Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma
Abstract
1. Introduction
2. Results
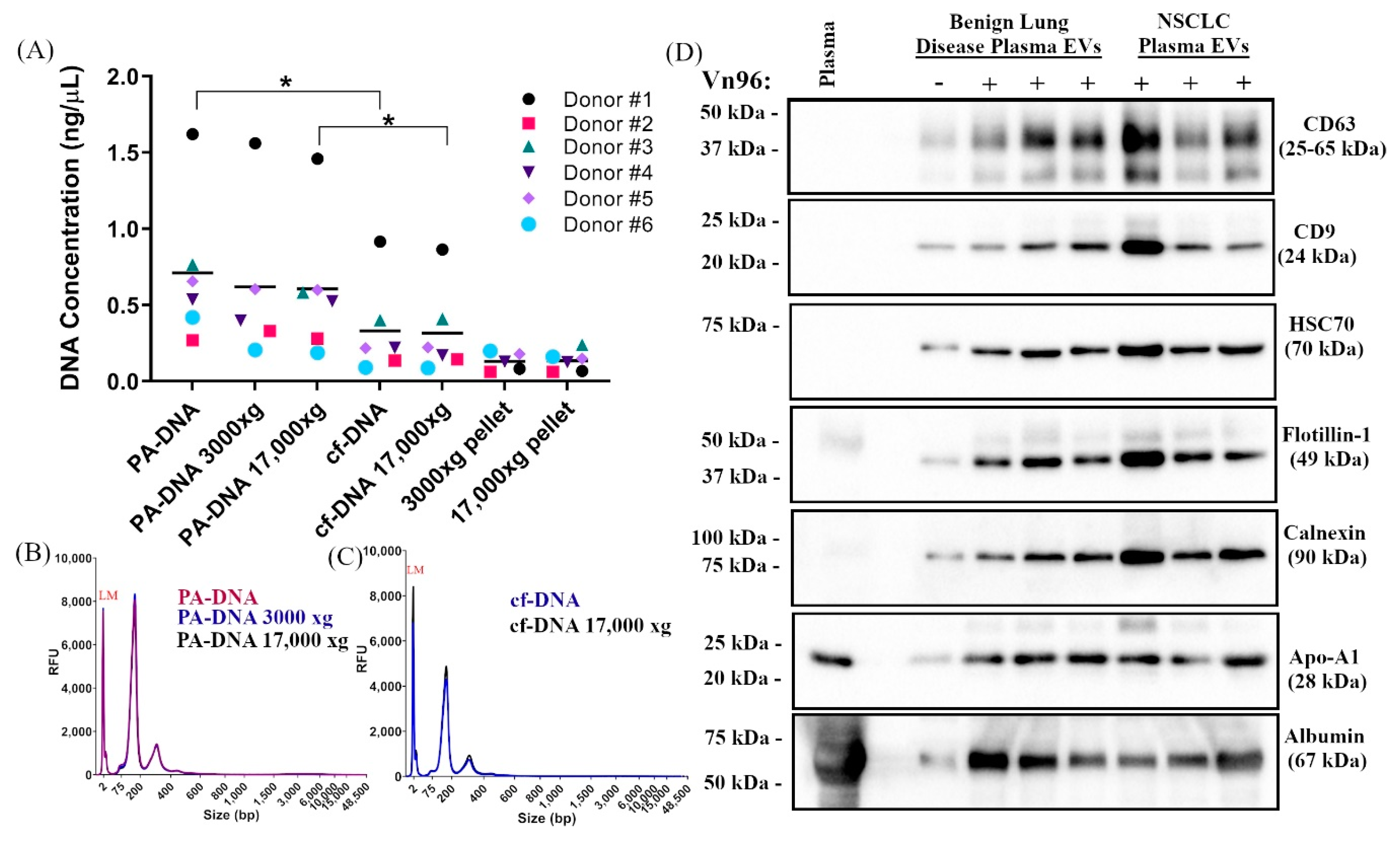
2.1. Centrifugation of Plasma Does Not Significantly Impact DNA Yield, Size, or SNV Detection
2.2. PA Precipitation Enriches for Extracellular Vesicles
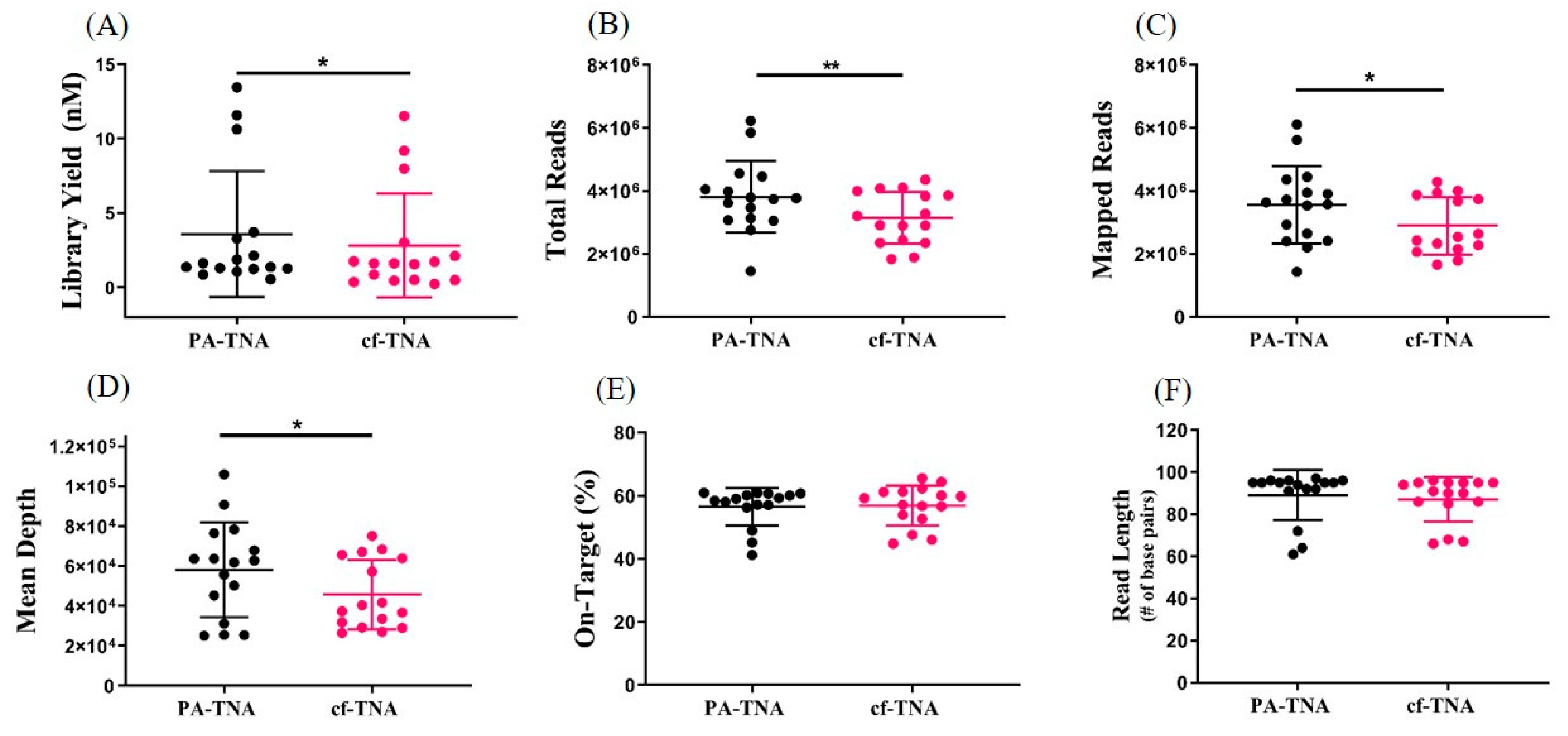
2.3. PA Improves Recovery of Nucleic Acids from NSCLC Patient Plasma
2.4. PA Improves NGS Performance
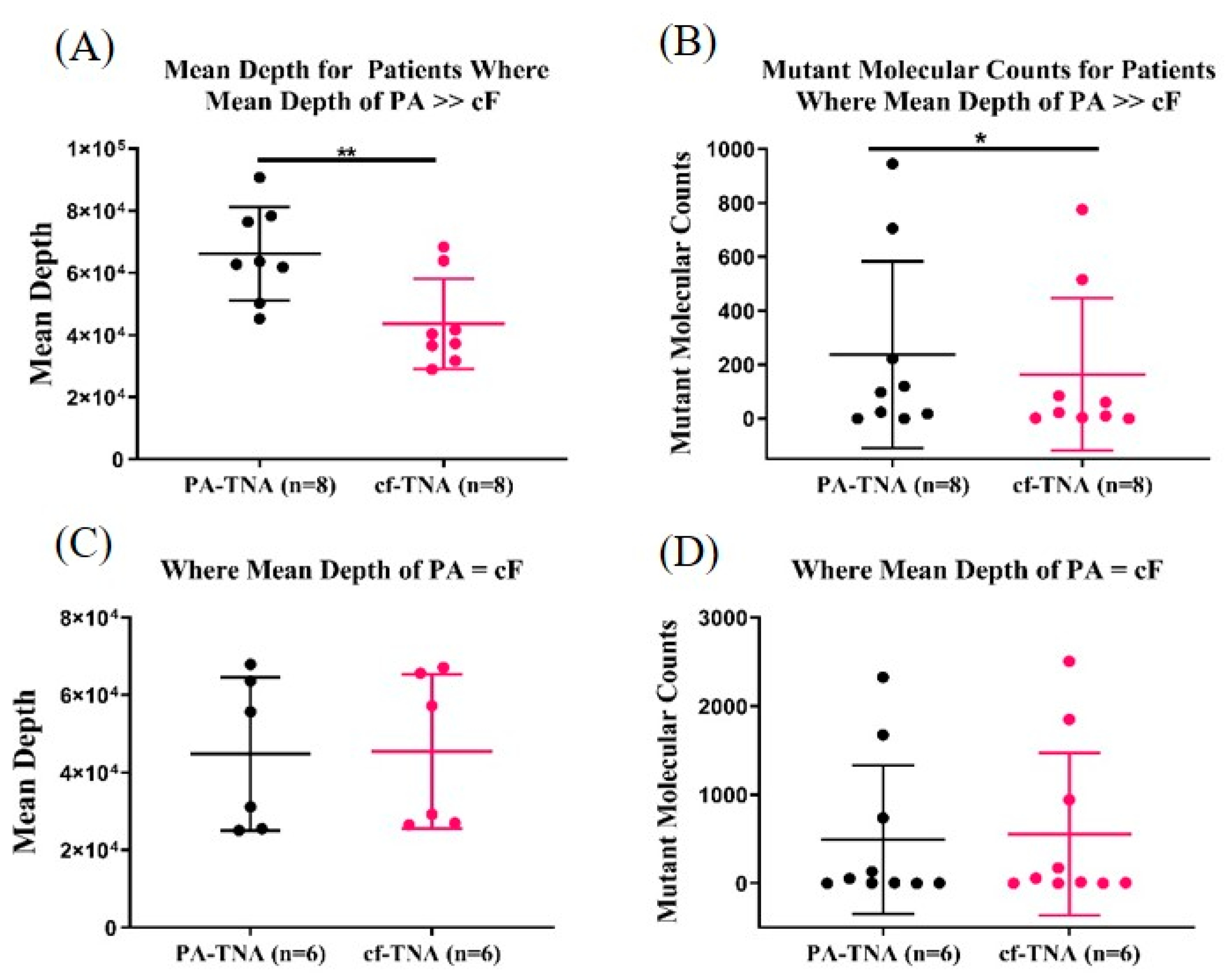
2.5. PA Improves Mutant Allele Detection
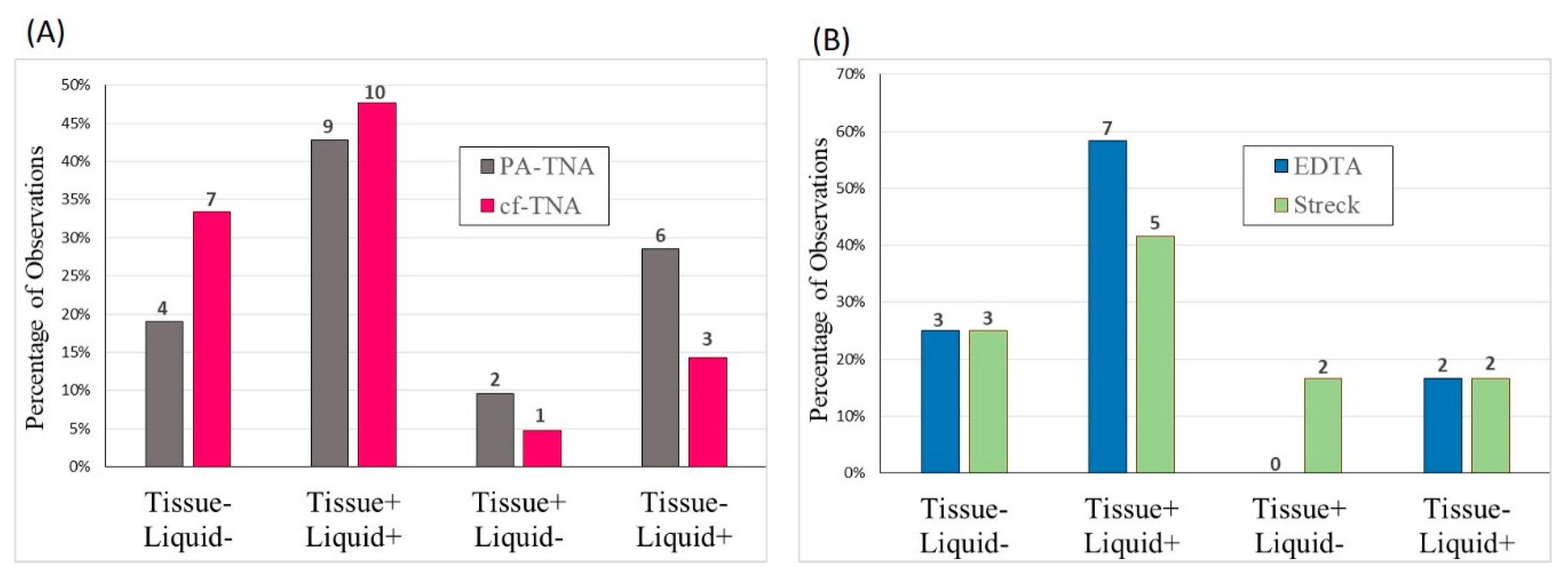
2.6. Concordance Observed between Liquid and Tissue Biopsies
2.7. High Specificity of EGFR Mutant Allele Detection Observed Using PA
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Plasma Processing
4.3. Peptide-Affinity EV and cf-DNA Precipitation
4.4. Western Blotting
4.5. Nucleic Acid Isolation
4.6. Fragment Analyzer
4.7. Droplet Digital PCR
4.8. Next-Generation Sequencing
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NSCLC | Non-Small Cell Lung Cancer |
| cf-DNA | cell-free DNA |
| EV | Extracellular vesicle |
| PA | Peptide-affinity |
| SNV | Single nucleotide variant |
| TNA | Total nucleic acid |
| cf-TNA | Cell-free total nucleic acid |
| PA-TNA | Peptide-affinity total nucleic acid |
| EGFR | Epidermal Growth Factor Receptor |
| NGS | Next-generation sequencing |
| ALK | Anaplastic lymphoma kinase |
| ctDNA | Circulating tumour DNA |
| S-EVs | Small extracellular vesicles |
| L-EVs | Large extracellular vesicles |
| HS | High sensitivity |
| PCR | Polymerase chain reaction |
| ddPCR | Droplet digital PCR |
| MAF | Mutant allele frequency |
| MW | Molecular weight |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| CNV | Copy number variant |
| EDTA | Ethylenediaminetetracetic acid |
| WBC | White blood cell |
