways | WP3661 | genetic interactions between sugar and hormone signaling | 12 | 1.67 | 2.15 × 10−2 | 2.90 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques, I.; Fernandes, I.; David, P.H.C.; Paulo, O.S.; Goulao, L.F.; Fortunato, A.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]. Int. J. Mol. Sci. 2020, 21, 9211. https://doi.org/10.3390/ijms21239211
Marques I, Fernandes I, David PHC, Paulo OS, Goulao LF, Fortunato AS, Lidon FC, DaMatta FM, Ramalho JC, Ribeiro-Barros AI. Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]. International Journal of Molecular Sciences. 2020; 21(23):9211. https://doi.org/10.3390/ijms21239211
Chicago/Turabian StyleMarques, Isabel, Isabel Fernandes, Pedro H.C. David, Octávio S. Paulo, Luis F. Goulao, Ana S. Fortunato, Fernando C. Lidon, Fábio M. DaMatta, José C. Ramalho, and Ana I. Ribeiro-Barros. 2020. "Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]" International Journal of Molecular Sciences 21, no. 23: 9211. https://doi.org/10.3390/ijms21239211
APA StyleMarques, I., Fernandes, I., David, P. H. C., Paulo, O. S., Goulao, L. F., Fortunato, A. S., Lidon, F. C., DaMatta, F. M., Ramalho, J. C., & Ribeiro-Barros, A. I. (2020). Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]. International Journal of Molecular Sciences, 21(23), 9211. https://doi.org/10.3390/ijms21239211












