Abstract
Abnormal activation of Toll-like receptor (TLRs) signaling can result in colon cancer development. The aim of this study was to investigate the expression of important TLRs in different histological types of colorectal polyps and evaluate their relationship with intestinal microbiota. The expression levels of TLR2, 3, 4, and 5 were analyzed in intestinal biopsy specimens of 21 hyperplastic polyp (HP), 16 sessile serrated adenoma (SSA), 29 tubular adenoma (TA), 21 villous/tubulovillous (VP/TVP) cases, and 31 normal controls. In addition, selected gut bacteria including Streptococcus bovis, Enterococcus faecalis, Enterotoxigenic Bacteroides fragilis (ETBF), Fusobacterium nucleatum, Porphyromonas spp., Lactobacillus spp., Roseburia spp., and Bifidobacterium spp. were quantified in fecal samples using absolute qRT PCR, and, finally, the association between TLRs and these gut microbiota- was evaluated by Spearman’s correlation coefficient. Higher expression of TLR2 and TLR4 in VP/TVP and TA, and lower expression levels of TLR3 and TLR5 in all type of polyps were observed. The differences in TLR expression patterns was not only dependent on the histology, location, size, and dysplasia grade of polyps but also related to the intestinal microbiota patterns. TLR2 and TLR4 expression was directly associated with the F. nucleatum, E. faecalis, S. bovis, Porphyromonas, and inversely to Bifidobacterium, Lactobacillus, and Roseburia quantity. Furthermore, TLR3 and TLR5 expression was directly associated with Bifidobacterium, Roseburia, and Lactobacillus quantity. Our results suggest a possible critical role of TLRs during colorectal polyp progression. An abnormal regulation of TLRs in relation to gut microbial quantity may contribute to carcinogenesis.
1. Introduction
Colorectal cancer (CRC) is one of the most common cancers, and accounts for almost half a million deaths annually worldwide [1,2]. CRC represents a heterogeneous group of cancers arising from at least two precursors, the conventional adenoma (CA) and the serrated polyp [3,4]. The majority of CRC cases (~60%) arise via the conventional pathway, with ~20% arising from the serrated pathway and ~20% from an alternate pathway [5]. These distinct molecular pathways dictate the different precursor lesions, such as the conventional pathway resulting in CA and the serrated pathway with sessile serrated adenomas (SSA) [4,5]. An additional serrated polyp type, the hyperplastic polyp (HP), has negligible malignant potential [6,7]. Previous evidence suggests that gut bacteria may be a major factor involved in colon cancer development [8], although distinct contributions through CAs or SSAs have not been studied simultaneously. Gut microbiota may play critical role in the progression of CRC via their metabolite or their structural components which interact with pathogen-associated molecular patterns (PAMP) and microbe-associated molecular pattern (MAMP) receptors such as Toll-like receptors (TLRs) [9,10]. TLRs are key molecules involved in inflammation and also contribute to carcinogenesis [11,12,13]. In addition to cancer development, TLRs are specifically involved in transduction of molecular signals guiding immune processes such as induction and regulation of both innate and adaptive immunity, production of cytokines, and recognition of specific molecular patterns on the surface of microorganisms [14,15,16]. In spite of the vast body of research surrounding TLRs, there is a lack of evidence on differentiating levels of TLR expression in different types of colorectal polyps as precursors of CRC [15,17].
The aim of our study was to evaluate the expression levels of TLR2, TLR3, TLR4, and TLR5 in intestinal biopsy specimens of patients with different histological colorectal polyp types including HP, SSA, tubular adenoma (TA), and villous/tubulovillous (VP/TVP) cases compared to normal controls. In addition, selected intestinal bacteria in matched fecal specimens from these participants were quantitatively analyzed to determine the association between TLRs and gut microbial patterns that may suggest a possible role of crosstalk between specific bacteria and TLRs in the progression of colon polyps to CRC.
2. Results
2.1. Characteristics of Study Groups
Demographic and clinical variables were evaluated between different categories of study groups. Participants’ characteristics with related P-value in normal, HP, SSA, VP/TVP, and TA types of polyps are presented in Table 1. Fortunately, the population studied was characterized by a similar distribution of age, gender, diabetes history, physical activity, GI disease history, alcohol consumption, and tumor location.

Table 1.
Patient demographics in normal, hyperplastic polyp (HP), sessile serrated adenoma (SSA), tubular adenoma (TA), and villous/tubulovillous (VP/TVP) groups.