| miRNA | microRNA |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global Cancer Statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Halliday, P.R.; Blakely, C.M.; Bivona, T.G. Emerging Targeted Therapies for the Treatment of Non-Small Cell Lung Cancer. Curr. Oncol. Rep. 2019, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Valentino, F.; Borra, G.; Allione, P.; Rossi, L. Emerging Targets in Advanced Non-Small-Cell Lung Cancer. Future Oncol. 2018, 14, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Aisner, D.L.; Allen, T.C.; Beasley, M.B.; Borczuk, A.C.; Cagle, P.T.; Capelozzi, V.; Dacic, S.; Hariri, L.; Kerr, K.M.; et al. Programmed Death Ligand-1 Immunohistochemistry—A New Challenge for Pathologists: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2016, 140, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Mathai, R.; Vidya, R.; Reddy, B.; Thomas, L.; Udupa, K.; Kolesar, J.; Rao, M. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J. Clin Med. 2019, 8, 373. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating Tumor DNA and Liquid Biopsy in Oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Weber, B.; Meldgaard, P.; Hager, H.; Wu, L.; Wei, W.; Tsai, J.; Khalil, A.; Nexo, E.; Sorensen, B.S. Detection of EGFR Mutations in Plasma and Biopsies from Non-Small Cell Lung Cancer Patients by Allele-Specific PCR Assays. BMC Cancer 2014, 14, 294. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-Free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Volik, S.; Alcaide, M.; Morin, R.D.; Collins, C. Cell-Free DNA (CfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol. Cancer Res. 2016, 14, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.C.; Yee, S.S.; Troxel, A.B.; Savitch, S.L.; Fan, R.; Balli, D.; Liberman, D.B.; Morrissette, J.D.; Evans, T.L.; Bauml, J.; et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin. Cancer Res. 2016, 22, 5772–5782. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.; Piscuoglio, S.; Aceto, N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Kim, C.-J.; Sunkara, V.; Kim, M.-H.; Cho, Y.-K. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and CtDNA). Micromachines 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular Vesicles as Emerging Intercellular Communicasomes. BMB Reports 2014, 47, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Voloshin, T.; Fremder, E.; Shaked, Y. Small But Mighty: Microparticles as Mediators of Tumor Progression. Cancer Microenviron. 2014, 7, 11–21. [Google Scholar] [CrossRef]
- Ciardiello, C.; Migliorino, R.; Leone, A.; Budillon, A. Large Extracellular Vesicles: Size Matters in Tumor Progression. Cytokine Growth Factor Rev. 2020, 51, 69–74. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Saha, B.; Kodys, K.; Catalano, D.; Satishchandran, A.; Szabo, G. Increased Number of Circulating Exosomes and Their MicroRNA Cargos Are Potential Novel Biomarkers in Alcoholic Hepatitis. J. Transl. Med. 2015, 13, 261. [Google Scholar] [CrossRef]
- Brinton, L.T.; Sloane, H.S.; Kester, M.; Kelly, K.A. Formation and Role of Exosomes in Cancer. Cell. Mol. Life Sci. 2015, 72, 659–671. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New Evidence That a Large Proportion of Human Blood Plasma Cell-Free DNA Is Localized in Exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, B.; Lei, H.; Zhang, B.; Huang, H.; Chen, S.; Feng, Y.; Zhu, L.; Gu, Y.; Zhang, Q.; et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell-free DNA for mutation detection in early-stage non-small-cell lung cancer. Ann. Oncol. 2018, 29, 2379–2383. [Google Scholar] [CrossRef]
- Allenson, K.; Castillo, J.; Lucas, F.A.S.; Scelo, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Funda, M.-B.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Hur, J.Y.; Kim, I.A.; Kim, H.J.; Choi, C.M.; Lee, J.C.; Kim, W.S.; Lee, K.Y. Liquid Biopsy Using the Supernatant of a Pleural Effusion for EGFR Genotyping in Pulmonary Adenocarcinoma Patients: A Comparison between Cell-Free DNA and Extracellular Vesicle-Derived DNA. BMC Cancer 2018, 18, 1236. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid Isolation of Extracellular Vesicles from Cell Culture and Biological Fluids Using a Synthetic Peptide with Specific Affinity for Heat Shock Proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Madala, S.K.; Trinath, J.; Reddy, G.B. Extracellular Small Heat Shock Proteins: Exosomal Biogenesis and Function. Cell Stress Chaperones 2018, 23, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Foroni, C.; Zarovni, N.; Bianciardi, L.; Bernardi, S.; Triggiani, L.; Zocco, D.; Venturella, M.; Chiesi, A.; Valcamonico, F.; Berruti, A. When Less Is More: Specific Capture and Analysis of Tumor Exosomes in Plasma Increases the Sensitivity of Liquid Biopsy for Comprehensive Detection of Multiple Androgen Receptor Phenotypes in Advanced Prostate Cancer Patients. Biomedicines 2020, 8, 131. [Google Scholar] [CrossRef]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P.; et al. Isolation of Extracellular Vesicles Improves the Detection of Mutant DNA from Plasma of Metastatic Melanoma Patients. Sci. Rep. 2020, 10, 15745. [Google Scholar] [CrossRef]
- Roy, J.; Taylor, C.; Beauregard, A.P.; Dhadi, S.R.; Ayre, D.C.; Fry, S.; Chacko, S.; Wajnberg, G.; Joy, A.; Mai-Thi, N.N.; et al. A multiparametric extraction method for the molecular characterization of Vn96-isolated plasma extracellular vesicles and cell free DNA that enables multi-omic profiling. Sci. Rep. (under review).