2.2. Expression of TLRs in HP, SSA, TA, VP/TVP, and Normal Controls
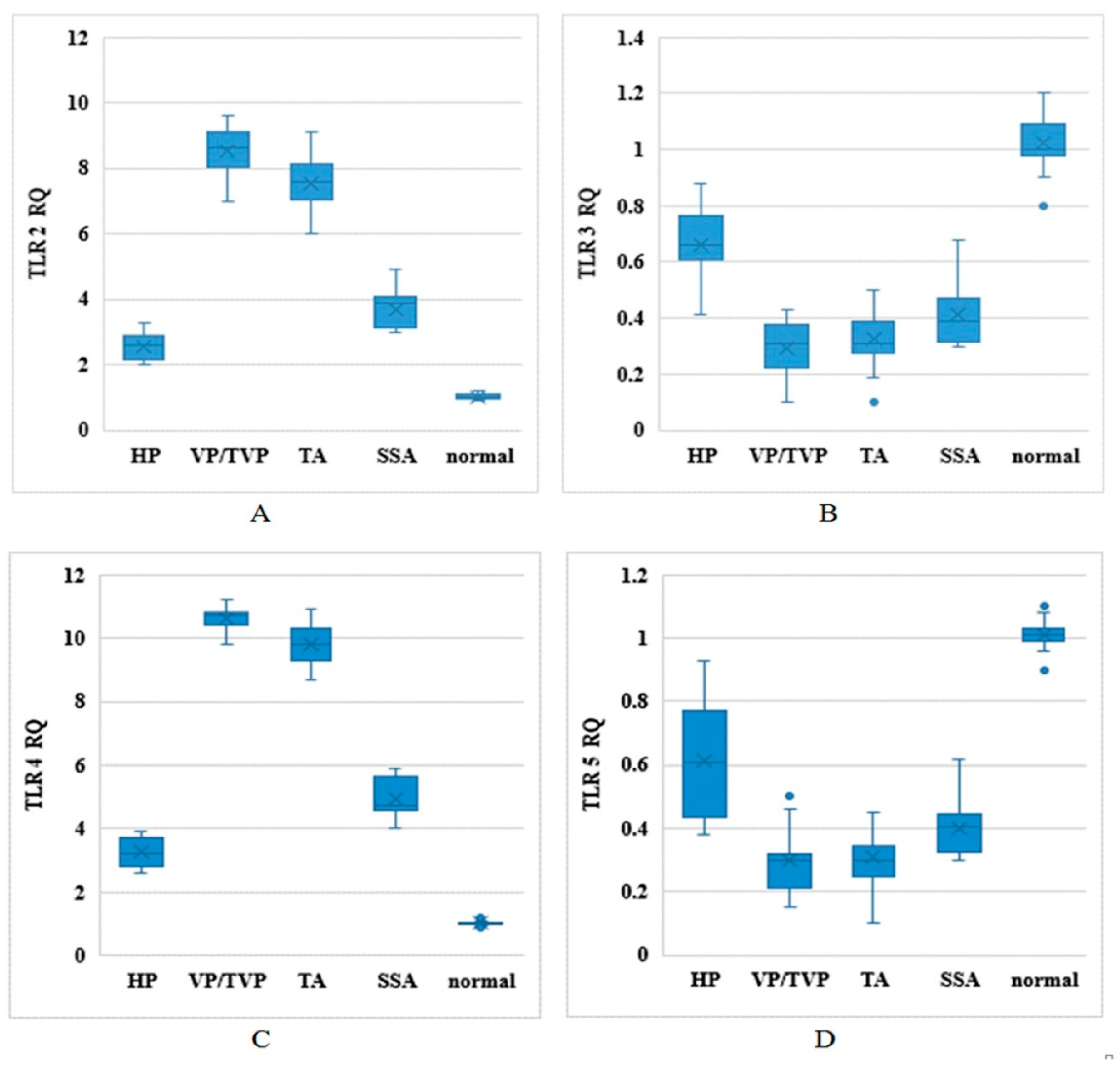
TLR2, TLR3, TLR4, and TLR5 mRNA expression levels were evaluated in clinical biospecimens of normal, HP, SSA, VP/TPV, and TA groups by relative qPCR and based on relative expression (RQ) levels, where normal tissue expression was set as the reference after all signals were normalized to β–2-microglobulin (Figure 1). Higher expression levels of TLR2 and TLR4 were detected in VP/TVP and TA compared to normal, HP, and SSA groups (p-value < 0.001). In addition, lower expression rates were observed for TLR3 and TLR5 in all polyp groups in contrast to normal individuals (p-value < 0.001). The noteworthy point was that the RQs of TLR2, TLR3, TLR4, and TLR5 were significantly different among histologically different colorectal polyp types compared to samples from normal participants (p-value < 0.001) (Table 2).
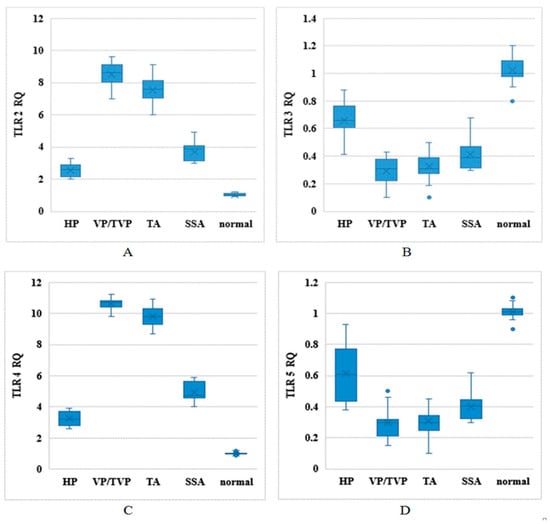
Figure 1.
Toll-like receptor (TLR) mRNA expression levels based on relative expression (RQ) in different types of histological polyps: Distribution of TLR2, TLR3, TLR4, and TLR5 mRNA expression levels between normal, hyperplastic (HP), sessile serrated adenoma (SSA), villous/tubulovillous (VP/TVP), and tubular adenoma (TA) polyps in horizontal axis, in vertical axis; (A) TLR2 RQ, (B) TLR3 RQ, (C) TLR4 RQ, and (D) TLR5 RQ.

Table 2.
Expression of candidate TLRs in normal, HP, SSA, VP/TPV, and TA groups.
We further classified colorectal polyps by location into proximal (n = 33) and distal (n = 54), by size either <0.5 mm (n = 51) or ≥0.5 mm (n = 35), as well as having low dysplasia (n = 33) or medium/high dysplasia (n = 54). The expression of targeted TLRs was then analyzed according to the polyp characterization described above and outlined in Table 3. A significant association between TLR2 and TLR4 expression levels and location of colorectal polyps was observed. The significant relationship between the polyp size and the RQ of TLR2, 3, 4, and, 5 was demonstrated. Finally, significant associations between the RQ of TLR2, TLR4, and TLR5 and grade of dysplasia were achieved.

Table 3.
Association between the relative expression of TLR2, 3, 4, and 5 and polyp size, location, and grade of dysplasia.
2.3. Bacterial Diversity across Different Types of Colon Polyps
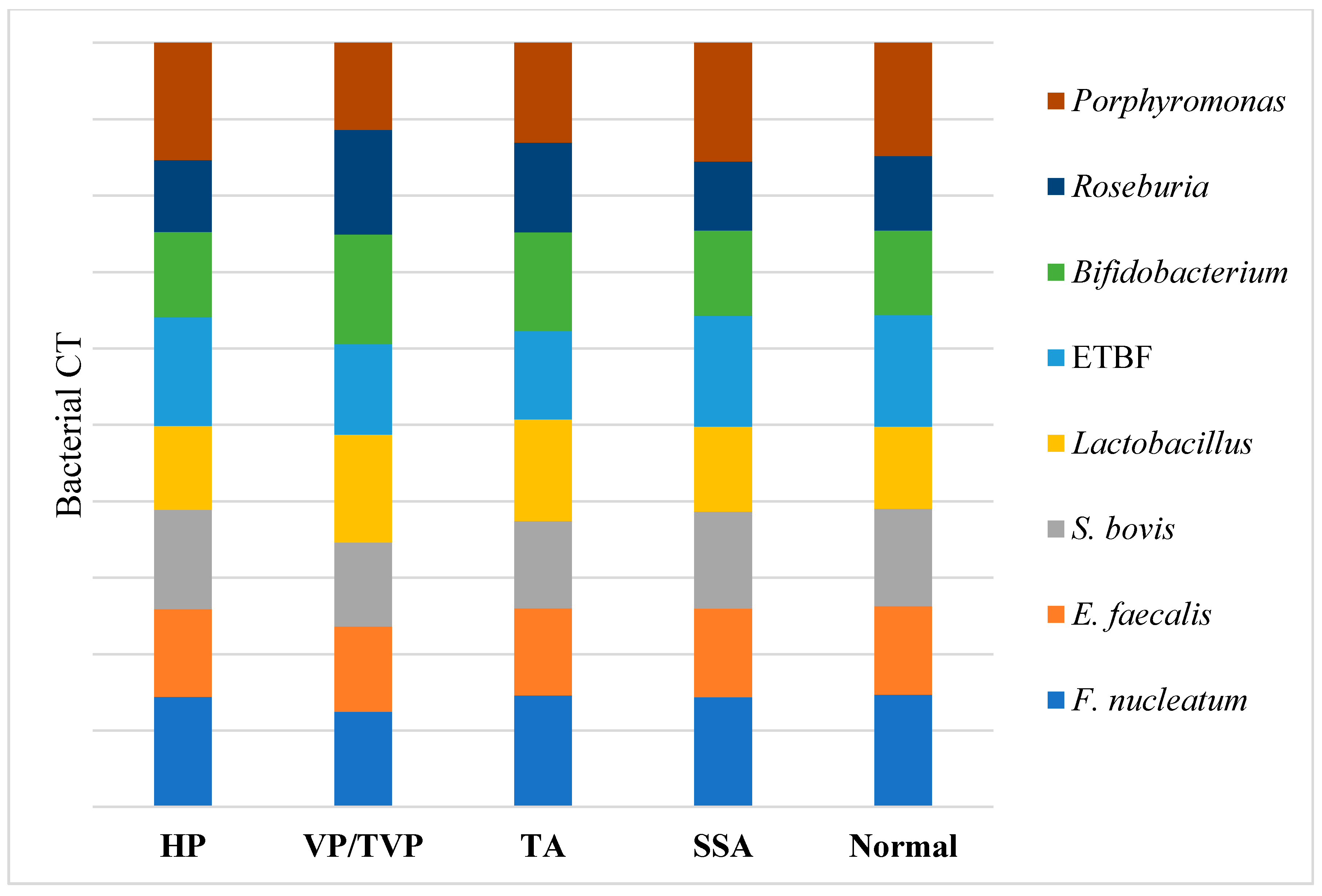