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large Extracellular Vesicles Carry Most of the Tumour DNA Circulating in Prostate Cancer Patient Plasma. J. Extracell. Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Sorber, L.; Zwaenepoel, K.; Jacobs, J.; De Winne, K.; Goethals, S.; Reclusa, P.; Van Casteren, K.; Augustus, E.; Lardon, F.; Roeyen, G.; et al. Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol. Cancers 2019, 11, 458. [Google Scholar] [CrossRef] [PubMed]
- Han, D.S.C.; Ni, M.; Chan, R.W.Y.; Chan, V.W.H.; Lui, K.O.; Chiu, R.W.K.; Lo, Y.M.D. The Biology of Cell-Free DNA Fragmentation and the Roles of DNASE1, DNASE1L3, and DFFB. Am. J. Hum. Genet. 2020, 106, 202–214. [Google Scholar] [CrossRef]
- Gutierrez, M.E.; Choi, K.; Lanman, R.B.; Licitra, E.J.; Skrzypczak, S.M.; Pe Benito, R.; Wu, T.; Arunajadai, S.; Kaur, S.; Harper, H.; et al. Genomic Profiling of Advanced Non–Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clin. Lung Cancer 2017, 18, 651–659. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Warton, K.; Yuwono, N.L.; Cowley, M.J.; McCabe, M.J.; So, A.; Ford, C.E. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell-Free Circulating DNA Studies in Plasma. Mol. Diagn. Ther. 2017, 21, 563–570. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Hoen, E.N.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; De Almagro, A.R.-V.M.; Dawson, S.-J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. J. Mol. Diagn. 2018, 20, 883–892. [Google Scholar] [CrossRef]
- Askeland, A.; Borup, A.; Østergaard, O.; Olsen, J.V.; Lund, S.M.; Christiansen, G.; Kristensen, S.R.; Heegaard, N.H.H.; Pedersen, S. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-Kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020, 8, 246. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of Exosomes as Liquid Biopsy in Clinical Diagnosis. Sig. Transduct. Target Ther. 2020, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K.; et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Kim, H.J.; Lee, J.S.; Choi, C.-M.; Lee, J.C.; Jung, M.K.; Pack, C.G.; Lee, K.Y. Extracellular Vesicle-Derived DNA for Performing EGFR Genotyping of NSCLC Patients. Mol. Cancer 2018, 17, 15. [Google Scholar] [CrossRef]
- Vasconcelos, M.H.; Caires, H.R.; Ābols, A.; Xavier, C.P.R.; Linē, A. Extracellular Vesicles as a Novel Source of Biomarkers in Liquid Biopsies for Monitoring Cancer Progression and Drug Resistance. Drug Resist. Updates 2019, 47, 100647. [Google Scholar] [CrossRef]
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 534. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, X.; Dong, Z.; Lu, Y.C.; Sun, Y.; Liu, Y.; McCormack, R.; Gu, Y.; Liu, X. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell–Free DNA from Patients with Advanced Non–Small Cell Lung Cancer. J. Mol. Diagn. 2015, 17, 265–272. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef]
- Soria-Comes, T.; Palomar-Abril, V.; Ureste, M.M.; Guerola, M.T.; Maiques, I.C.M. Real-World Data of the Correlation between EGFR Determination by Liquid Biopsy in Non-Squamous Non-Small Cell Lung Cancer (NSCLC) and the EGFR Profile in Tumor Biopsy. Pathol. Oncol. Res. 2020, 26, 845–851. [Google Scholar] [CrossRef]
- Luchini, C.; Veronese, N.; Nottegar, A.; Cappelletti, V.; Daidone, M.G.; Smith, L.; Parris, C.; Brosens, L.A.A.; Caruso, M.G.; Cheng, L.; et al. Liquid Biopsy as Surrogate for Tissue for Molecular Profiling in Pancreatic Cancer: A Meta-Analysis Towards Precision Medicine. Cancers 2019, 11, 1152. [Google Scholar] [CrossRef]
- Wyatt, A.W.; Annala, M.; Aggarwal, R.; Beja, K.; Feng, F.; Youngren, J.; Foye, A.; Lloyd, P.; Nykter, M.; Beer, T.M.; et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J. Natl. Cancer Inst. 2017, 109, 109. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Li, Z.; Zhou, B.; Tian, G.; Zeng, L.; Dai, H.; Li, X.; Liu, C.; Lu, S.; Xu, F.; et al. Statistical Analysis of Mutant Allele Frequency Level of Circulating Cell-Free DNA and Blood Cells in Healthy Individuals. Sci. Rep. 2017, 7, 7526. [Google Scholar] [CrossRef] [PubMed]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef] [PubMed]