According to the results visualized in Figure 2 and Table S1, higher quantities of Fusobacterium nucleatum, Enterococcus faecalis, Streptococcus bovis/gallolyticus, Enterotoxigenic Bacteroides fragilis, and Porphyromonas spp. were detected in patients’ samples with TAs and VP/TVPs, compared to the normal fecal samples from subjects that were controls and patients with HPs and SSAs (p-value < 0.001). An opposite proportion was observed for the quantification of Lactobacillus spp., Roseburia spp., and Bifidobacterium spp., where greater numbers of bacteria were detected in normal, HP, and SSA polyp groups, compared to the TA and VP/TVP cases (p-value < 0.001).
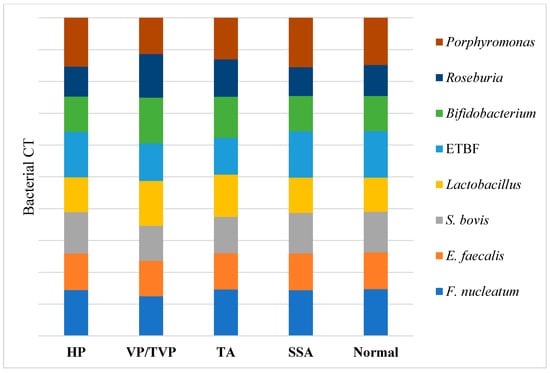
Figure 2.
Distribution of targeted bacterial markers between hyperplastic polyp (HP), villous/tubulovillous (VP/TVP), tubular adenoma (TA), sessile serrated adenoma (SSA), and normal groups were depicted in horizontal axis. In addition, bacterial CT of Porphyromonas spp., Roseburia spp., Bifidobacterium spp., Enterotoxigenic Bacteroides fragilis (ETBF), Lactobacillus spp., Streptococcus bovis, Enterococcus faecalis, and Fusobacterium nucleatum were demonstrated in vertical axis. Higher amounts of F. nucleatum, E. faecalis, S. bovis, ETBF, and Porphyromonas spp. based on CT analysis were detected in patients’ samples with TAs and VP/TVPs (p-value < 0.001), while higher abundance of Lactobacillus, Roseburia, and Bifidobacterium were reported in normal, HP, and SSA (p-value < 0.001).
Lastly, we analyzed the association between the quantity of these targeted gut bacteria (based on their real-time CT) and the RQ of the main TLRs being studied (TLR2, TLR3, TLR4, and TLR5). Table 4 shows a significant correlation between all selected bacteria (S. bovis, E. faecalis, ETBF, F. nucleatum, Porphyromonas spp., Lactobacillus spp., Roseburia spp., and Bifidobacterium spp.) and expression levels of all TLRs. We found that the TLR2 and TLR4 expression rates were directly associated with the quantity of F. nucleatum, E. faecalis, S. bovis, and Porphyromonas spp., but inversely related to the Bifidobacterium, Lactobacillus, and Roseburia spp. quantity. In addition, we observed that TLR3 and TLR5 expression rates were directly associated with Bifidobacterium spp., Roseburia spp., and Lactobacillus spp. quantity.

Table 4.
Association between candidate bacteria and the relative expression of TLR2, TLR3, TLR4, and TLR5.
3. Discussion
This study focused on TLR expression in different histological types of colorectal polyps and their relationship with some selected gut bacteria. We found differences in TLR expression patterns depending on the histological type of polyps, their size, location, and grade of dysplasia, and further differentiated polyp types by fecal bacterial quantity. To the best of our knowledge, there have been limited studies investigating the expression of TLRs across different types of polyps as early precursors of CRC [18,19]. We observed significant differential mRNA expression of intestinal TLR2, 3, 4, and 5 in patents with TA, VP/TVP, HP, and SSA compared to normal individuals. In fact, TLR3 and TLR5 expression levels were higher in normal individuals, compared to different types of progressive colorectal polyps. These TLRs (3 and 5) might be essential for maintaining intestinal balance and homeostasis, suggesting a possible protective role against malignant transformation of the colorectal mucosa. In our study, TLR2 and TLR4 overexpression was observed in subjects with TA and VP/TVP polyps. Our findings that TLR3 and TLR5 are constitutively expressed in the healthy gut, while TLR2 and TLR4 appear to be overexpressed in unhealthy gut, confirmed earlier observations in studies by Yang et al., Kelly et al., and Kutikhin et al. [20,21,22]. Moossavi et al. [17] declared more evidence is required to fully determine the effect of TLRs on intestinal tumorigenesis before being able to translate and generalize the results to clinical practice. For instance, based on our previous study, we observed that aberrant surface expression of TLR9 on tumor cells may promote the growth and invasion of colorectal polyps [18], while Gao et al. [23] indicated that TLR9 signaling activation participated in the clinical process of CRC and influenced NF-kappaB expression. Bednarczyk et al. further observed a correlation in the expression levels of TLR7 and TLR9 with colorectal polyp progression to CRC [15]. These study differences could be explained by the limitations of the methods employed and lack of attention to the spatiotemporal variation in TLR expression patterns [17]. We also found that intestinal TLR2, 3, 4, and 5 expression levels were associated with location, size, and dysplasia grade of different types of colorectal polyps. In fact, our results indicated intestinal TLR expression levels may play important roles in the development of site-specific histological types of colorectal polyps.
Moreover, we have investigated the relationship between several fecal bacteria, including F. nucleatum, E. faecalis, S. bovis, Lactobacillus, ETBF, Bifidobacterium, Roseburia, and Porphyromonas, and intestinal TLRs and observed significant association between TLR mRNA expression levels and select microbial species abundance. An overpopulation of stool bacteria, consisting of F. nucleatum, E. faecalis, S. bovis, and Porphyromonas in relation to TLR2 and TLR4 expression level was shown in TA and VP/TVP cases, while Roseburia spp., Lactobacillus spp., and Bifidobacterium spp. were decreased in these cases. These results are strongly consistent with previous studies that demonstrated the interactions between gut microbiota and TLRs that impact homeostasis and immune responses or gut microbiota associated with immunological regulation and inflammation [24,25]. In fact, commensal microbiota and their structural components and products such as LPS, flagella, and DNA or RNA can be recognized by TLRs and these interactions trigger responses that help to maintain the homeostasis of intestinal immunity [26]. Overall, these findings confirm the potential utility of TLR expression patterns in relation to gut microbiota for discriminating polyp cases from normal group samples, and suggest that changes in microbial abundance in combination with specific TLR relative levels may represent early events in the pathway(s) leading to colorectal polyp. Although our results are promising, a limitation of our study is that we only profiled eight selected bacterial species, which may not represent the full biodiversity within our patient population. Future studies will take a more comprehensive approach to microbial profiling as well as seek to correlate both this biodiversity with a specific set of inflammation markers regulated through NF-kappaB.