- Que, D.; Xiao, H.; Zhao, B.; Zhang, X.; Wang, Q.; Xiao, H.; Wang, G. EGFR Mutation Status in Plasma and Tumor Tissues in Non-Small Cell Lung Cancer Serves as a Predictor of Response to EGFR-TKI Treatment. Cancer Biol. Ther. 2016, 17, 320–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reclusa, P.; Taverna, S.; Pucci, M.; Durendez, E.; Calabuig, S.; Manca, P.; Serrano, M.J.; Sober, L.; Pauwels, P.; Russo, A.; et al. Exosomes as Diagnostic and Predictive Biomarkers in Lung Cancer. J. Thorac. Dis. 2017, 9, S1373–S1382. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Katsuda, T.; Ochiya, T. Extracellular Vesicles and Encapusulated MiRNAs as Emerging Cancer Biomarkers for Novel Liquid Biopsy. Jpn. J. Clin. Oncol. 2018, 48, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Baek, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.H.; Rasmussen, T.R.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal Proteins as Prognostic Biomarkers in Non-Small Cell Lung Cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef]

| NSCLC Patient Characteristics (n = 20) | Overall | |
|---|---|---|
| Age in years | Mean | 65.6 |
| Median | 67 | |
| Range | 37–86 | |
| Gender | Male (%) | 12/20 (60%) |
| Female (%) | 8/20 (40%) | |
| Cancer Type | NSCLC | 20/20 (100%) |
| Adenocarcinoma (%) | 18/20 (90%) | |
| Adenosquamous (%) | 1/20 (5%) | |
| Sarcamatoid (%) | 1/20 (5%) | |
| Disease Stage | Stage III | 4/20 (20%) |
| Stage IV | 16/20 (80%) | |
| Treatment History | Newly Diagnosed | 16/20 (80%) |
| Enrolled After Recurrence | 4/20 (20%) | |
| Smoking | History of Smoking | 17/20 (85%) |
| Non-Smoker | 2/20 (10%) | |
| Unknown | 1/20 (5%) | |
| Patient Molecular Profiles | Overall |
|---|---|