In conclusion, the present study has shown significant difference in the expression levels of intestinal TLR2, 3, 4, and 5, in patients with TA, VP/TVP, HP, and SSA polyps versus healthy controls. Based on current findings, TLR2 and TLR4 may be upregulated in the process of polyp formation and progression. In contrast, TLR3 and TLR5 were downregulated in all patient polyp types compared with normal tissue samples. Hence, they might be essential for maintaining balance and homeostasis in a healthy gut and have a possible protective role against malignant transformation of the colorectal mucosa. In addition, we found that intestinal TLR2, 3, 4, and 5 expression levels associate with location, size, and dysplasia grade of colorectal polyps and their expression levels may play important roles in the development of site-specific histological types of colorectal polyps. Moreover, there was a clear correlation between TLR mRNA expression levels and specific fecal bacterial species, suggesting an abnormal activation or regulation of TLRs in relation to gut microbiota which may promote disease progression. Finally, more evidence is required to fully appreciate the effect of differential TLR expression on intestinal tumorigenesis, and data on larger cohorts are necessary to validate and translate the results into clinical practice. Further studies on TLR expression patterns and gut microbial interactions in colorectal polyps can improve strategies in CRC prevention and earlier detection.
4. Materials and Methods
4.1. Sample Collection and Storage
One-hundred and eighteen colonic biopsy and fecal specimens, including samples from 31 normal controls, 21 HP, 16 SSA, 29 TA, and 21 VP/TVP, were collected from candidate individuals at Taleghani Hospital, Tehran, Iran between 2016 and 2018. Eligible participants were individuals 50–80 years old and scheduled to have a colonoscopy for routine screening. Additional demographic data characterizing the study cohort are summarized in Table 1. Colonic tissue samples included in the study were consented from these participants at the time of colonoscopy and provided during the procedure. All fresh stool samples were collected three days to two weeks before colonoscopy and bowel cleansing procedures associated with the routine screen [27,28]. All colonic biopsy samples were classified after colonoscopy and confirmed by an expert pathologist. Fecal and biopsy samples were stored at −80 °C immediately until further analysis. The case-control study was approved by the Clinical Research Ethics Committee of the Shahid Beheshti University of Medical Sciences and the Ethics Committee of Taleghani Hospital, Tehran, Iran (No. 851; 2016-12-7).
4.2. RNA Extraction from Colon-Derived Tissues
RNA was extracted from the colonic biopsy and normal tissues using the TRIzol reagent (ThermoFisher Scientific, Massachusetts, United States) in accordance with the manufacturer’s guidelines. After final precipitation, the DNA was resuspended in Tris-EDTA (TE) buffer and stored at −20 °C for further analysis.
4.3. DNA Extraction from Fecal Specimens
Genomic DNA was extracted from frozen preserved fecal samples using the QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s instructions.
4.4. TLR mRNA Expression Levels in Extracted RNA from Colonic Tissues
TLR2, TLR3, TLR4, and TLR5 mRNA expression levels were evaluated in extracted RNA from tissue samples using the Premix Ex Taq SYBR (Takara Bio, Kusatsu, Japan) and the relative qRT-PCR technique in accordance with the manufacturer’s guidelines. Specific primers were selected from previously published studies [29,30,31]. Amplification signals for samples were normalized by β–2-microglobulin [32] and relative fold change of TLRs genes expression was evaluated by the 2−ΔΔCT method [18,32].
4.5. Quantification of Bacterial Species by 16SrRNA in Extracted DNA from Fecal Samples
Bacterial 16SrRNA quantification for several target gut bacteria including S. bovis, E. faecalis, ETBF, F. nucleatum, Porphyromonas spp., Lactobacillus spp., Roseburia spp., and Bifidobacterium spp. was precisely determined in fecal samples by absolute qRT PCR using the SYBR Green detection system, as described in our previous study [27]. The association between mRNA expression levels of TLR2, TLR3, TLR4, and TLR5 and the quantification of the selected gut microbiota was evaluated by statistical analysis and Spearman’s correlation coefficient.
4.6. Statistical Analysis
The descriptive statistics were expressed for numerical and categorical variables using mean (standard deviation (SD)/median (interquartile range (IQR)) and frequency (percentage), respectively. The association between categorical variables and polyp groups was assessed by Pearson chi-square and Fisher exact test. In addition, analysis of variance (ANOVA) was used to compare the mean numerical variables (including demographic and TLRs) between polyp types. The assumption of normality was checked for all numeric variables and the non-parametric test were used when the data were not normally distributed. Accordingly, the distribution of tumor size was compared between polyp types using the Kruskal–Wallis test. The Mann–Whitney U test was used to compare the difference in TLRs between each level of polyp location, polyp size, and grade of dysplasia. The association between TLRs and candidate bacteria was evaluated using Spearman’s correlation coefficient. A box plot and bar plot were used to indicate the distribution of TLR mRNA expression levels based on RQ and bacterial species in different colon polyp types. All analyses were performed in R (version 4.02) and SPSS (version 26) and a p-value of less than 0.05 was regarded as statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/23/8968/s1.