| No mutation or fusion | 4/20 (20%) |
| kRAS mutation | 7/20 (35%) |
| PIK3CA mutation | 4/20 (20%) |
| TP53 mutation | 6/20 (30%) |
| EGFR mutation or deletion | 6/20 (30%) |
| Patient n | Mutation Observed | Tissue (MAF) | PA-TNA (MAF) |
|---|---|---|---|
| 1 | EGFR-L858R | 22 % | 0.7 % |
| 2 | EGFR-L858R | 30 % | Not detected |
| 3 | EGFR-E746A_A750del | 25.8 % | 8.2 % |
| 4 | EGFR-E746A_A750del | 65 % | 7.3 % |
| 5 | EGFR-L858R | 78.8 % | 16.8 % |
| 6 | EGFR-L858R | 24.7 % | 0.5 % |
| EGFR Detection NSCLC Patients Using PA | |
|---|---|
| Sensitivity | 83.3% (5/6) |
| Specificity | 100% (14/14) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taylor, C.; Chacko, S.; Davey, M.; Lacroix, J.; MacPherson, A.; Finn, N.; Wajnberg, G.; Ghosh, A.; Crapoulet, N.; Lewis, S.M.; et al. Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma. Int. J. Mol. Sci. 2020, 21, 9083. https://doi.org/10.3390/ijms21239083
Taylor C, Chacko S, Davey M, Lacroix J, MacPherson A, Finn N, Wajnberg G, Ghosh A, Crapoulet N, Lewis SM, et al. Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma. International Journal of Molecular Sciences. 2020; 21(23):9083. https://doi.org/10.3390/ijms21239083
Chicago/Turabian StyleTaylor, Catherine, Simi Chacko, Michelle Davey, Jacynthe Lacroix, Alexander MacPherson, Nicholas Finn, Gabriel Wajnberg, Anirban Ghosh, Nicolas Crapoulet, Stephen M. Lewis, and et al. 2020. "Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma" International Journal of Molecular Sciences 21, no. 23: 9083. https://doi.org/10.3390/ijms21239083
APA StyleTaylor, C., Chacko, S., Davey, M., Lacroix, J., MacPherson, A., Finn, N., Wajnberg, G., Ghosh, A., Crapoulet, N., Lewis, S. M., & Ouellette, R. J. (2020). Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma. International Journal of Molecular Sciences, 21(23), 9083. https://doi.org/10.3390/ijms21239083