Author Contributions
Conceptualization, S.R., E.N.M., and D.S.; methodology, S.R., E.N.M., and H.A.A.; software, S.R. and M.A.L.; validation, S.R., R.G., and A.Y.; formal analysis, S.R. and M.A.L.; investigation, S.R., E.N.M., and H.A.A.; resources, S.R. and H.A.A.; data curation, E.N.M. and R.G.; writing—original draft preparation, S.R.; writing—review and editing, S.R., E.N.M., R.G., and D.S.; visualization, A.Y. and R.G.; supervision, H.A.A. and M.R.Z.; project administration, E.N.M. and H.A.A.; funding acquisition, H.A.A., M.R.Z., and D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant#: RIGLD 851).
Acknowledgments
The authors would like to thank all the staff of Cancer Department in Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
| TLRs | Toll-like receptors |
| HP | hyperplastic polyp |
| SSA | sessile serrated adenoma |
| TA | tubular adenoma |
| VP/TVP | villous/tubulovillous polyp |
| ETBF | Enterotoxigenic Bacteroides fragilis |
| CRC | colorectal cancer |
| RQ | relative expression |
References
- Wong, M.C.; Ding, H.; Wang, J.; Chan, P.S.; Huang, J. Prevalence and risk factors of colorectal cancer in Asia. Intest. Res. 2019, 17, 317–329. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Gastroenterol. Rev. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Dominianni, C.; Shapiro, J.A.; Church, T.R.; Wu, J.; Miller, G.; Yuen, E.; Freiman, H.; Lustbader, I.; Salik, J.; et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 2016, 4, 1–14. [Google Scholar] [CrossRef]
- Jass, J.R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007, 50, 113–130. [Google Scholar] [CrossRef]
- Langner, C. Serrated and Non-Serrated Precursor Lesions of Colorectal Cancer. Dig. Dis. 2015, 33, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Strum, W.B. Colorectal Adenomas. N. Engl. J. Med. 2016, 374, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Keku, T.O.; Dulal, S.; Deveaux, A.; Jovov, B.; Han, X. The gastrointestinal microbiota and colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G351–G363. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Kitazawa, H.; Van Wees, S.C.M.; Pieterse, C.M.; Takahashi, H. Receptors and Signaling Pathways for Recognition of Bacteria in Livestock and Crops: Prospects for Beneficial Microbes in Healthy Growth Strategies. Front. Immunol. 2018, 9, 2223. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Asadzadeh-Aghdaei, H.; Nazemalhosseini-Mojarad, E.; Dabiri, H.; Ghanbari, R.; Zali, M.R. Gut microbiota, epigenetic modification and colorectal cancer. Iran. J. Microbiol. 2017, 9, 55–63. [Google Scholar]
- Rakoff-Nahoum, S.; Medzhitov, R. Toll-like receptors and cancer. Nat. Rev. Cancer 2009, 9, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-T.; Ogino, S.; Qian, Z.R. Toll-like receptor signaling in colorectal cancer: Carcinogenesis to cancer therapy. World J. Gastroenterol. 2014, 20, 17699–17708. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Jezierka, A.; Kolosova, I.A.; Verin, A.D. Toll Like Receptors Signaling Pathways as a Target for Therapeutic Interventions. Curr. Signal Transduct. Ther. 2011, 6, 428–440. [Google Scholar] [CrossRef]
- Bednarczyk, M.; Muc-Wierzgoń, M.; Walkiewicz, K.; Kokot, T.; Fatyga, E.; Mazurek, U. Profile of gene expression of TLR-signaling pathways in colorectal cancer tissues. Int. J. Immunopathol. Pharmacol. 2017, 30, 322–326. [Google Scholar] [CrossRef]
- Kirill, V.K.; Kamar-Sulu, N.; Atretkhany, K.-S.N.; Drutskaya, M.; Grivennikov, S.I.; Kuprash, D.V.; Nedospasov, S.A. TLR-signaling and proinflammatory cytokines as drivers of tumorigenesis. Cytokine 2017, 89, 127–135. [Google Scholar] [CrossRef]
- Moossavi, S.; Rezaei, N. Toll-like receptor expression pattern: Clinical application. J. Clin. Immunol. 2012, 32, 1421–1422. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Khatibi, S.; Pezeshkiyan, Z.; Mojarad, E.N.; Sharafkhah, M.; Sadeghi, A.; Aghdaei, H.A.; Zali, M.R. Investigating the TLR9 mRNA Expression Level in Different Histological Types of Colorectal Polyps. Asian Pac. J. Cancer Prev. 2019, 20, 2299–2302. [Google Scholar] [CrossRef]
- Eiro, N.; Gonzalez, L.; Gonzalez, L.O.; Andicoechea, A.; Fernández-Díaz, M.; Altadill, A.; Vizoso, F.J. Study of the Expression of Toll-Like Receptors in Different Histological Types of Colorectal Polyps and Their Relationship with Colorectal Cancer. J. Clin. Immunol. 2012, 32, 848–854. [Google Scholar] [CrossRef]
- Yang, T.; Owen, J.L.; Lightfoot, Y.L.; Kladde, M.P.; Zadeh, M.; Lightfooot, Y.L. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol. Med. 2013, 19, 714–725. [Google Scholar] [CrossRef]
- Kelly, D.; Mulder, I.E. Microbiome and immunological interactions. Nutr. Rev. 2012, 70, S18–S30. [Google Scholar] [CrossRef]
- Kutikhin, A.G.; Yuzhalin, A.E. Editorial: Pattern Recognition Receptors and Cancer. Front. Immunol. 2015, 6, 481. [Google Scholar] [CrossRef]
- Gao, C.; Qiao, T.; Zhang, B.; Yuan, S.; Zhuang, X.; Luo, Y. TLR9 signaling activation at different stages in colorectal cancer and NF-kappaB expression. OncoTargets Ther. 2018, 11, 5963–5971. [Google Scholar] [CrossRef]
- Inoue, R.; Yajima, T.; Tsukahara, T. Expression of TLR2 and TLR4 in murine small intestine during postnatal development. Biosci. Biotechnol. Biochem. 2016, 81, 350–358. [Google Scholar] [CrossRef][Green Version]
- Zou, S.; Fang, L.; Lee, M.-H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 2018, 6, 1–12. [Google Scholar] [CrossRef]
- Yiu, J.H.; Dorweiler, B.; Woo, C.W. Interaction between gut microbiota and toll-like receptor: From immunity to metabolism. J. Mol. Med. 2017, 95, 13–20. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Aghdaei, H.A.; Dabiri, H.; Sepahi, A.A.; Modarressi, M.H.; Mojarad, E.N. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Sharafkhah, M.; Aghdaei, H.A.; Mojarad, E.N.; Dabiri, H.; Sepahi, A.A.; Modarressi, M.H.; Feizabadi, M.M.; Zali, M.R. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J. Microbiol. Methods 2018, 155, 82–88. [Google Scholar] [CrossRef]
- Kim, T.W.; Lee, S.-J.; Oh, B.M.; Lee, H.; Uhm, T.G.; Min, J.-K.; Park, Y.-J.; Yoon, S.R.; Kim, B.-Y.; Kim, J.W.; et al. Epigenetic modification of TLR4 promotes activation of NF-κB by regulating methyl-CpG-binding domain protein 2 and Sp1 in gastric cancer. Oncotarget 2015, 7, 4195–4209. [Google Scholar] [CrossRef]
- Li, H.; Yang, T.; Li, F.-Y.; Ning, Q.; Sun, Z.-M. TLR4 Overexpression Inhibits Endothelial PAS Domain-Containing Protein 1 Expression in the Lower Respiratory Tract of Patients with Chronic COPD. Cell. Physiol. Biochem. 2016, 39, 685–692. [Google Scholar] [CrossRef]
- Allhorn, S.; Böing, C.; Koch, A.A.; Kimmig, R.; Gashaw, I. TLR3 and TLR4 expression in healthy and diseased human endometrium. Reprod. Biol. Endocrinol. 2008, 6, 40. [Google Scholar] [CrossRef]
- Peyravian, N.; Gharib, E.; Moradi, A.; Mobahat, M.; Tarban, P.; Azimzadeh, P.; Nazemalhosseini-Mojarad, E.; Aghdaei, H.A. Evaluating the expression level of co-stimulatory molecules CD 80 and CD 86 in different types of colon polyps. Curr. Res. Transl. Med. 2018, 66, 19–25. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).